Abstract
Microscale technologies are emerging as powerful tools for tissue engineering and biological studies. In this review, we present an overview of these technologies in various tissue engineering applications, such as for fabricating 3D microfabricated scaffolds, as templates for cell aggregate formation, or for fabricating materials in a spatially regulated manner. In addition, we give examples of the use of microscale technologies for controlling the cellular microenvironment in vitro and for performing high-throughput assays. The use of microfluidics, surface patterning, and patterned cocultures in regulating various aspects of cellular microenvironment is discussed, as well as the application of these technologies in directing cell fate and elucidating the underlying biology. Throughout this review, we will use specific examples where available and will provide trends and future directions in the field.
Keywords: microfabrication, review, soft lithography, stem cells, microfluidics
Each year in the U.S., millions of people suffer from a variety of diseases that could be aided from therapies such as organ transplantation. However, despite the widespread need for transplantable tissues, many patients die while waiting for donor organs. It is from this need that the field of tissue engineering has emerged. Tissue engineering is an interdisciplinary field that involves the use of biological sciences and engineering to develop tissues that restore, maintain, or enhance tissue function (1). Tissue engineering has particular advantages over other therapies such as drugs because it can provide a permanent solution to the problem of organ failure. In general, there are three main approaches to tissue engineering: (i) to use isolated cells or cell substitutes as cellular replacement parts, (ii) to use acellular biomaterials capable of inducing tissue regeneration, and (iii) to use a combination of cells and materials (typically in the form of scaffolds) (Fig. 1) (2).
Fig. 1.
Tissue engineering approaches. Tissue engineering approaches are classified into three categories: (i) cells alone, (ii) cells with scaffolds, and (iii) scaffolds alone. Each one of these approaches can be enhanced by in vitro microenvironmental factors before application as a tissue substitute.
Despite significant advances in tissue engineering (3), which have resulted in successful engineering of organs such as skin and cartilage, there are a number of challenges that remain in making off-the-shelf tissue-engineered organs. These barriers include the lack of a renewable source of functional cells that are immunologically compatible with the patient; the lack of biomaterials with desired mechanical, chemical, and biological properties; and the inability to generate large, vascularized tissues that can easily integrate into the host’s circulatory system with the architectural complexity of native tissues.
Microscale technologies are potentially powerful tools for addressing some of the challenges in tissue engineering (4). MEMS (microelectromechanical systems), which are an extension of the semiconductor and microelectronics industries, can be used to control features at length scales from <1 μm to >1 cm (5). These techniques are compatible with cells and are now being integrated with biomaterials to facilitate fabrication of cell–material composites that can be used for tissue engineering. In addition, microscale technologies allow for an unprecedented ability to control the cellular microenvironment in culture and miniaturize assays for high-throughput applications (Fig. 2).
Fig. 2.
Microscale technologies for tissue engineering. (A) Microtechnologies can be used directly to fabricate improved scaffolds and bioreactors or indirectly to study cellular behavior in controlled conditions or through the use of high-throughput experimentation. (B) A PDMS microfabricated tissue engineering scaffold with the vasculature directly embedded into the scaffold (17). (C) Various microscale techniques used to control different aspects of cell–microenvironment interactions are shown.
In the past few years, microfabrication has been increasingly used in biomedical and biological applications, partly because of the emergence of techniques such as soft lithography to fabricate microscale devices without the use of expensive “clean rooms” and photolithographic equipment (5). Soft lithography is a set of microfabrication techniques that use elastomeric stamps fabricated from patterned silicon wafers to print or mold materials at resolutions as low as several hundred nanometers (6–9). Therefore, soft lithography can be used to control the topography and spatial distribution of molecules on a surface, as well as the subsequent deposition of cells (10, 11). Soft lithographic methods can also be used to fabricate microfluidic channels and scaffolds for tissue engineering in a convenient, rapid, and inexpensive manner (5, 12). In addition, photolithography, a technique in which microscale features are fabricated based on selective exposure of a material to light, can also be used for microfabrication of tissue engineering structures.
In this review, we will discuss the use of microfabrication technologies as they relate to tissue engineering in addressing challenges such as vascularizing engineered tissues, fabricating 3D structures, directing stem cell fate, and controlling cellular microenvironment. In addition, we will discuss the use of these technologies for providing significant insights into cellular behavior in vitro and for creating physiological microenvironments in culture. We will first discuss the use of microscale approaches as they apply directly to tissue engineering in fabricating scaffolds and bioreactors. We will then discuss the use of these technologies as they apply to tissue engineering indirectly through their use in controlling cellular microenvironment.
Microscale Approaches for Tissue Engineering
Cell-Seeded, Microfabricated Scaffolds.
In many tissue engineering applications, scaffolds are used to provide cells with a suitable growth environment, optimal oxygen levels, and effective nutrient transport as well as mechanical integrity (13). Scaffolds aim to provide 3D environments to bring cells in close proximity so that they can assemble to form tissues. Ideally, as the scaffold is degraded, the cells deposit their own extracellular matrix (ECM) molecules and eventually form 3D structures that closely mimic the native tissue architecture. Currently, tissue engineering scaffolds are prepared by using a variety of techniques, such as solvent casting and particulate leaching (14). However, scaffold properties such as pore geometry, size, interconnectivity, and spatial distribution depend on the fabrication process rather than design. The inability to generate desired scaffolds has hindered the construction of engineered tissues that are larger than a few hundred micrometers due to oxygen diffusion limitations (15, 16).
Microfabrication approaches have been used to engineer the desired microvasculature directly into the tissue engineering scaffolds (17, 18). Initial experiments used micromachining technologies on silicon surfaces to generate vascularized systems. Subsequent work on the replica molding of biocompatible polymers from patterned silicon wafers has resulted in the fabrication of biocompatible scaffolds (Fig. 2B). Originally, such scaffolds were fabricated from poly(dimethylsiloxane) (PDMS) (17), a transparent, biocompatible elastomer that is widely used in replica molding (5, 19). More recently, microfabricated capillary networks have been fabricated out of biodegradable elastomers such as poly(dl-lactide-co-glycolide) (PLGA) (20) and poly(glycerol sebacate) (PGS) (21). PLGA has been used to generate biodegradable microfluidic systems and microfabricated scaffolds by superpositioning and stacking multiple layers of fabricated structures (20, 21). These artificial capillary networks were coated with fibronectin and seeded with endothelial cells, which grew to confluence within a few days. However, there are potential disadvantages with the use of PLGA, including its rigid mechanical properties (22) and bulk degradation kinetics (23). Thus, to improve the mechanical properties and to allow for surface erosion instead of bulk erosion of biodegradable scaffolds, other polymers such as PGS (23) have been used (24). PGS films were patterned with the desired microvasculature and bonded to a flat film to create a capillary network.
Layer-by-layer microfluidic patterning has also been used to generate scaffolds (25–27). In this method, cells and matrix biopolymers were flowed through channels with controlled flow rates. By sequential deposition of cells and matrix on particular regions within the microchannels, 3D structures were generated with controlled location of the deposited cells.
Alternative methods of fabricating scaffolds with microscale and nanoscale resolution include 3D printing, microsyringe deposition, tissue spin casting, and electrospinning of nanofibers. In 3D printing, polymer particles and salt are printed by using a bonding agent, which forms a porous scaffold once the salt is dissolved (28, 29). More recently, rapid prototyping methods such as soft lithography and microsyringe deposition have been used to fabricate PLGA (21) and polyurethane (30) scaffolds composed of multilayer structures with controlled resolution. Although a comprehensive review of these technologies is beyond the scope of this report (for detailed reviews, see refs. 31–33), they are emerging as powerful methods of fabricating tissue engineering scaffolds.
Spatially Regulated Hydrogels and Scaffolds.
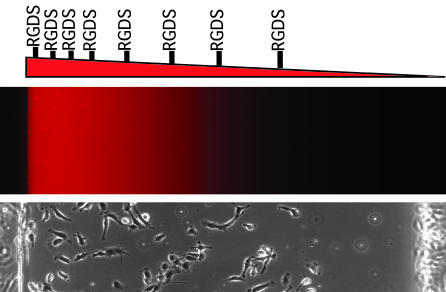
Many biological processes are regulated by spatially dependent signals. For example, gradients of molecules are commonly used in the body to regulate cell migration, axon extension, angiogenesis, and differentiation. Therefore, controlling the spatial location of molecules on a surface or throughout a material could be potentially beneficial for tissue engineering (34). A common approach to generate gradients is to release molecules from a source to form a concentration gradient over time as the molecule diffuses away from the source (35). However, these gradients are unstable, and it is difficult to control their shapes. Therefore, methods for conjugating biomolecules to materials have been used to increase the stability of signaling molecules in a spatially controlled manner. For example, by using lasers, specific regions within an agarose gel were tethered with RGD (Arg–Gly–Asp) peptide, which allowed for neurite extension within peptide-modified regions (36). By using similar techniques, multicomponent, spatially patterned, photocrosslinkable hydrogels were fabricated to localize growth factors within hydrogels (37). Microfabrication approaches such as microfluidics provide an attractive alternative to these technologies because of their availability and cost effectiveness. Recently, the ability to pattern fluids within microchannels has been merged with photopolymerization chemistry to form spatially oriented hydrogels (38, 39). As a result, gradients of the photocrosslinkable monomers were formed within microfluidic channels and subsequently gelled by exposure to UV light. Hydrogels were synthesized with gradients of signaling or adhesive molecules or with varying crosslinking density across the material (Fig. 3) with the ability to direct cell behavior such as migration, adhesion, and differentiation (39, 40).
Fig. 3.
Gradient hydrogels for tissue engineering. (Top) Hydrogels can be fabricated with control over the spatial properties of the materials by embedding a gradient of materials, such as RGD peptide, directly into the material. (Middle and Bottom) The shape of the gradient can be visualized by using fluorescent molecules (Middle), and its function can observed by imaging endothelial cell adhesion after a few hours on the gels (Bottom) (39).
In addition to bulk characteristics of scaffold materials, surface properties also have a significant effect on cellular behavior such as cell adhesion and migration. Microcontact printing has been used to modify surfaces of scaffold materials such as PLGA (41, 42), poly(lactic acid) (42), and chitosan (43) with variations of polyethylene glycol (PEG) or other biological ligands, such as RGD or biotin (44). In addition, other techniques such as microfluidic patterning have been used to generate patterns on various tissue engineering surfaces such as PLGA (45). In most cases, these techniques have been limited to 2D substrates, and their incorporation to porous scaffolds remains a challenge.
Microfabrication approaches have also been used to control substrate topography with resolutions as small as 50 nm to control various cell fate decisions. For example, it has been seen that surface topography of tissue culture substrates can significantly affect cardiomyocyte orientation. In these cultures, topographical features that were a few microns across were used to orient cells and enhance their function (46). Furthermore, because microtextured and nanotextured substrates have been shown to significantly influence cell adhesion, gene expression (47–49), and migration (50), such features can be incorporated into microfabricated tissue engineering scaffolds to provide functional cues to cells. For example, topographically patterned PLGA surfaces have been shown to induce alignment and elongation of smooth muscle cells (51) and to enhance the adhesion of several cell types such as endothelial cells and smooth muscle cells (52, 53). Although these approaches have not been easily incorporated into 3D PLGA scaffolds, it may be possible to stack PLGA layers with the desired topographical features to induce specific cell responses.
Controlled Microbioreactors.
Bioreactors could be useful in tissue engineering either for growing cells and tissues or as extracorporeal devices for liver and kidney diseases. Microfluidic bioreactors are potentially advantageous for cellular applications because they provide a large surface-area-to-volume ratio, as well as many other biomimetic properties. Recently, microreactors have shown promise in applications where conventional bioreactors have failed to provide the cells with adequate nutrients and oxygen. For example, multilayer PDMS microfluidic networks have been used to culture hepatocytes in vitro (54, 55). Inside these reactors, hepatocytes were maintained for many days as they spread and grew to confluency within the channels. In addition, at least 10 layers have been stacked, indicating that such technology is scalable. In another approach, the combination of 3D architecture and fluid perfusion has been used to mimic liver sinusoids (56). Silicon microfluidic chips with holes through the plates were placed on a membrane. The medium was flowed through each hole in the membrane as the cells were retained inside each well. Inside these wells, cells formed spheroids and maintained elevated liver function. Microchannels have also been used as improved versions of flat plate bioreactors for hepatocyte culture with the ability to control parameters such as shear stress, contact with parenchymal cells, and oxygenation (57, 58).
Future generations of microfluidic reactors provide powerful means of exposing cells to various physiological stimuli. For example, a Braille system has been developed to create physiological flow conditions such as pulsatile flow. In this scheme, computer-actuated metal pins were used to deform PDMS channels to pump and regulate the flow of the fluid within the microchannels (59, 60). In addition, complex reactors have been fabricated with the aim of recapitulating the multiple organ interactions of the body. For example, compartmentalized microreactors have been generated to mimic the key functions of the lungs, liver, and fat tissues (61). These animal-on-a-chip devices could be used to test the pharmacological and toxicity effects of drugs in a controlled manner.
Cell Assembly for Tissue Engineering.
Artificial microtissues can also be fabricated by inducing the reaggregation of one or more cell types (62). These microtissues may be beneficial for applications such as pancreatic, liver, vascular, and cardiac tissue engineering, as well as drug discovery. Layering of cells has been used to engineer myocardial tissues by assembling multiple sheets of cardiomyocytes (63) or to engineer blood vessels by fabricating cylindrically rolled sheets of endothelial cells (64). Although such approaches may be suitable for some tissue engineering applications, they lack the complexity associated with the architecture of more complex organs. Microscale approaches may provide a solution to this challenge as templates to generate microtissues in a reproducible manner. For example, by using a combination of microcontact printing and micromachining, hepatocyte spheroids have been formed (65). More recently, nonadhesive PEG microwells have been used as templates for formation of aggregates of various cell types, including ES cells (66) (Fig. 4). This approach aims to overcome the disadvantages associated with the hanging drop and suspension culture methods (62) by providing control over the size, shape, and other features of the cellular assembly in a scalable manner. The controlled formation of embryoid bodies may also be important in generating more homogeneous cultures that are capable of directing the differentiation of ES cells. In addition, template-based assembly of cells could be used to organize multiple cell types into specific geometries relative to each other within these aggregated tissue sections. It is envisioned that with the integration of such technologies with biomaterials such as photocrosslinkable gels and microfluidics, more complex tissue sections for therapeutic applications can be fabricated.
Fig. 4.
Microscale tissue engineering using template-based cell assembly. (A) A schematic diagram of the template-based assembly method. PEG microwells were fabricated so that cells could dock within the low-shear-stress regions generated within the microstructures. Once cells had immobilized within the microwells, other cells were washed away, and the cells within the microwells formed aggregates of controlled properties. (B) A light microscope image of 100-μm PEG wells that were seeded with ES cells and washed. (C) A scanning electron micrograph of cells within PEG microwells (A.K., J. Yeh, G. Eng, J. Fukuda, O. Farokhzad, J. J. Cheng, J. Bumbling, and R.L., unpublished data).
Controlling the Cellular Microenvironment in Vitro
A major challenge to the clinical feasibility of tissue engineering is a viable cell source. Cells have been traditionally derived from autografts (from the same patient), allografts (from others), or xenografts (from other species). With the emergence of adult stem cells and ES cells, potentially powerful cell sources have emerged. One of the major challenges associated with the use of stem cells is to understand the microenvironmental cues that regulate their fate. Microscale approaches can also be used to control culture conditions and perform high-throughput experimentation, hence providing a suitable tool to study cell–microenvironment interactions in vitro. In most tissue culture systems, the cellular microenvironment is vastly different relative to in vivo conditions. In the body, cells are exposed to a controlled microenvironment that is tightly regulated with respect to interactions with the surrounding cells, soluble factors, and ECM molecules. In particular, the spatial and temporal distribution of these signals is tightly controlled and unique to each organ. Furthermore, cells in the body are exposed to a 3D environment instead of the 2D environment experienced by cells in traditional culture dishes.
Extracellular cues are important in regulating adult stem cell and ES cell fate decisions. Adult stem cells are present within unique stem cell niches that regulate their self-renewal and differentiation (67–71). Engineering artificial stem cell niches, potentially through microscale techniques, is a potent bioengineering approach to regulate stem cell fates in vitro (72). Furthermore, ES cells differentiate based on a series of spatially and temporally regulated signals. Therefore, given the proper series of signals, it may be possible to differentiate ES cells into any desired cell type efficiently and reproducibly (73, 74). Microscale technologies provide a powerful tool to investigate the extracellular signals that regulate cell fate, because they can control cell–microenvironment interactions and be merged with high-throughput technologies to test many environmental factors simultaneously. In general, microscale approaches to control cell–microenvironment interactions can be separated into cell–cell, cell–matrix, and cell-soluble factor components (Fig. 5), which are described below.
Fig. 5.
Microscale approaches for controlling the in vitro cellular microenvironment. (A) Light microscope images of ES cells patterned on PEG-coated substrates as an example of surface patterning for regulating cell–ECM interactions. (B) A fluorescent image of patterned cocultures depicting control over the degree of heterotypic and homotypic interactions between ES cells and fibroblasts (95). (C) An image of a cell and a schematic diagram of microfluidic methods of regulating cell-soluble signal interactions by flowing two parallel streams of fluids on an individual cell (81).
Arrays of Cells for Clonal Analysis and Controlling Cell Shape.
Cell arrays have been used to pattern stem cells on 2D substrates (Fig. 5A). Arrays of cells can be used to localize and track individual cells, enabling clonal analysis of stem cell fates (75, 76). For example, clonal populations of neural stem cells were immobilized within microfabricated structures, and their progeny was tracked by using real-time microscopy, yielding information about cellular kinetics and cell fate decisions in a high-throughput manner (76) (Fig. 6). By using this approach, it is possible to study the response of individual stem cells to various microenvironmental signals.
Fig. 6.
Stem cell arrays for tracking cell fates in culture. (A) Cell microarrays used to track clonal populations of stem cells after 4 days. Green boxes had proliferating cells, whereas red boxes did not contain proliferating cells (76). (B) Fluorescent images of the differentiating cells inside a microfabricated array that were stained with Tuj1 (red) and glial fibrillary acidic protein (blue) (76). (Scale bar, 100 μm.)
Cell patterning on geometrically defined shapes has been used to study the effects of cell shape on various cell fate decisions. As cells adhere onto micropatterned substrates, they align themselves to the shape of the underlying adhesive region. This shape change induces changes in the cytoskeletal features and has been shown to influence apoptosis and proliferation (77). More recently, it has been determined that cell shape can also control stem cell differentiation. When human mesenchymal stem cells were patterned on fibronectin islands of various sizes, cells on large islands adhered and flattened, whereas cells on small islands generated spherically shaped cells. As these cells were stained for differentiation markers, it was observed that the spread cells generated osteoblasts, whereas spherical cells gave rise to adipocytes (78) (Fig. 7). Further elucidation of the mechanism indicated that cell shape regulated the activation of the RhoA pathway, demonstrating that mechanical stresses experienced during differentiation can be crucial for directing stem cell differentiation. Therefore, controlling the cellular microenvironment through micropatterning may be used for directing cell fate for tissue engineering applications.
Fig. 7.
Light microscope images of human mesenchymal stem cells on small and large fibronectin islands after 1 week of culture. The images indicate that cells on the small islands stained for lipids, thus differentiating into adipogenic fates, whereas cells on large islands stained for alkaline phosphatase and differentiated into osteoblasts (78). (Scale bars, 50 μm.)
Microfluidics for Regulating Cell-Soluble Factor Interactions.
The ability to laminarly flow fluids within microchannels can be used to control the spatial positioning of soluble factors relative to cells. Laminar flows form within microchannels because of low convective mixing, which limits molecular transport to diffusion. Laminarly flowing fluids have been used to pattern cells and their microenvironments (79, 80). Furthermore, an individual cell can be exposed to multiple microenvironments simultaneously by being placed at the interface between two or more adjacent streams (81). One can treat distinct parts of a cell or cell aggregate to probe the influence of asymmetrically oriented signals on cell behavior or to stain specific regions of a cell for the study of intracellular molecular kinetics. The partial treatment of cell aggregates has already been used to perturb embryonic development and to study the effects of changing the temperature of part of an embryo (82). In addition, microfluidic gradient generators that are fabricated by using the same principles have been used to study the effects of concentration gradients on cells. Linear or complex soluble gradients are generated by sequential merging, mixing, and splitting of two or more inlet streams, each of which may contain a particular environmental stimulus (83, 84) (Fig. 5C). These stable gradients have been used to study complex biological systems. For example, cell chemotaxis in response to soluble chemoattractants has been studied to yield insights into neutrophils migration in response to various shape gradients of IL-8 (85). Microfluidic gradients have also been used to study neural axon extensions (86, 87) and neural stem cell differentiation (88). It is important to consider design parameters such as shear stress, transport phenomenon, and material–cell interactions within these devices (12); however, with the explosion of research in this area in the past few years, such parameters are continuously optimized and modified to attain optimum conditions for various cell types.
Patterned Cocultures for Controlling Cell–Cell Interactions.
Inside the body, cells lie in contact or in close proximity to other cell types in a tightly controlled architecture. Tissue engineering constructs that aim to reproduce the architecture and the geometry of tissues will benefit from methods of controlling cell–cell interactions within these tissues. Patterned cocultures are a useful tool for tissue engineered constructs and for studying cell–cell interactions in vitro, because they can be used to control the degree of homotypic and heterotypic cell–cell contact. Some of the pioneering work in this area has been performed by studying the interaction of hepatocytes and nonparenchymal fibroblasts in cocultures (89). By using these techniques, specific information about the nature of hepatoycte and fibroblast interaction in culture has been elucidated (90–92). Although the original methods of fabricating patterned coculture required selective adhesion of each cell types to a particular material on a patterned region, subsequent methods have been developed that allow a wider array of cell types to be tested. The recently developed methods are based on thermally reversible polymers (93, 94), layer-by-layer deposition of ionic polymers (95) (Fig. 5B), microfluidic deposition (96), and molding of hydrogels (97). Through these approaches, other coculture techniques have been used to probe the interaction of a number of other cell types, including ES cells and other developmentally relevant cell types (95).
High-Throughput Assays for Tissue Engineering.
Material arrays.
As methods to develop small-molecule (98), polymeric (99), and genome-based libraries become more widespread and available, there is a great need to test these libraries by using high-throughput assays. Such libraries are useful for tissue engineering and have already yielded candidates that have been shown to induce osteogenesis (100) and cardiomyogenesis (101) from ES cells as well as the dedifferentiation of committed cells (102). Microscale technologies can miniaturize assays and facilitate high-throughput experimentation and therefore provide a potential tool for screening libraries. Recently, robotic spotters capable of dispensing and immobilizing nanoliters of material have been used to fabricate microarrays, where cell–matrix interactions can be tested and optimized in a high-throughput manner. For example, synthetic biomaterial arrays have been fabricated to test the interaction of stem cells with various extracellular signals (103) (Fig. 8). In this approach, thousands of polymeric materials were synthesized, and their effect on differentiation of human ES cells (103) and human mesenchymal stem cells (104) was evaluated. These interactions have led to unexpected cell–material interactions. Although the molecular mechanisms associated with the biological responses have yet to be clarified, such technology may be widely applicable in cell–microenvironment studies and in the identification of cues that induce desired cell responses. Also, the materials that yield the desired responses could be used as templates for tissue engineering scaffolds. Such an approach is a radical change from traditional methods of developing new biomaterials, where polymers have been individually developed and tested for their effect on cells.
Fig. 8.
Schematic diagram of the procedure used to fabricate arrays of polymeric biomaterials (103).
In addition to analyzing synthetic material libraries, the effect of natural ECM molecules on cell fate can be evaluated in a high-throughput manner. In one example, combinatorial matrices of various natural ECM proteins were tested for their ability to maintain the function of differentiated hepatocytes and to induce hepatic differentiation from murine ES cells (105).
Microfluidic systems.
Microfluidic systems are ideal for performing cell-based high-throughput experiments because they are cheap and can be used to miniaturize assays, reduce expensive reagent volumes, and reduce cell numbers (19). Microfluidic arrays can be used to perform screening experiments, to test drug toxicity, and to optimize culture conditions for inducing specific cell fates. To create such systems, it is important to be able to culture cells inside microfluidic devices in a reliable and long-term manner and to be able to combine these devices in such a way as to test many conditions simultaneously. Many experiments have localized cells within microfluidic channels; however, the long-term survival of cells inside these channels has not been demonstrated (75, 87). In addition, it is important to localize the cells in particular regions of microfluidic channels by using substrate patterning or topographical structures (66, 106). Also, it is important to interface the microscale channels to the outside world. Current techniques include the use of complicated valves and pumps to fabricate large arrays of microchannels that can be individually addressed (107). The ability to test various combinations and doses of drugs can be useful for performing factorial experiments, which can optimize culture conditions for inducing various biological fates (108). An alternative approach for cell-based molecular screening is to use multiphenotype cell arrays (109, 110). In these systems, the effect of a soluble factor can be tested on many cell types simultaneously. Incorporating multiphenotype cell arrays within microfluidic arrays has shown promise as a tool for fabricating high-throughput assays (110).
Future Directions
The synthesis of new materials and the incorporation of improved fabrication strategies have led to dramatic enhancements in the complexity and biomimecry of today’s tissue engineering constructs. In particular, the use of microscale technologies in tissue engineering, either directly as transplantable constructs or indirectly as a tool for understanding the biology of organs, has pushed the field closer to clinical therapies. However, significant challenges need to be addressed. These barriers include the lack of suitable materials with the desired degradation rates, products, and suitable mechanical properties for the desired tissue. Another challenge that must be addressed is the optimization of scaffold architecture, including pore size, morphology, surface topography, and bioactivity. Also, new and optimized processing methods must be developed to address issues related to cell seeding, vascularization, and fabrication of 3D tissues without cumbersome methods of assembly. Furthermore, more research is required in testing and validating the in vivo functionality of microfabricated constructs and in assessing the performance of these constructs against competing technologies.
In addition, microscale tools have provided much insight into the fundamental biology of how cells interact with the surrounding components, such as cell–cell, cell-soluble factors, and cell–ECM molecules. This knowledge should be used to direct cell fates as a cell source or be incorporated into tissue engineering scaffolds. Thus, the continued merger of engineering, medicine, materials, and biological sciences as mediated by microscale approaches in tissue engineering and biology will enhance our ability to create in vivo-like physiological models that can be used for fabricating tissues or for understanding fundamental biology.
Conclusions
The widespread use and availability of lithographic technologies have made microscale engineering a powerful tool for tissue engineering and biological applications. Microfabrication techniques and engineered biomaterials are being used for tissue engineering in a variety of applications: for example, by fabricating scaffolds with control over features such as shape and pore architecture, as templates for microtissue formation, or as improved bioreactors. In addition, microscale control of cellular environments has been used to probe the influence of the spatial and temporal effects of specific cell–cell, cell–ECM, and cell-soluble factor interactions on cell fate. Finally, the ability to simultaneously test many environmental factors on cell behavior has been used to optimize culture conditions and material–cell interactions.
Acknowledgments
This work was supported by National Institutes of Health Grants HL060435, DE013023, and DE16516; the National Science Laboratory; The Charles Stark Draper Laboratory; Center for Integration of Medicine and Innovative Technology Cooperative Agreement DAMD-02-2-0006; and Institute for Soldier Nanotechnology Grant DAAD-19-02-D-002.
Abbreviations
- ECM
extracellular matrix
- PDMS
poly(dimethylsiloxane)
- PLGA
poly(dl-lactide-co-glycolide)
- PEG
polyethylene glycol.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Langer R., Vacanti J. P. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Griffith L. G., Naughton G. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 3.Niklason L. E., Langer R. J. Am. Med. Assoc. 2001;285:573–576. doi: 10.1001/jama.285.5.573. [DOI] [PubMed] [Google Scholar]
- 4.Andersson H., van den Berg A. Lab Chip. 2004;4:98–103. doi: 10.1039/b314469k. [DOI] [PubMed] [Google Scholar]
- 5.Whitesides G. M., Ostuni E., Takayama S., Jiang X. Y., Ingber D. E. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 6.Ostuni E., Chen C. S., Ingber D. E., Whitesides G. M. Langmuir. 2001;17:2828–2834. [Google Scholar]
- 7.Ostuni E., Yan L., Whitesides G. M. Colloids Surf. B Biointerfaces. 1999;15:3–30. [Google Scholar]
- 8.Kane R. S., Takayama S., Ostuni E., Ingber D. E., Whitesides G. M. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 9.Xia Y. N., Whitesides G. M. Angew. Chem. Int. Ed. 1998;37:551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 10.Khademhosseini A., Jon S., Suh K. Y., Tran T. N. T., Eng G., Yeh J., Seong J., Langer R. Adv. Mater. 2003;15:1995–2000. [Google Scholar]
- 11.Suh K. Y., Khademhosseini A., Yang J. M., Eng G., Langer R. Adv. Mater. 2004;16:584–588. [Google Scholar]
- 12.Walker G. M., Zeringue H. C., Beebe D. J. Lab Chip. 2004;4:91–97. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 13.Cohen S., Bano M. C., Cima L. G., Allcock H. R., Vacanti J. P., Vacanti C. A., Langer R. Clin. Mater. 1993;13:3–10. doi: 10.1016/0267-6605(93)90082-i. [DOI] [PubMed] [Google Scholar]
- 14.Karp J., Dalton P., Shoichet M. MRS Bull. 2003;28:301–306. [Google Scholar]
- 15.Folkman J., Hochberg M. J. Exp. Med. 1973;138:745–753. doi: 10.1084/jem.138.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutmacher D. W., Sittinger M., Risbud M. V. Trends Biotechnol. 2004;22:354–362. doi: 10.1016/j.tibtech.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Borenstein J. T., Terai H., King K. R., Weinberg E. J., Kaazempur-Mofrad M. R., Vacanti J. P. Biomed. Microdevices. 2002;4:167–175. [Google Scholar]
- 18.Kaihara S., Borenstein J., Koka R., Lalan S., Ochoa E. R., Ravens M., Pien H., Cunningham B., Vacanti J. P. Tissue Eng. 2000;6:105–117. doi: 10.1089/107632700320739. [DOI] [PubMed] [Google Scholar]
- 19.Sia S. K., Whitesides G. M. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 20.King K., Wang C., Kaazempur-Mofrad M., Vacanti J., Borenstein J. Adv. Mater. 2004;16:2007–2012. [Google Scholar]
- 21.Vozzi G., Flaim C., Ahluwalia A., Bhatia S. Biomaterials. 2003;24:2533–2540. doi: 10.1016/s0142-9612(03)00052-8. [DOI] [PubMed] [Google Scholar]
- 22.Leatrese D. H., Kim B. S., Mooney D. J. J. Biomed. Mater. Res. 1998;42:396–402. doi: 10.1002/(sici)1097-4636(19981205)42:3<396::aid-jbm7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Ameer G. A., Sheppard B. J., Langer R. Nat. Biotechnol. 2002;20:602–606. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 24.Fidkowski C., Kaazempur-Mofrad M. R., Borenstein J., Vacanti J. P., Langer R., Wang Y. Tissue Eng. 2005;11:302–309. doi: 10.1089/ten.2005.11.302. [DOI] [PubMed] [Google Scholar]
- 25.Tan W., Desai T. A. Tissue Eng. 2003;9:255–267. doi: 10.1089/107632703764664729. [DOI] [PubMed] [Google Scholar]
- 26.Tan W., Desai T. A. Biomaterials. 2004;25:1355–1364. doi: 10.1016/j.biomaterials.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Tan W., Desai T. A. J. Biomed. Mater. Res. A. 2005;72:146–160. doi: 10.1002/jbm.a.30182. [DOI] [PubMed] [Google Scholar]
- 28.Griffith L. G., Wu B., Cima M. J., Powers M. J., Chaignaud B., Vacanti J. P. Ann. N.Y. Acad. Sci. 1997;831:382–397. doi: 10.1111/j.1749-6632.1997.tb52212.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim S. S., Utsunomiya H., Koski J. A., Wu B. M., Cima M. J., Sohn J., Mukai K., Griffith L. G., Vacanti J. P. Ann. Surg. 1998;228:8–13. doi: 10.1097/00000658-199807000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folch A., Mezzour S., During M., Hurtado O., Toner M., Muller R. Biomed. Microdevices. 2000;2:207–214. [Google Scholar]
- 31.Yeong W. Y., Chua C. K., Leong K. F., Chandrasekaran M. Trends Biotechnol. 2004;22:643–652. doi: 10.1016/j.tibtech.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia S. N., Chen S. C. Biomed. Microdevices. 1999;2:131–144. [Google Scholar]
- 33.Tsang V. L., Bhatia S. N. Adv. Drug Delivery Rev. 2004;56:1635–1647. doi: 10.1016/j.addr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Kapur T. A., Shoichet M. S. J. Biomed. Mater. Res. A. 2004;68:235–243. doi: 10.1002/jbm.a.10168. [DOI] [PubMed] [Google Scholar]
- 35.Richardson T. P., Peters M. C., Ennett A. B., Mooney D. J. Nat. Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 36.Luo Y., Shoichet M. S. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 37.Mapili G., Lu Y., Chen S., Roy K. J. Biomed. Mater. Res. B. 2005;75:414–424. doi: 10.1002/jbm.b.30325. [DOI] [PubMed] [Google Scholar]
- 38.Koh W. G., Revzin A., Pishko M. V. Langmuir. 2002;18:2459–2462. doi: 10.1021/la0115740. [DOI] [PubMed] [Google Scholar]
- 39.Burdick J. A., Khademhosseini A., Langer R. Langmuir. 2004;20:5153–5156. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 40.Zaari N., Rajagopalan S. K., Kim S. K., Engler A. J., Wong J. Y. Adv. Mater. 2004;16:2133–2137. [Google Scholar]
- 41.Lu L., Nyalakonda K., Kam L., Bizios R., Gopferich A., Mikos A. G. Biomaterials. 2001;22:291–297. doi: 10.1016/s0142-9612(00)00179-4. [DOI] [PubMed] [Google Scholar]
- 42.Lin C. C., Co C. C., Ho C. C. Biomaterials. 2005;26:3655–3662. doi: 10.1016/j.biomaterials.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 43.Kumar G., Wang Y. C., Co C. C., Ho C. C. Langmuir. 2003;19:10550–10556. [Google Scholar]
- 44.Lee K. B., Kim D. J., Lee Z. W., Woo S. I., Choi I. S. Langmuir. 2004;20:2531–2535. doi: 10.1021/la036209i. [DOI] [PubMed] [Google Scholar]
- 45.Liu V. A., Jastromb W. E., Bhatia S. N. J. Biomed. Mater. Res. 2002;60:126–134. doi: 10.1002/jbm.10005. [DOI] [PubMed] [Google Scholar]
- 46.Deutsch J., Motlagh D., Russell B., Desai T. A. J. Biomed. Mater. Res. 2000;53:267–275. doi: 10.1002/(sici)1097-4636(2000)53:3<267::aid-jbm12>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 47.den Braber E. T., de Ruijter J. E., Ginsel L. A., von Recum A. F., Jansen J. A. J. Biomed. Mater. Res. 1998;40:291–300. doi: 10.1002/(sici)1097-4636(199805)40:2<291::aid-jbm14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 48.Walboomers X. F., Croes H. J., Ginsel L. A., Jansen J. A. J. Biomed. Mater. Res. 1999;47:204–212. doi: 10.1002/(sici)1097-4636(199911)47:2<204::aid-jbm10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 49.van Kooten T. G., Whitesides J. F., von Recum A. J. Biomed. Mater. Res. 1998;43:1–14. doi: 10.1002/(sici)1097-4636(199821)43:1<1::aid-jbm1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 50.Teixeira A. I., Abrams G. A., Bertics P. J., Murphy C. J., Nealey P. F. J. Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thakar R. G., Ho F., Huang N. F., Liepmann D., Li S. Biochem. Biophys. Res. Commun. 2003;307:883–890. doi: 10.1016/s0006-291x(03)01285-3. [DOI] [PubMed] [Google Scholar]
- 52.Miller D. C., Thapa A., Haberstroh K. M., Webster T. J. Biomaterials. 2004;25:53–61. doi: 10.1016/s0142-9612(03)00471-x. [DOI] [PubMed] [Google Scholar]
- 53.Thapa A., Webster T. J., Haberstroh K. M. J. Biomed. Mater. Res. A. 2003;67:1374–1383. doi: 10.1002/jbm.a.20037. [DOI] [PubMed] [Google Scholar]
- 54.Leclerc E., Sakai Y., Fujii T. Biotechnol. Prog. 2004;20:750–755. doi: 10.1021/bp0300568. [DOI] [PubMed] [Google Scholar]
- 55.Leclerc E., Sakai Y., Fujii T. Biomed. Microdevices. 2003;5:109–114. [Google Scholar]
- 56.Powers M. J., Domansky K., Kaazempur-Mofrad M. R., Kalezi A., Capitano A., Upadhyaya A., Kurzawski P., Wack K. E., Stolz D. B., Kamm R., Griffith L. G. Biotechnol. Bioeng. 2002;78:257–269. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- 57.Park J., Berthiaume F., Toner M., Yarmush M. L., Tilles A. W. Biotechnol. Bioeng. 2005;90:632–644. doi: 10.1002/bit.20463. [DOI] [PubMed] [Google Scholar]
- 58.Tilles A. W., Berthiaume F., Yarmush M. L., Tompkins R. G., Toner M. J. Hepatobiliary Pancreat. Surg. 2002;9:686–696. doi: 10.1007/s005340200095. [DOI] [PubMed] [Google Scholar]
- 59.Song J. W., Gu W., Futai N., Warner K. A., Nor J. E., Takayama S. Anal. Chem. 2005;77:3993–3999. doi: 10.1021/ac050131o. [DOI] [PubMed] [Google Scholar]
- 60.Gu W., Zhu X., Futai N., Cho B. S., Takayama S. Proc. Natl. Acad. Sci. USA. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viravaidya K., Sin A., Shuler M. L. Biotechnol. Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 62.Kelm J. M., Fussenegger M. Trends Biotechnol. 2004;22:195–202. doi: 10.1016/j.tibtech.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Shimizu T., Yamato M., Kikuchi A., Okano T. Biomaterials. 2003;24:2309–2316. doi: 10.1016/s0142-9612(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 64.L’Heureux N., Paquet S., Labbe R., Germain L., Auger F. A. FASEB J. 1998;12:47–56. doi: 10.1096/fasebj.12.1.47. [DOI] [PubMed] [Google Scholar]
- 65.Fukuda J., Mizumoto H., Nakazawa K., Kajiwara T., Funatsu K. Int. J. Artif. Organs. 2004;27:1091–1099. doi: 10.1177/039139880402701213. [DOI] [PubMed] [Google Scholar]
- 66.Khademhosseini A., Yeh J., Jon S., Eng G., Suh K. Y., Burdick J. A., Langer R. Lab Chip. 2004;4:425–430. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 67.Zhang J., Niu C., Ye L., Huang H., He X., Tong W. G., Ross J., Haug J., Johnson T., Feng J. Q., et al. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 68.Calvi L. M., Adams G. B., Weibrecht K. W., Weber J. M., Olson D. P., Knight M. C., Martin R. P., Schipani E., Divieti P., Bringhurst F. R., et al. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 69.Lemischka I. R. Stem Cells. 1997;15:63–68. doi: 10.1002/stem.5530150810. [DOI] [PubMed] [Google Scholar]
- 70.Benitah S. A., Frye M., Glogauer M., Watt F. M. Science. 2005;309:933–935. doi: 10.1126/science.1113579. [DOI] [PubMed] [Google Scholar]
- 71.Watt F. M., Hogan B. L. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 72.Semino C. E. J. Biomed. Biotechnol. 2003;2003:164–169. doi: 10.1155/S1110724303208019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zandstra P. W., Nagy A. Annu. Rev. Biomed. Eng. 2001;3:275–305. doi: 10.1146/annurev.bioeng.3.1.275. [DOI] [PubMed] [Google Scholar]
- 74.Czyz J., Wobus A. Differentiation. 2001;68:167–174. doi: 10.1046/j.1432-0436.2001.680404.x. [DOI] [PubMed] [Google Scholar]
- 75.Tourovskaia A., Figueroa-Masot X., Folch A. Lab Chip. 2005;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 76.Chin V. I., Taupin P., Sanga S., Scheel J., Gage F. H., Bhatia S. N. Biotechnol. Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 77.Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 78.McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 79.Takayama S., Ostuni E., Qian X. P., McDonald J. C., Jiang X. Y., LeDuc P., Wu M. H., Ingber D. E., Whitesides G. M. Adv. Mater. 2001;13:570–574. [Google Scholar]
- 80.Takayama S., McDonald J. C., Ostuni E., Liang M. N., Kenis P. J. A., Ismagilov R. F., Whitesides G. M. Proc. Natl. Acad. Sci. USA. 1999;96:5545–5548. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D. E., Whitesides G. M. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 82.Lucchetta E. M., Lee J. H., Fu L. A., Patel N. H., Ismagilov R. F. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeon N. L., Dertinger S. K. W., Chiu D. T., Choi I. S., Stroock A. D., Whitesides G. M. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 84.Dertinger S. K. W., Chiu D. T., Jeon N. L., Whitesides G. M. Anal. Chem. 2001;73:1240–1246. [Google Scholar]
- 85.Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van de Water L., Toner M. Nat. Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 86.Dertinger S. K. W., Jiang X. Y., Li Z. Y., Murthy V. N., Whitesides G. M. Proc. Natl. Acad. Sci. USA. 2002;99:12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor A. M., Blurton-Jones M., Rhee S. W., Cribbs D. H., Cotman C. W., Jeon N. L. Nat. Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chung B. G., Flanagan L. A., Rhee S. W., Schwartz P. H., Lee A. P., Monuki E. S., Jeon N. L. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 89.Bhatia S. N., Balis U. J., Yarmush M. L., Toner M. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 90.Bhatia S. N., Balis U. J., Yarmush M. L., Toner M. Biotechnol. Prog. 1998;14:378–387. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- 91.Bhatia S. N., Balis U. J., Yarmush M. L., Toner M. J. Biomater. Sci. Polym. Ed. 1998;9:1137–1160. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- 92.Bhatia S. N., Yarmush M. L., Toner M. J. Biomed. Mater. Res. 1997;34:189–199. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 93.Yamato M., Konno C., Utsumi M., Kikuchi A., Okano T. Biomaterials. 2002;23:561–567. doi: 10.1016/s0142-9612(01)00138-7. [DOI] [PubMed] [Google Scholar]
- 94.Hirose M., Yamato M., Kwon O. H., Harimoto M., Kushida A., Shimizu T., Kikuchi A., Okano T. Yonsei Med. J. 2000;41:803–813. doi: 10.3349/ymj.2000.41.6.803. [DOI] [PubMed] [Google Scholar]
- 95.Khademhosseini A., Suh K. Y., Yang J. M., Eng G., Yeh J., Levenberg S., Langer R. Biomaterials. 2004;25:3583–3592. doi: 10.1016/j.biomaterials.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 96.Chiu D. T., Jeon N. L., Huang S., Kane R. S., Wargo C. J., Choi I. S., Ingber D. E., Whitesides G. M. Proc. Natl. Acad. Sci. USA. 2000;97:2408–2413. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang M. D., Golden A. P., Tien J. J. Am. Chem. Soc. 2003;125:12988–12989. doi: 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- 98.Ding S., Gray N. S., Wu X., Ding Q., Schultz P. G. J. Am. Chem. Soc. 2002;124:1594–1596. doi: 10.1021/ja0170302. [DOI] [PubMed] [Google Scholar]
- 99.Lynn D. M., Anderson D. G., Putnam D., Langer R. J. Am. Chem. Soc. 2001;123:8155–8156. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 100.Wu X., Ding S., Ding Q., Gray N. S., Schultz P. G. J. Am. Chem. Soc. 2002;124:14520–14521. doi: 10.1021/ja0283908. [DOI] [PubMed] [Google Scholar]
- 101.Wu X., Ding S., Ding G., Gray N. S., Schultz P. G. J. Am. Chem. Soc. 2004;126:1590–1591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- 102.Chen S. B., Zhang Q. S., Wu X., Schultz P. G., Ding S. J. Am. Chem. Soc. 2004;126:410–411. doi: 10.1021/ja037390k. [DOI] [PubMed] [Google Scholar]
- 103.Anderson D. G., Levenberg S., Langer R. Nat. Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 104.Anderson D. G., Putnam D., Lavik E. B., Mahmood T. A., Langer R. Biomaterials. 2005;26:4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 105.Flaim C. J., Chien S., Bhatia S. N. Nat. Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 106.Khademhosseini A., Suh K. Y., Jon S., Chen G., Eng G., Yeh J., Langer R. Anal. Chem. 2004;76:3675–3681. doi: 10.1021/ac035415s. [DOI] [PubMed] [Google Scholar]
- 107.Thorsen T., Maerkl S. J., Quake S. R. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 108.Prudhomme W., Daley G. Q., Zandstra P., Lauffenburger D. A. Proc. Natl. Acad. Sci. USA. 2004;101:2900–2905. doi: 10.1073/pnas.0308768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koh W. G., Itle L. J., Pishko M. V. Anal. Chem. 2003;75:5783–5789. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- 110.Khademhosseini A., Yeh J., Eng G., Kazi H., Borenstein J., Karp J. M., Farokhzad O., Langer R. Lab Chip. 5:1380–1386. doi: 10.1039/b508096g. [DOI] [PubMed] [Google Scholar]