Abstract
Ischemia-reperfusion (IR) injury induces endoplasmic reticulum (ER) stress and cell death. Bax Inhibitor-1 (BI-1) is an evolutionarily conserved ER protein that suppresses cell death and that is abundantly expressed in both liver and kidney. We explored the role of BI-1 in protection from ER stress and IR injury by using bi-1 knockout mice, employing models of transient hepatic or renal artery occlusion. Compared to wild-type bi-1 mice, bi-1 knockout mice subjected to hepatic IR injury exhibited these characteristics: (i) increased histological injury; (ii) increased serum transaminases, indicative of more hepatocyte death; (iii) increased percentages of TUNEL-positive hepatocytes; (iv) greater elevations in caspase activity; and (v) more activation of ER stress proteins inositol-requiring enzyme 1 and activating transcription factor 6 and greater increases in expression of ER stress proteins C/EBP homologous protein and spliced XBP-1 protein. Moreover, hepatic IR injury induced elevations in bi-1 mRNA in wild-type liver, suggesting a need for bi-1 gene induction to limit tissue injury. Similar sensitization of kidney to ER stress and IR injury was observed in bi-1−/− mice. We conclude that bi-1 provides endogenous protection of liver and kidney from ER stress and IR injury. Analysis of components of the bi-1-dependent pathway for protection from IR injury may therefore reveal new strategies for organ preservation.
Keywords: apoptosis, oxygen-glucose deprivation, C/EBP homologous protein, inositol-requiring enzyme 1, activating transcription factor 6
Ischemia reperfusion (IR) injury occurs during transient arterial occlusion, cardiac arrest, and traumatic blood loss, resulting in cell death and tissue destruction. Cell death induced in the setting of IR injury can involve necrosis, apoptosis, or both (reviewed in refs. 1 and 2). Recent evidence strongly suggests involvement of endoplasmic reticulum (ER) stress as an initiator of the cell death mechanisms activated during hypoxia and IR insults (3, 4). For example, ATP deficiency caused by hypoxia can promote escape of Ca2+ from ER into cytosol, triggering several downstream pathways that promote cell death (3, 4). Reduced ER Ca2+ also alters the activity of intra-ER molecular chaperones (Grp78 and Grp94), thereby initiating several signal transduction pathways, including some that have been linked to cell death induction (reviewed in ref. 3). In addition, hypoxia and hypoglycemia, which occur during arterial occlusion or cardiac arrest, are both known to initiate protein misfolding in the ER, thereby diverting resident ER chaperones and freeing transmembrane signal transducing proteins of the ER to initiate programs for cell death. Also, reperfusion triggers oxidative stress, with production of nitric oxide (NO) and reactive oxygen species, altering cellular redox-dependent reactions, interfering with protein disulfide bonding, and resulting in protein misfolding in the ER (3, 4).
The unfolded protein response is characterized by coordinated activation of multiple ER proteins, including inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase, and activating transcription factor (ATF) 6 (reviewed in ref. 5). IRE1, PKR-like ER kinase, and ATF6 are transmembrane proteins that are normally held in inactive states in ER membranes by binding to intra-ER chaperones, particularly Grp78. In response to stimuli that divert ER chaperones to misfolded proteins, these transmembrane proteins either oligomerize or move to other cellular locations, initiating signal transduction processes to promote expression of genes required for folding of newly synthesized proteins and degradation of the unfolded proteins in an effort to reestablish homeostasis and normal ER function. However, when injury is excessive, these same ER stress signal transduction pathways also can stimulate cell death.
Recently, the anti-apoptotic protein, BI-1 (Bax Inhibitor-1) (6), has been linked to protection from apoptosis induced by ER stress. BI-1 contains several transmembrane domains, localizes to ER membranes, and its protective function is conserved in both animal and plant species (7–9). Cells isolated from bi-1−/− mice exhibit selective hypersensitivity in vitro to apoptosis induced by ER stress triggered by chemical agents that impact the ER (Thapsigargin, Tunicamycin, and Brefeldin A) (9). Moreover, bi-1−/− mice were shown to be more sensitive to renal tubule cell death induced by tunicamycin, a pharmacological inducer of ER stress, and to stroke injury, by using a model of transient cerebral artery occlusion (6). Conversely, BI-1 overexpression selectively protects cells in vitro against apoptosis induced by ER stress inducers (6, 7, 9). In mammals, the bi-1 gene is abundantly expressed in liver and kidney. Given that oxidative stress is important in IR injury, and based on findings linking oxidative injury to ER stress, we explored the role of bi-1 in protection from liver and kidney ER stress and tissue injury induced by IR.
Results
BI-1-Deficient Hepatocytes Are More Sensitive to Cell Death Caused by Oxygen-Glucose Deprivation (OGD).
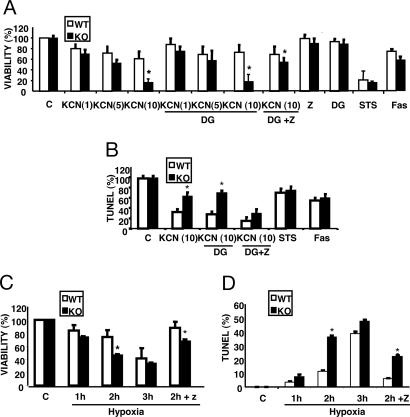
To explore the effects of BI-1 deficiency on the sensitivity of hepatocytes to IR, we used in vitro models of OGD. The first experimental paradigm includes an initial short phase of OGD created by culturing cells in deoxyglucose (DG) and KCN for 1 h, followed by a restoration phase. The second model involves placing the cells in a hypoxia chamber (1% O2) for 1–3 h in media without glucose, followed by a restoration phase. Accordingly, hepatocytes were isolated from bi-1−/− and bi-1+/+ livers (10) and subjected to OGD, then placed into fresh medium for 1 day, before assessing viability of hepatocytes by MTT assay (Fig. 1A and C). Apoptosis was also monitored by TUNEL assay (Fig. 1 B and D).
Fig. 1.
BI-1 deficiency sensitizes to cell death induced by OGD. Hepatocytes were isolated from adult wild-type (+/+) and knockout (−/−) mice. (A and B) Hepatocytes were cultured for 60 min in normal medium (C) or in medium containing 20 mM deoxyglucose (DG), 2.5, 5, or 10 mM KCN, or the combination (OGD), then washed and cultured 1 day in normal medium with or without 50 μM benzoyl-Val-Ala-Asp-fluoromethyl-ketone (z). As positive controls, hepatocytes were also treated with 10 μM staurosporine (STS) or 500 ng/ml anti-Fas (Fas) for 24 h. After treatments, the percentages of viable cells (A) (relative to control) (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay) and TUNEL-positive cells (B) were quantified (mean ± SD; n = 5). (C and D) Hepatocytes were cultured for 1–3 h in medium lacking glucose in 1% O2, then washed and switched to normal medium and atmosphere, with or without 50 μM z. The percentages of viable (C) and TUNEL-positive (D) cells were quantified (mean ± SD; n = 4).
Compared to wild-type hepatocytes, BI-1-deficient cells displayed significantly increased sensitivity to culture conditions that mimic IR injury, including KCN alone and KCN plus DG (Fig. 1 A and B). Also, bi-1 hepatocytes were more sensitive to hypoxia/glucose deprivation, with data for 2 h treatment reaching statistical significance (P ≤ 0.05) (Fig. 1 C and D). Cell death induced by OGD was partially suppressed by benzoyl-Val-Ala-Asp-fluoromethyl-ketone (z), suggesting involvement of caspases. In contrast, BI-1 deficiency did not alter sensitivity to staurosporine (STS) or anti-Fas antibody (Fig. 1 A and B).
BI-1 Deficiency Increases Hepatic IR Injury.
We tested the effects of BI-1 deficiency on sensitivity of liver to IR injury, employing a hepatic artery occlusion model (11). Using the Suzuki scoring method (0–4 scale) (12), histological analysis of livers from bi-1−/− and bi-1+/+ animals revealed more severe injury in BI-1-deficient animals (5.5 ± 0.5 vs. 3 ± 0.4; P < 0.05) (Supporting Text, Table 1, and Fig. 6, which are published as supporting information on the PNAS web site). In particular, the degree of necrosis was significantly higher in bi-1−/− mice than bi-1+/+ animals (P < 0.05). Histological lesions were absent in sham-operated animals.
We also evaluated the serum levels of enzymes released from dying hepatocytes in animals subjected to IR. At 90 min after hepatic ischemia, followed by 6 h of reperfusion, serum levels of aspartate (AST) and alanine (ALT) aminotransferases, as well as lactate dehydrogenase (LDH), were significantly higher in bi-1−/− mice compared to bi-1+/+ animals (AST, 4,712 ± 756 vs. 1,396 ± 183 units/liter, P < 0.05; ALT, 3,996 ± 795 vs. 1,251 ± 223 units/liter, P < 0.05; LDH, 12,727 ± 791 vs. 5,078 ± 80 units/liter, P < 0.05) (Supporting Text and Fig. 6). Thus, bi-1−/− mice demonstrate increased sensitivity to liver IR injury as determined by enzyme markers.
Finally, we evaluated neutrophil accumulation in the post-IR livers of bi-1−/− and bi-1+/+ mice as another indicator of the extent of injury. As measured by myeloperoxidase activity, neutrophil deposition 6 h after reperfusion in ischemic liver lobes was higher in bi-1−/− compared to bi-1+/+ mice, increasing from 0.04 ± 0.001 unit/g in sham controls to 0.74 ± 0.07 unit/g in bi-1+/+ mice and 1.88 ± 0.36 units/g in bi-1−/− mice (P < 0.05) (Supporting Text and Fig. 6).
BI-1 Deficiency Sensitizes Hepatocytes to IR-Induced Apoptosis.
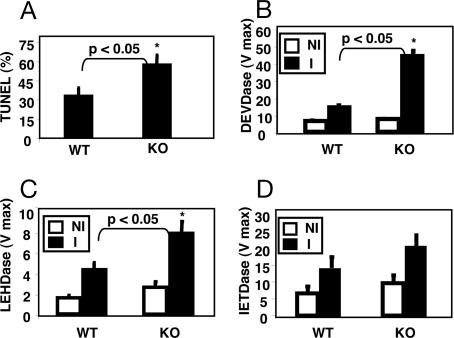
To evaluate the effect of BI-1 deficiency on apoptosis in the IR model, TUNEL assays were performed by using liver sections from mice taken 6 h after reperfusion, detecting in situ cells with fragmented DNA indicative of apoptotic demise. The ischemic liver lobes of bi-1−/− mice contained statistically more TUNEL-positive hepatocytes (60 ± 9%) compared to wild-type animals (40 ± 9%) (P < 0.05) (Fig. 2A). In contrast, virtually no TUNEL-positive liver cells were observed in non-ischemic liver lobes or in sham-operated animals (data not shown).
Fig. 2.
Increased apoptosis and caspase activity in post-IR livers of bi-1 knockout mice. (A) TUNEL Assay. Apoptotic mouse hepatocytes were visualized by TUNEL assay in tissue sections from livers of bi-1−/− (KO) and bi-1+/+ (WT) mice (mean ± SD; n = 5–8). (B–D) Caspase activities. Liver homogenates from ischemic (I) and nonischemic (NI) lobes were prepared from bi-1−/− (KO) and bi-1+/+ (WT) mice after IR injury, normalized for total protein content, and caspase protease activities were measured by using fluorigenic tetrapeptide substrates: Ac-DEVD-AFC (B); Ac-LEHD-AFC (C) and Ac-IETD-AFC (D). Data represent Vmax (mean ± SD; n = 5–8). Statistical significance (∗) was determined by the Mann–Whitney U test (P < 0.05, KO versus WT).
Because effector caspases are required for the execution phases of apoptosis, we investigated the activity of these proteases in ischemia-damaged liver 6 h after reperfusion. Asp-Glu-Val-Asp (DEVD)-hydrolyase activity in tissue lysates provides a measure of effector proteases such as caspases-3 and -7 (13). The rate of Asp-Glu-Val-Asp-amino-4-trifluoromethyl coumarin cleavage was much higher in homogenates of liver lobes (normalized for total protein content) derived from bi-1−/− mice compared to bi-1+/+ after IR injury (Fig. 2B).
Caspase-3 is typically activated by proteolytic processing mediated by upstream apoptotic proteases such as caspase-8 and caspase-9. Caspase-8 represents the apical caspase in the death receptor pathway (14), whereas caspase-9 serves as the initiator caspase of the mitochondrial pathway (15). The preferred tetrapeptide substrates of caspases-8 and -9 are IETD and LEHD, respectively (16). We therefore evaluated levels of IETD and LEHD hydrolyase activity in liver from bi-1−/− and bi-1+/+ mice after IR injury. IR induced marked increases in the levels of both caspase-8 and -9 activity, as measured in liver homogenates (Fig. 2 C and D). The levels of caspase-9 activity, however, were significantly higher in liver of bi-1−/− compared to bi-1+/+ mice (Fig. 2C). In contrast, caspase-8 activity was slightly higher in the IR-damaged livers of bi-1−/− mice, but the results did not reach statistical significance (Fig. 2D).
Analysis of Hepatic Post-IR Events Associated with ER Stress and Apoptosis.
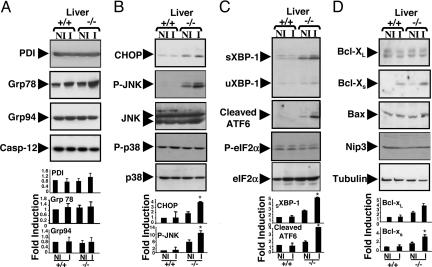
It is known that IR injury promotes ER stress. During ER stress, increases occur in the expression of protective intra-ER molecular chaperones, such as protein disulfide isomerase, Grp78, and Grp94, intended to compensate for damage (4). To determine whether the protection afforded by BI-1 maps upstream or downstream of these events, we monitored expression of these proteins by immunoblotting in post-IR liver (Fig. 3A). IR injury induced comparable increases in Grp78 protein levels in both bi-1−/− and bi-1+/+ mice, indicative of ER stress. Protein disulfide bonding and Grp94 levels, however, were unchanged.
Fig. 3.
Analysis of molecular markers of ER stress. Mice were subjected to 90 min of hepatic ischemia, followed by 6 h of reperfusion. The ischemic (I) and normal liver lobes (NI) were then collected, and total proteins were extracted. The expression of ER stress and apoptotic proteins was compared in bi-1+/+ and bi-1−/− livers by immunoblot analysis. Examples of representative blots are shown at the top. Quantification of the most relevant data were performed by scanning densitometry and is shown at the bottom (mean ± SEM; n = 3–7). Statistical significance was determined by t test and is denoted by asterisks (P ≤ 0.05).
ER stress triggers activation of ER-resident protease, caspase-12, in some scenarios (4). We therefore evaluated the levels and proteolytic processing of caspase-12 by immunoblotting but observed no changes (Fig. 3A), although many hepatocytes underwent apoptosis as measured by TUNEL assay (see above). Thus, it appears unlikely that caspase-12 plays a major role in the apoptotic program activated by IR injury in liver.
The C/EBP homologous protein (CHOP) is a transcription factor whose expression is induced during ER stress and that participates in ER-mediated apoptosis (reviewed in ref. 17). Deregulated CHOP activity compromises cell viability, and cells lacking chop are significantly protected from the lethal consequences of ER stress (18). When assessed by immunoblot analysis in livers of control and BI-1-deficient mice, CHOP protein was elevated in untreated liver of bi-1−/− compared to control bi-1+/+ animals. After IR injury, CHOP protein became further elevated in bi-1−/− mice but was barely detectable in bi-1+/+ mice (Fig. 3B). Thus, BI-1-deficient mice produce more proapoptotic CHOP protein compared to normal animals before and after IR injury.
c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase phosphorylate and enhance CHOP transcription and proapoptotic activity (17). We therefore examined the status of JNK and p38 in livers of bi-1−/− and bi-1+/+ mice before and after IR injury by using phospho-specific antibodies that serve as surrogate markers of kinase activity. Before IR injury, higher levels of phospho-JNK were detected in livers of bi-1−/− mice compared to bi-1+/+ animals (Fig. 3B). After IR injury, levels of phospho-JNK rose further in BI-1-deficient liver, whereas little phospho-JNK was detected in livers of wild-type mice. In contrast, no differences in phospho-p38 levels were observed in livers of either bi-1−/− or bi-1+/+ mice before or after IR injury. The bi-1+/+ and bi-1−/− mice also did not differ in their total levels of JNK and p38 proteins, as revealed by phospho-independent antibodies (Fig. 3B).
XBP-1, ATF4, and ATF6 are transcription factors known to induce chop during ER stress (17, 19). Activated IRE1 initiates unconventional splicing of the mRNA encoding an isoform of XBP-1 protein that induces expression of chop (17). Activated PKR-like ER kinase phosphorylates eIF2α, on serine-51A, which results in translational induction of ATF4 (20). ATF6 is cleaved during ER stress, and its cytosolic domain [ATF6(N)] translocates to the nucleus. We therefore examined the status of XBP-1, ATF4, and ATF6 in livers of bi-1−/− and bi-1+/+ mice before and after IR injury. Levels of spliced XBP-1 protein and cleaved ATF6 were higher in livers of bi-1−/− mice before and after IR injury (Fig. 3C). After IR injury, sXBP-1 protein levels in bi-1−/− mice approximately doubled but remained undetectable in bi-1+/+ animals. In contrast, differences in eIF2α phosphorylation were not detected in bi-1−/− and bi-1+/+ mice before or after IR injury by using phospho-specific antibodies (Fig. 3C). Consistent with this result, levels of ATF4 remained unchanged after IR in both bi-1+/+ and bi-1−/− mice (data not shown). These observations suggest that BI-1 deficiency enhances ATF6 processing and increases selected IRE1 activities (ribonuclease activity but not caspase-12 activation), whereas activation of PKR-like ER kinase pathway is not affected by loss of BI-1. Portions of these data were confirmed by using cultured hepatocytes subjected to hypoxia/glucose deprivation, revealing constitutively elevated levels of CHOP, P-JNK, and ATF6 (but not Grp78, Grp94, or ATF4) in bi-1 cells (Fig. 7, which is published as supporting information on the PNAS web site).
Members of Bcl-2 family play important roles in the regulation of apoptosis and cell death, and some of these proteins target not only mitochondrial but also ER membranes (reviewed in ref. 4). We therefore analyzed by immunoblotting the expression of selected Bcl-2-family proteins in liver before and after IR injury. Because the antiapoptotic protein Bcl-2 is not constitutively expressed in mouse liver (10), we studied other family members. Bcl-XL is the major antiapoptotic protein expressed in hepatocytes (21) and, thus, its expression was evaluated. In addition, we evaluated expression of Bcl-XS, a proapoptotic product of the bcl-X gene that arises through alternative mRNA splicing (22) by using an antibody that preferentially reacts with this isoform of Bcl-X (23). Furthermore, because hypoxia, IR injury, or ER stress has been reported to induce expression of proapoptotic Bcl-2-family proteins Nip3, Bax, and PUMA (24), we also examined these proteins by immunoblotting. Levels of antiapoptotic Bcl-XL and proapoptotic Nip3, Bax, and PUMA were not detectably different in the livers of bi-1−/− and bi-1+/+ mice before or after IR injury. In contrast, IR injury induced increases in the proapoptotic Bcl-XS protein in livers of both bi-1+/+ and bi-1−/− mice after IR injury, with the extent of induction significantly higher in BI-1-deficient animals compared to wild-type mice (Fig. 3D). Thus, although BI-1 deficiency has no effect on expression of most Bcl-2-family proteins examined, BI-1 deficiency may promote increased induction of Bcl-Xs during liver IR injury.
IR Injury Induces bi-1 Gene Expression.
To determine whether bi-1 gene expression is affected by IR injury, we evaluated intra-hepatic bi-1 mRNA levels in wild-type mice, contrasting nonischemic (NI) and ischemic (I) areas. After IR, livers of mice contained significantly increased hepatic bi-1 mRNA levels, as compared with nonischemic controls (390 ± 77 vs. 200 ± 61 arbitrary units (AU); P < 0.05) (Fig. 8, which is published as supporting information on the PNAS web site). In contrast, levels of STAT3 and RelA mRNA were not substantially different, serving as a specificity control. Thus, bi-1 may be a cytoprotective component of the cellular response to hypoxia adaptation, making it a candidate mediator of preconditioning.
Recent data suggest that endothelial nitric oxide synthase (eNOS) is important in limiting the extent of postischemic liver injury, whereas inducible NOS (iNOS) exacerbates it (25). We therefore compared expression of constitutive (eNOS) and inducible (iNOS) mRNA levels after hepatic IR injury in bi-1+/+ and bi-1−/− mice. Hepatic expression of iNOS was higher in bi-1−/− livers subjected to IR injury (1,090 ± 313 vs. 201 ± 50 a.u.; P < 0.05), whereas eNOS mRNA levels were comparable in bi-1+/+ and bi-1−/− animals (3,913 ± 1,077 vs. 3,040 ± 705 a.u.; P = nonsignificant) (Fig. 9, which is published as supporting information on the PNAS web site). We conclude, therefore, that the increased sensitivity of bi-1−/− mice to IR injury cannot be explained by deficient eNOS expression. In contrast, the higher iNOS expression in postischemic bi-1−/− livers correlates with greater tissue damage in these mice but may be a secondary consequence of the more extensive cytokine elaboration that occurs in BI-1-deficient mice (Fig. 9) as a result of more extensive necrosis and inflammatory cell infiltration (Fig. 6).
BI-1 Deficiency Increases Kidney Sensitivity to IR Injury.
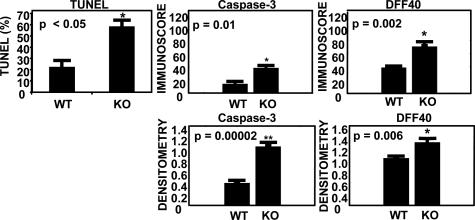
We previously demonstrated that bi-1−/− mice display increased sensitivity in vivo to tunicamycin-induced renal tubular toxicity, consistent with heightened sensitivity to ER stress-induced cell death in this organ (6). To extend these results, we determined the effect of BI-1 deficiency on sensitivity to IR in the kidney, employing a renal artery occlusion model. At 45 min after renal ischemia, followed by 24 h of reperfusion, serum levels of creatinine and blood urea nitrogen (BUN) were significantly higher in bi-1−/− compared to bi-1+/+ animals (creatinine: 1.95 ± 0.26 vs. 0.43 ± 0.09 mg/dl, P < 0.005; BUN, 149.8 ± 10.4 vs. 48.3 ± 13.4 mg/dl, P < 0.0017). Thus, bi-1−/− mice demonstrate increased sensitivity to renal IR injury as determined by renal function markers. Using TUNEL assays to evaluate cell death in tissue sections, we observed greater sensitivity of bi-1−/− kidneys to IR injury, as revealed by a significant increase in the prevalence of TUNEL-positive cells in ischemic kidneys from bi-1−/− compared to bi-1+/+ mice (Fig. 4; see also Fig. 10, which is published as supporting information on the PNAS web site). Evidence of elevated apoptosis in bi-1−/− kidneys subjected to IR injury was also obtained by immunohistochemical analysis by using epitope-specific antibodies that recognized the cleaved (active) forms of caspase-3 and the caspase-3 substrate DNA fragmentation factor-45 (DFF45) (Figs. 4 and 9).
Fig. 4.
Analysis of apoptosis markers in kidney from bi-1−/− and bi-1+/+ mice. (Upper Left) TUNEL-positive nuclei were quantified in tissue sections from kidney of bi-1−/− (KO) and bi-1+/+ (WT) mice and expressed as a percentage of total nuclei counted (mean ± SD; n = 4). The histograms summarize the expression levels of active caspase-3 and cleaved DFF40 as determined by immunoscore (Upper Center and Upper Right) and immunohistochemical densitometry (Lower).
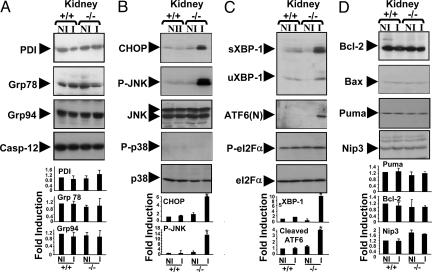
Analysis of IR-Associated Changes in ER Stress Proteins in Kidney.
Because elevated levels of CHOP, sXBP-1, ATF6(N), and phospho-JNK were observed in livers of bi-1−/− mice, we analyzed the levels of these proteins in the kidneys from bi-1−/− versus bi-1+/+ mice before and after IR. Before IR injury, levels of these proteins were similar in the kidneys of bi-1−/− and bi-1+/+ animals (Fig. 5). After IR, marked increases in CHOP, sXBP-1, ATF6(N), and phospho-JNK were found in kidneys of bi-1−/− but not bi-1+/+ mice. In contrast, differences in phospho-p38 and phospho-eIF2α were not observed in bi-1−/− and bi-1+/+ animals. We conclude therefore that BI-1 is required for suppression of ER stress pathways that lead to expression of CHOP and sXBP-1 proteins and that are responsible for activation of ATF6, IRE1, and JNK in both kidney and liver.
Fig. 5.
BI-1 deficiency results in expression of ER stress proteins in kidney. Total proteins were extracted from ischemic (I) and nonischemic (NI) kidneys of bi-1+/+ and bi-1−/− mice. The expression of ER and apoptotic proteins was compared by immunoblotting. Examples of representative blots are shown at top. Data were quantified by scanning densitometry at bottom (mean ± SEM; n = 3–4). Statistical significance was determined by t test (∗, P ≤ 0.05).
Discussion
The data reported here show that loss of bi-1 sensitizes both liver and kidney to IR injury, thus establishing an important in vivo role for this gene in intrinsic resistance to IR-induced cell loss and tissue damage in these organs. Although we favor a direct cytoprotective role for BI-1, we cannot exclude the possibility that secondary factors contribute to the increased sensitivity of liver and kidney to IR injury in vivo. For example, the post-IR livers of bi-1−/− mice accumulated more inflammatory cells and more intrahepatic production of TNFα and IL-6 mRNA was detected, along with higher levels of circulating TNFα and IL-6 in serum. Thus, it is conceivable that bi-1 is required for suppressing inflammatory responses in vivo, and absence of this gene therefore results in unrestrained inflammatory cell activity. However, it is more likely that the increased inflammatory response observed in bi-1−/− mice after IR injury is a consequence of more tissue injury, given the increased release of liver enzymes, more extensive necrosis, and increased incidence of TUNEL-positive hepatocytes seen in bi-1−/− mice within hours after IR injury. Also, the results obtained by using cultured bi-1−/− hepatocytes showing increased sensitivity to OGD also argue for a cell autonomous effect of BI-1 deficiency.
The mechanism by which the BI-1 protein protects hepatocytes from IR injury remains to be clarified. IR injury induces ER stress. The induction of ER chaperones was not significantly different in bi-1−/− versus bi-1+/+ mice, indicating that the increased sensitivity of bi-1−/− mice cannot be explained by differences in this particular ER stress response. However, BI-1-deficient liver and kidney showed evidence of activation of ER stress pathways under basal conditions, with further rises in pathway activation after IR injury, as manifested by increases in the levels of CHOP, sXBP-1, ATF6(N), and phospho-JNK (Fig. 11, which is published as supporting information on the PNAS web site). The sXBP-1 protein is produced as a downstream consequence of activation of the ribonuclease activity of IRE1 during ER stress, coupled with ATF6-mediated increases in XBP-1 mRNA production (26). Therefore, the increases in sXBP-1 observed in liver and kidney of bi-1−/− mice could be a reflection of both ATF6 and IRE1 activation. These results suggest that absence of BI-1 protein predisposes to activation of ATF6 and IRE1 pathways during liver and kidney I/R injury. Activation of ATF6 and IRE1 is known to increase expression of chop (gadd153), a member of the C/EBP-family of basic leucine zipper transcription factors that induces apoptosis (reviewed in ref. 17). In addition to inducing sXBP-1 production, IRE1 activation also induces JNK activation as a downstream consequence of recruiting TRAF2 and activating Ask1 (27). Thus, the increases in phospho-JNK observed in liver and kidney of bi-1−/− mice could be another reflection of IRE1 activation. Interestingly, JNK is speculated to promote CHOP activity at a posttranscriptional level by increasing its transcriptional activity through phosphorylation (17), thereby potentially reinforcing the sXBP-1-dependent pathway that increases CHOP production. How loss of BI-1 encourages activation of the IRE1 and ATF6 pathways remains to be determined. Among the possibilities are effects of BI-1: (i) on ER Ca2+-affecting Ca2+-regulated ER chaperones that suppresses IRE1 and ATF6, and (ii) direct interactions of BI-1 with IRE1 and ATF6 proteins complexes in ER membranes. These and other possible mechanisms require future experimental investigation.
Interestingly, we observed that IR injury induces bi-1 expression in normal liver. We hypothesize, therefore, that induction of endogenous bi-1 gene expression represents an important adaptive response that limits tissue injury during IR. This finding raises the possibility that bi-1 could play a role in the phenomenon of preconditioning, where transient episodes of reversible ischemia provide protection from subsequent hypoxic events. Further experimentation is required to test this hypothesis.
In summary, we have demonstrated the bi-1 gene is required for intrinsic resistance to IR injury in the mammalian liver and kidney in vivo. These data thus expand recent evidence that bi-1 is required for intrinsic resistance to stroke injury (9), showing a broad role for this cytoprotective gene in protection from IR injury. Moreover, we show here that BI-1 deficiency results in an ER stress phenotype, as manifested by induction of CHOP, sXBP-1, ATF6(N), and phospho-JNK production and activation of IRE1. Analysis of components of the BI-1-dependent pathway for protection from ER stress and IR injury may reveal new strategies for organ preservation.
Materials and Methods
Mice.
Mice with targeted disruption of the bi-1 gene (9) were used at 8 to 10 weeks of age, representing bi-1−/− and bi-1+/+ littermates on a C57BL/6 background.
IR Injury Models.
A warm hepatic IR model was used as described in ref. 11 with minor modifications. Arterial and portal venous blood supply were interrupted to the median and left lobes of the liver for 90 min, which was before reperfusion. Sham controls underwent the same surgical procedure but without vascular occlusion. Animals were killed 6 h after reperfusion. The renal IR injury model involved occlusion of the right renal artery and vein for 45 min (28). Animals were killed 24 h after surgery.
Hepatocyte Cultures.
Hepatocytes were isolated from mouse liver and cultured as described in ref. 21. Hepatocytes were seeded into 24-well plates and cultured in normal medium or in medium containing 20 mM deoxyglucose and either 2.5 or 10 mM KCN to achieve OGD, then washed and cultured for 1 day in normal medium before performing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assays (29). Alternatively, cells were cultured with staurosporine (10 μM), anti-Fas mAb (500 ng/ml), or benzoyl-Val-Ala-Asp-fluoromethyl-ketone (50 μM) (Sigma Chemicals). For some experiments, hepatocytes were cultured in 60-mm plates (21) in Hanks’ Balanced Salt Solution (with NaHCO3) in a hypoxia chamber (1% O2) for 1–3 h, then washed and switched to normal media for 1 day in regular atmosphere of 95% air and 5% CO2.
Biomarkers and Immunohistochemical Assays.
Methods for measurement of serum markers of liver and kidney injury and function, and assays for immunohistochemical analysis of apoptosis markers in tissues are provided in Supporting Text.
Histological Evaluation.
Tissue specimens were fixed in 10% buffered formalin, embedded in paraffin, sectioned (4-μm thick), stained with hematoxylin/eosin, and then analyzed blindly. The severity of hepatic IR injury was graded by using Suzuki’s criteria (ref. 12; Table 1). In this classification, sinusoidal congestion, hepatocyte necrosis, and ballooning degeneration are graded from 0 to 4.
Immunoblotting and Caspase Activity Assays.
Detailed methods for immunoblot analysis and caspase activity assays are provided as Supporting Text. Briefly, liver and kidney homogenates were normalized for total protein content and either subjected to SDS/PAGE and immunoblotting by using various primary antibodies in conjunction with an enhanced chemiluminescence detection procedure or diluted into a caspase reaction buffer containing fluorigenic caspase substrate peptides, with subsequent measurement of protease activity by spectrofluorimetry.
TUNEL Assay.
Reperfused tissue specimens were frozen in OCT compound and sectioned at 5 μm for processing by the TUNEL method by using a commercial kit, employing DAB peroxidase substrate (Roche Molecular Biochemicals) and counterstained with 0.5% (wt/vol) methyl green. Specimens were evaluated by UV microscopy at high power magnification (×400) in a blinded fashion. A total of 30 random fields were counted for each TUNEL-stained tissue sample.
Statistics.
The data are expressed as mean ± standard deviation (SD) from a minimum of three determinations. Statistical significance of differences between various samples was determined by Mann–Whitney U test and t test.
Supplementary Material
Acknowledgments
We thank C. L. Kress, M. Thomas, and the personnel of the animal facility at the Burnham Institute for Medical Research for technical assistance; J. Chipuk (La Jolla Institute for Allergy and Immunology) for the anti-PUMA antibody; and D. Ron for valuable discussions. This work was supported by the Californian Breast Cancer Research Program, Fondation pour La Recherche Medicale, Philippe Fondation, and National Institutes of Health Grant AG15393.
Abbreviations
- ATF
activating transcription factor
- BI-1
Bax Inhibitor-1
- CHOP
C/EBP homologous protein
- ER
endoplasmic reticulum
- IR
ischemia reperfusion
- IRE1
inositol-requiring enzyme-1
- OGD
oxygen-glucose deprivation
- sXBP
spliced XBP protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fondevila C., Busuttil R. W., Kupiec-Weglinski J. W. Exp. Mol. Pathol. 2003;74:86–93. doi: 10.1016/s0014-4800(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 2.Cursio R., Gugenheim J., Ricci J. E., Crenesse D., Rostagno P., Maulon L., Saint-Paul M. C., Ferrua B., Auberger A. P. FASEB J. 1999;13:253–261. doi: 10.1096/fasebj.13.2.253. [DOI] [PubMed] [Google Scholar]
- 3.Orrenius S., Zhivotovsky B., Nicotera P. Nat. Rev. Mol. Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- 4.Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C. Oncogene. 2003;22:8608–8618. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]
- 5.Ron D. J. Clin. Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Q., Reed J. C. Mol. Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 7.Bolduc N., Ouellet M., Pitre F., Brisson L. F. Planta. 2003;216:377–386. doi: 10.1007/s00425-002-0879-1. [DOI] [PubMed] [Google Scholar]
- 8.Chae H. J., Ke N., Kim H. R., Chen S., Godzik A., Dickman M., Reed J. C. Gene. 2003;323:101–113. doi: 10.1016/j.gene.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., et al. Mol. Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 10.Bailly-Maitre B., de Sousa G., Boulukos K., Gugenheim J., Rahmani R. Cell Death Differ. 2001;8:279–288. doi: 10.1038/sj.cdd.4400815. [DOI] [PubMed] [Google Scholar]
- 11.Zwacka R. M., Zhang Y., Halldorson J., Schlossberg H., Dudus L., Engelhardt J. F. J. Clin. Invest. 1997;100:279–289. doi: 10.1172/JCI119533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki S., Toledo-Pereyra L. H., Rodriguez F. J., Cejalvo D. Transplantation. 1993;55:1265–1272. doi: 10.1097/00007890-199306000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson D. W., Thornberry N. A. Trends Biochem. Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 14.Varfolomeev E. E., Schuchmann M., Luria V., Chiannilkulchai N., Beckmann J. S., Mett I. L., Rebrikov D., Brodianski V. M., Kemper O. C., Kollet O., et al. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 15.Hakem R., Hakem A., Duncan G. S., Henderson J. T., Woo M., Soengas M. S., Elia A., de la Pompa J. L., Kagi D., Khoo W., et al. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 16.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 17.Oyadomari S., Mori M. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 18.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ron D. J. Clin. Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 21.Bailly-Maitre B., de Sousa G., Zucchini N., Gugenheim J., Boulukos K. E., Rahmani R. Cell Death Differ. 2002;9:945–955. doi: 10.1038/sj.cdd.4401043. [DOI] [PubMed] [Google Scholar]
- 22.Boise L. H., Gonzalez-Garcia M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nunez G., Thompson C. B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 23.Krajewski S., Krajewska M., Shabaik A., Wang H. G., Irie S., Fong L., Reed J. C. Cancer Res. 1994;54:5501–5507. [PubMed] [Google Scholar]
- 24.Krajewski S., Mai J. K., Krajewska M., Sikorska M., Mossakowski M. J., Reed J. C. J. Neurosci. 1995;15:6364–6376. doi: 10.1523/JNEUROSCI.15-10-06364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee V. G., Johnson M. L., Baust J., Laubach V. E., Watkins S. C., Billiar T. R. Shock. 2001;16:355–360. doi: 10.1097/00024382-200116050-00006. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 27.Nishitoh H., Saitoh M., Mochida Y., Takeda K., Nakano H., Rothe M., Miyazono K., Ichijo H. Mol. Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 28.Blydt-Hansen T. D., Katori M., Lassman C., Ke B., Coito A. J., Iyer S., Buelow R., Ettenger R., Busuttil R. W., Kupiec-Weglinski J. W. J. Am. Soc. Nephrol. 2003;14:745–754. doi: 10.1097/01.asn.0000050760.87113.25. [DOI] [PubMed] [Google Scholar]
- 29.Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.