Abstract
Backbone–backbone hydrogen-bonding interactions are a ubiquitous and highly conserved structural feature of proteins that adopt the same fold (i.e., have the same overall backbone topology). This work addresses the question of whether or not this structural conservation is also reflected as a thermodynamic conservation. Reported here is a comparative thermodynamic analysis of backbone hydrogen bonds in two proteins that adopt the same fold but are unrelated at the primary amino acid sequence level. With amide-to-ester bond mutations introduced by total chemical synthesis methods, the thermodynamic consequences of backbone–backbone hydrogen-bond deletions at five different structurally equivalent positions throughout the β-α-α fold of Arc repressor and CopG were assessed. The ester bond-containing analogues all folded into native-like three-dimensional structures that were destabilized from 2.5 to 6.0 kcal/(mol dimer) compared with wild-type controls. Remarkably, the five paired analogues with amide-to-ester bond mutations at structurally equivalent positions were destabilized to exactly the same degree, regardless of the degree to which the mutation site was buried in the structure. The results are interpreted as evidence that the thermodynamics of backbone–backbone hydrogen-bonding interactions in a protein fold are conserved.
Keywords: protein mutagenesis, chemical synthesis
The 30,000-plus high-resolution protein structures that have been solved to date can be grouped, according to their overall backbone topology, into a remarkably small number of protein folds (≈800) (1, 2). The apparent existence of such a small number of naturally occurring protein folds has meant that a surprisingly large number of proteins, which are unrelated at the amino acid sequence level, adopt the same protein fold (i.e., fold into three-dimensional structures with the same backbone topology). Thus, nature appears to have used a number of different combinations of chemical interactions to stabilize its protein folds. However, one set of chemical interactions that are structurally conserved within a protein fold is the set of backbone–backbone hydrogen-bonding interactions that help define it. This structural conservation raises a fundamental question of whether or not the thermodynamics of backbone–backbone hydrogen-bonding interactions are also conserved in a protein fold. Such fundamental knowledge about the detailed molecular interactions that guide protein-folding reactions stands to impact a wide variety of research areas from drug discovery to theories of human evolution.
The relative contributions of different backbone–backbone hydrogen-bonding interactions in protein-folding reactions have been explored in a series of recent studies on several different protein systems (3–12). These studies have typically relied on total chemical synthesis methods (13) or specialized in vitro translation techniques (14) to incorporate amide-to-ester bond mutations into the polypeptide backbone of proteins and modulate the hydrogen-bonding properties of a protein’s polypeptide backbone. The structural and thermodynamic consequences of introducing such amide-to-ester-bond mutations into β-sheet and α-helical regions of different protein folds have been reported (3–12). In most cases reported to date, the amide-to-ester bond mutation has not significantly altered the three-dimensional structure of the protein under study; however, it has been observed that the thermodynamic stabilities of protein analogues containing ester-bond mutations in the β-sheet and α-helical regions of different protein folds are generally reduced 0.5–3.0 kcal/mol per ester bond compared with the wild-type protein.
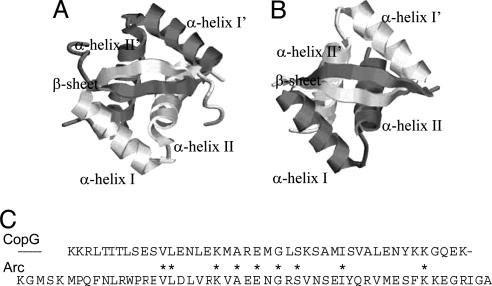
The structural and thermodynamic properties of some 20 different ester bond-containing analogues have been studied to date. However, there are no comparative studies of amide-to-ester bond mutations in proteins with the same protein fold and different primary amino acid sequences. Here we report on such a study using the model protein systems, the Arc repressor and CopG, which are both homodimeric DNA-binding proteins that share the same ribbon–helix–helix fold. The high-resolution x-ray crystallographic data available on these proteins indicate that the homodimeric structure of each protein consists of four α-helices packed against an antiparallel β-sheet (see Fig. 1) (15, 16). Despite their folding into higher-order structures with essentially identical backbone topologies, the primary amino acid sequence homology between Arc and CopG is relatively low (≈20%) (16). An alignment of the two amino acid sequences indicates that only nine positions are occupied by the same amino acid residue (Fig. 1).
Fig. 1.
Arc and CopG structures. (A and B) Schematic representation of the three-dimensional folded structures of wild-type Arc repressor (A) and wild-type CopG (B). The ribbon diagrams were generated in protein explorer by using x-ray crystallographic data for Arc (15) and CopG (16). (C) The primary amino acid sequences of CopG and Arc are shown with positions occupied by the same residue noted by ∗, according to the sequence alignment in ref. 16.
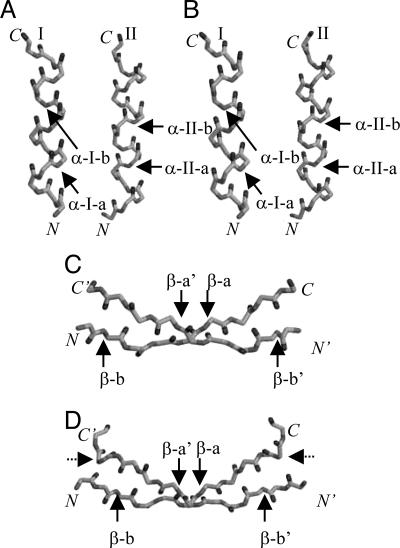
In this work, amide-to-ester bond mutations, introduced by total chemical synthesis methods, were used in a series of unnatural amino acid mutagenesis experiments to assess the thermodynamic consequences of backbone–backbone hydrogen-bond deletions at five different structurally equivalent positions throughout the β-α-α fold of Arc repressor and CopG (see Fig. 2). The structural properties of each ester bond-containing analogue were analyzed by far-UV CD spectroscopy, and the thermodynamic consequences of each amide-to-ester bond mutation were measured in guanidinium (Gdm)Cl-induced equilibrium unfolding studies.
Fig. 2.
Schematic representation of the backbone atoms in the α-helical and β-sheet regions of the polypeptide chains of Arc* and CopG. (A and B) The backbone atoms in the two α-helical regions of the Arc* (A) and CopG (B) structures are shown. (C and D) The backbone atoms in the intersubunit β-sheet regions in the Arc* (C) and CopG (D) structures are shown. The positions where amide-to-ester bond mutations have been incorporated into the backbone for our comparative analyses are indicated by solid arrows. The abbreviations used in the text to describe each position (e.g., α-I-a, α-I-b, etc.) are associated with each arrow. The dotted arrows in D represent the position of the amide-to-ester bond mutation in CopG*, the CopG control construct used here.
Results and Discussion
Although a large majority of the amino acid side-chain interactions that define the three-dimensional structures of Arc and CopG are clearly different, the network of backbone–backbone hydrogen bonds in the three-dimensional structure of each protein is very similar. The numbers of backbone–backbone hydrogen bonds in the α-helical regions of wild-type Arc and CopG are exactly the same (a total of 24 per subunit in each case), and the number of backbone–backbone hydrogen bonds in the intersubunit, β-sheet regions of wild-type Arc and CopG are 6 and 10, respectively. However, the Arc and CopG constructs used as the wild-type controls in this work (referred to hereafter as Arc* and CopG*) were engineered to have the same number of backbone–backbone hydrogen bonds (i.e., eight) in their intersubunit, β-sheet region.
The Arc* construct in this work contains a proline-to-leucine mutation at the eighth position in the naturally occurring protein’s 53-aa polypeptide chain. This mutation was previously shown to increase the number of backbone–backbone hydrogen bonds in the β-sheet region of Arc from six to eight and increase the protein’s folding free energy by 2.5 kcal/(mol dimer) (17). The CopG* construct in this work contains an amide-to-ester bond mutation at the 11th position in the naturally occurring protein’s 45-aa polypeptide chain. This mutation was designed to delete the outermost two backbone–backbone hydrogen bonds in CopG’s intersubunit, β-sheet region. The CopG* construct also contained a Glu-to-Ala mutation at position 11 to facilitate its synthesis (see Materials and Methods). It also turned out that the Arc* and CopG* constructs had essentially the same overall thermodynamic stabilities, 12.8 ± 0.2 and 13.2 ± 0.1 kcal/mol, respectively.
The structural and thermodynamic consequences of amide-to-ester bond mutations at five different positions throughout the β-α-α fold of Arc* and CopG* were compared. These positions included: one position (β-a) in the β-strand, two positions (α-I-a and α-I-b) in the first α-helix, and two positions (α-II-a and α-II-b) in the second α-helix (see Fig. 2). This work involved the total chemical synthesis (see Materials and Methods) and thermodynamic analysis (see Materials and Methods) of the 10 ester bond-containing Arc and CopG analogues and four wild-type controls listed in Table 1.
Table 1.
Thermodynamic parameters for the GdmCl-induced equilibrium unfolding of the Arc and CopG constructs compared in this work
| Analogue | Ester-bond position† | m value,‡ kcal/mol·M | ΔGf, kcal/(mol dimer) |
|---|---|---|---|
| Arc* | n/a (wt-control-1) | 3.2 ± 0.1§ | −12.8 ± 0.2§ |
| Arc** | n/a (wt-control-2) | 3.5 ± 0.1 | −13.3 ± 0.1 |
| (OL12)Arc* | Middle of β-sheet (β-a) | 3.2 ± 0.1 | −10.0 ± 0.2 |
| (OA20)Arc** | End of α-helix I (α-I-a) | 4.0 ± 0.1 | −10.8 ± 0.1 |
| (OA26)Arc* | Middle of α-helix I (α-I-b) | 4.7 ± 0.1 | −8.7 ± 0.2 |
| (OI37)Arc* | End of α-helix II (α-II-a) | 4.3 ± 0.1 | −9.6 ± 0.2 |
| (OV41)Arc* | Middle of α-helix II (α-II-b) | 3.8 ± 0.1¶ | −6.9 ± 0.2¶ |
| CopG* | End of β-sheet (wt-control-1) | 2.1 ± 0.1 | −13.2 ± 0.1 |
| CopG** | Same as CopG* (wt-control-2) | 2.2 ± 0.1 | −13.6 ± 0.1 |
| (OI7)CopG* | Middle of β-sheet (β-a) | 2.1 ± 0.1 | −10.3 ± 0.1 |
| (OA15)CopG** | End of α-helix I (α-I-a) | 2.6 ± 0.1 | −11.0 ± 0.1 |
| (OA21)CopG* | Middle of α-helix I (α-I-b) | 3.1 ± 0.1 | −8.9 ± 0.2 |
| (OI32)CopG* | End of α-helix II (α-II-a) | 2.8 ± 0.1 | −9.7 ± 0.2 |
| (OL36)CopG* | Middle of α-helix II (α-II-b) | 2.6 ± 0.1¶ | −7.2 ± 0.2¶ |
†Note that each ester-bond mutation appeared twice in each analogue’s homodimeric structure (only one of the two positions is noted). n/a, not applicable.
‡Values were obtained by fitting the chemical denaturation curve data to a two-state model involving native dimer and unfolded monomer (see Materials and Methods). Reported values are the average and standard deviation of the values extracted from three separate denaturation curves.
§Values are taken from ref. 17.
¶Values were determined in the presence of TMAO (see Materials and Methods and Fig. 5).
Both CopG and Arc are homodimers, therefore the amide-to-ester bond mutations in the polypeptide chains of analogues studied here appeared twice in each protein’s folded, three-dimensional structure, and in each case the mutation affected multiple backbone–backbone hydrogen bonds. In the case of the ester-bond mutation in the β-sheet position (i.e., the β-a position noted in Fig. 2) two N H groups (one per subunit) are replaced with O atoms in the middle of the β-sheet, effectively deleting the two middle backbone–backbone hydrogen bonds in the intersubunit β-sheet region of the β-α-α fold of CopG* and Arc*. In the case of the ester-bond mutations in the four α-helical positions (i.e., the α-I-a, α-I-b, α-II-a, and α-II-b positions noted in Fig. 2), two backbone hydrogen bonds per subunit (a total of four backbone–backbone hydrogen-bonding interactions) are affected in the folded, three-dimensional structure of each homodimer. Two hydrogen bonds are deleted because the C
H groups (one per subunit) are replaced with O atoms in the middle of the β-sheet, effectively deleting the two middle backbone–backbone hydrogen bonds in the intersubunit β-sheet region of the β-α-α fold of CopG* and Arc*. In the case of the ester-bond mutations in the four α-helical positions (i.e., the α-I-a, α-I-b, α-II-a, and α-II-b positions noted in Fig. 2), two backbone hydrogen bonds per subunit (a total of four backbone–backbone hydrogen-bonding interactions) are affected in the folded, three-dimensional structure of each homodimer. Two hydrogen bonds are deleted because the C O and N
O and N H groups of the amide bonds at these α-helical positions are hydrogen-bond acceptors and donors, respectively, in two separate backbone–backbone hydrogen bonds. Thus, the lack of a hydrogen-bond donor in the ester bond (i.e., there is an O atom in place of an N
H groups of the amide bonds at these α-helical positions are hydrogen-bond acceptors and donors, respectively, in two separate backbone–backbone hydrogen bonds. Thus, the lack of a hydrogen-bond donor in the ester bond (i.e., there is an O atom in place of an N H group) effectively deletes one of these two hydrogen bonds per subunit, and the lower pKa of the ester-bond carbonyl compared with the amide-bond carbonyl (18) makes it a much poorer hydrogen-bond acceptor, significantly reducing the strength of the other two hydrogen bonds per subunit.
H group) effectively deletes one of these two hydrogen bonds per subunit, and the lower pKa of the ester-bond carbonyl compared with the amide-bond carbonyl (18) makes it a much poorer hydrogen-bond acceptor, significantly reducing the strength of the other two hydrogen bonds per subunit.
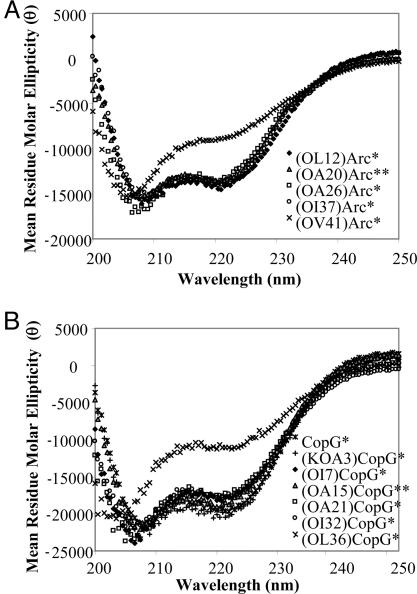
The synthetic polypeptide chains of all of the ester bond-containing protein analogues in this work folded into native-like three-dimensional structures as judged by far-UV CD spectroscopy with the exception of the (OV41)Arc* and (OL36)CopG* analogues (see Fig. 3). § Far-UV CD analysis of these two analogues, which contained amide-to-ester bond mutations at structurally equivalent positions in α-helix II, revealed that these proteins were only ≈60% folded after their dissolution in folding buffer as determined by the magnitude of their CD signals at 222 nm. However, the addition of trimethylamine N-oxide (TMAO), an osmolyte known to stabilize proteins (19, 20), to the folding buffer promoted the native-like folding of these analogues as determined by their CD signals at 222 nm. The CD signals at 222 nm obtained for (OV41)Arc* and (OL36)CopG* in folding buffer containing as little as 1.50 and 1.75 M TMAO, respectively, were indistinguishable from such signals obtained for the wild-type controls, Arc* and CopG*, in the absence of TMAO (data not shown).
Fig. 3.
Far-UV CD spectra recorded for the ester bond-containing Arc (A) and CopG (B) analogues.¶ Note that far-UV CD spectra recorded for wild-type Arc and CopG are not shown. However, such spectra have been reported (7, 24), and they are essentially identical to all of the spectra shown, with the exception of the spectra shown for the (OV41)Arc* and (OL36)CopG* analogues.
The chemical denaturant-induced equilibrium unfolding properties of each ester bond-containing analogue were determined by using far-UV CD spectroscopy as a structural probe and guanidine as the denaturant (see Figs. 4 and 5, which are published as supporting information on the PNAS web site). The protein folding free energies (i.e., ΔGf values) and m values (i.e., δΔG/δ[GdmCl]) calculated in these equilibrium unfolding experiments are summarized in Table 1. The m values determined for the ester bond-containing analogues were on average ≈20% larger than the values determined for the wild-type controls. Such increased m values of ester bond-containing analogues have been previously observed in other protein systems (3, 5, 7, 9, 10). It is particularly noteworthy in this work that similarly increased m values were observed at each ester-bond position. For example, the m values obtained for the Arc and CopG analogues with an ester bond in the α-II-a position increased 34% and 33%, respectively, as compared with their wild-type controls. Such closely matched increases were observed at each ester-bond position.
The ΔGf values summarized in Table 1 indicate that the amide-to-ester bond mutations in all 10 protein analogues were destabilizing. The magnitudes of the measured destabilization (i.e., the ΔΔGf value determined for each ester-bond analogue) are summarized in Table 2, and they ranged from 2.5 to 6.0 kcal/(mol dimer). The range of ΔΔGf values observed in Table 2 corresponds to a range from 1.3 to 3.0 kcal/mol per ester-bond mutation and a range from 0.7 to 1.5 kcal/mol per hydrogen bond affected (see above discussion of the number of hydrogen bonds affected per ester-bond mutation). The ΔΔGf values reported in Table 2 are all within the range of those previously reported for ester bond-containing protein analogues when the number of hydrogen bonds affected is considered. The ΔΔGf values determined for the backbone–backbone hydrogen bonds are also consistent with the destabilizing effects previously noted for side-chain–backbone and side-chain–side-chain hydrogen bonds in other protein folding studies (21–23).
Table 2.
Comparative analysis of the Arc and CopG analogues in this work
| Ester-bond position† | Analogues | Buried surface area,‡ % | ΔΔGf,§ kcal/(mol dimer) |
|---|---|---|---|
| β-a | (OL12)Arc* vs. (OI7)CopG* | 51 vs. 53 | 2.8 vs. 2.9 |
| α-I-a | (OA20)Arc** vs. (OA15)CopG** | 100 vs. 100 | 2.5 vs. 2.6¶ |
| α-I-b | (OA26)Arc* vs. (OA21)CopG* | 100 vs. 100 | 4.1 vs. 4.3 |
| α-II-a | (OI37)Arc* vs. (OI32)CopG* | 100 vs. 100 | 3.2 vs. 3.5 |
| α-II-b | (OV41)Arc* vs. (OL36)CopG* | 97 vs. 100 | 5.9 vs. 6.0 |
†Note that each ester-bond mutation appeared twice in each analogue’s homodimeric structure (only one of the two positions is noted).
‡The x-ray crystallographic data available for Arc* (16) and wild-type CopG (17) were used for all calculations.
§The standard error associated with the reported values ranged from 0.1 to 0.3 kcal/mol. This standard error was calculated by using the standard deviations of the ΔGf values in Table 1.
¶First value is relative to Arc**; second value is relative to CopG**.
The ΔΔGf values in Table 2 certainly include the thermodynamic contributions of the backbone–backbone hydrogen bonds deleted (or altered as in the case of those involving the CO group as a hydrogen-bond acceptor) in the ester bond-containing Arc and CopG analogues studied here. However, such ΔΔGf values are difficult to equate with backbone–backbone hydrogen-bond strengths (i.e., the free energy associated with the electrostatic interactions between desolvated peptide amide C O and N
O and N H groups). In addition to the hydrogen bond strength, it is generally accepted that these values contain energetic contributions from the electrostatic repulsions and the solvation differences that can result from amide-to-ester bond mutations in the polypeptide backbones of proteins (12). For example, in recent work by Deechongkit et al. (12), an electrostatic correction term as large as 0.7 kcal/mol was suggested, and a solvation correction of 0 to 1.3 kcal/mol per hydrogen bond was proposed depending on the solvent accessibility of the deleted hydrogen bond.
H groups). In addition to the hydrogen bond strength, it is generally accepted that these values contain energetic contributions from the electrostatic repulsions and the solvation differences that can result from amide-to-ester bond mutations in the polypeptide backbones of proteins (12). For example, in recent work by Deechongkit et al. (12), an electrostatic correction term as large as 0.7 kcal/mol was suggested, and a solvation correction of 0 to 1.3 kcal/mol per hydrogen bond was proposed depending on the solvent accessibility of the deleted hydrogen bond.
Of particular note here is that the solvent accessibility of the mutation site was essentially identical for each of the five pairs of ester bond-containing Arc and CopG analogues reported in Table 2. Thus, the solvation correction terms for the paired CopG and Arc analogues reported on here are likely to be the same and not likely to impact the comparative analyses described here. Interestingly, all but two (one per subunit) of the 56 backbone–backbone hydrogen bonds (28 per subunit) that define the β-α-α fold of Arc* and CopG* are similarly buried in the two structures (i.e., have buried surface areas that are within 10% of each other) (data not shown). Only the outermost two backbone–backbone hydrogen bonds in the intersubunit β-sheet region of the three-dimensional structures of Arc* and CopG* (the β-b position in Fig. 2) have significantly different solvent exposure. The buried surface area of the amide groups in this β-b position were significantly different (i.e., 73% and 44%, respectively). Interestingly, ester-bond mutations in this position resulted in ΔΔGf values of 2.4 and ≈0 kcal/mol for Arc* and CopG*, respectively [see (OL8)Arc* data in ref. 7 and (KOA3)CopG*¶ ].
Clearly, the physical environment (e.g., buried surface area) surrounding backbone–backbone hydrogen bonds can influence their apparent “strength,” or measured ΔΔGf value. This is likely one contributing factor to the variability of the measured ΔΔGf values determined for the ester bond-containing analogues in this study and other studies (3–12). For example, in this study the ΔΔGf value observed in the β-a position, which is only partially buried, was smaller than the values observed in the α-I-b, α-II-a, and α-II-b positions, which are all completely buried in the structure. However, it should be noted that the buried surface area is not the only physical factor contributing to the variability of measured ΔΔGf values. For example, in this work the ΔΔGf values for the ester-bond mutations in the four α-helical positions ranged from 2.5 to 6.0 kcal/(mol dimer), even though the mutation sites were all essentially buried in the structure. This and other recent data (12) suggest that amide-to-ester bond mutations at certain positions in a protein’s structure are more destabilizing than others, even when buried surface area is taken into consideration.
What is most remarkable about the ΔΔGf values in Table 2 is that all five pairs of the Arc and CopG analogues with amide-to-ester bond mutations at structurally analogous positions yielded ΔΔGf values that were essentially identical considering the errors (typically 0.1–0.3 kcal/mol) associated with our measurements. This result was despite the relatively wide range of the measured ΔΔGf values [2.5 to 6.0 kcal/(mol dimer)], and it was despite the buried surface area of one site being 50% and the other four sites being 100%. Assuming that the ΔΔGf values observed for the paired Arc and CopG analogues can be equated with hydrogen-bond strength by using similar correction terms for each pair, our results suggest that the thermodynamic contributions of backbone–backbone hydrogen-bonding interactions are conserved in the β-α-α fold of Arc* and CopG*. The above assumption is likely a valid one considering the electrostatic and solvation correction terms that have been proposed (12).
The results reported here constitute experimental evidence that the thermodynamics of backbone–backbone hydrogen-bonding interactions are conserved in a protein fold. The generality of our results to other protein folds remains to be explored. However, the results of our studies on Arc and CopG suggest that the conservation of backbone hydrogen-bond thermodynamics in a protein fold may be an important general principle of protein-folding reactions.
Materials and Methods
Synthesis and Purification of Protein Analogues.
The polypeptide chains of the CopG and Arc constructs were assembled by highly optimized, stepwise solid-phase synthesis using tert-butoxycarbonyl chemistry according to previously described synthetic protocols for the wild-type proteins (7, 24). Ester bonds were incorporated into the polypeptide chain of each analogue by coupling the appropriate α-hydroxy acid [either l-lactic acid for alanine, (2S,3S)-2-hydroxy-3-methyl pentanoic acid for isoleucine, (S)-(−)-2-hydroxy-isocaproic acid for leucine, or (S)-(+)-2-hydroxy-3-methylbutyric acid for valine] at the desired position, according to a previously described protocol (25). In several cases, which included the (OA20)Arc**, (OA15)CopG**, and (KOA3)CopG* analogues, there was not an alanine, isoleucine, or valine in the wild-type sequence C-terminal to the mutation site. In these cases, the C-terminal residue at the amide-to-ester bond mutation site [i.e., Asp, Glu, or Lys for (OA20)Arc**, (OA15)CopG**, and (KOA3)CopG*, respectively] was mutated to Ala to facilitate the incorporation of the ester-bond mutation by using total chemical synthesis as the α-hydroxy acid of alanine, l-lactic acid, is commercially available and the α-hydroxy acids of Asp, Glu, and Lys are not. Note that the Ala mutations had minimal impact on each protein’s thermodynamic stability and each protein’s three-dimensional structure (see data in Table 1).
The crude polypeptide product obtained from each synthesis was lyophilized and purified by using preparative RP-HPLC, as described for the wild-type proteins (7, 24), and the purity of each analogue was confirmed by RP-HPLC (see Figs. 6 and 7, which are published as supporting information on the PNAS web site). The pure, lyophilized, synthetic polypeptide chains of each analogue were folded in buffer at concentrations of ≈10 mg/ml over 30 min. The buffer used to fold all of the CopG and Arc analogues was 20 mM Tris·HCl (pH 7.5), 100 mM KCl, and 0.2 mM EDTA. Ultimately, the solutions of folded protein were centrifuged to pellet any precipitate that formed during the folding reaction. The supernatants in the resulting protein solutions were used for subsequent analyses. Protein concentrations for all of the CopG analogues were determined by using the Waddell method (26). Protein concentrations for all of the Arc analogues were determined by using absorbance measurements at 280 nm and the reported molar extinction coefficient for Arc at 280 nm, 6,756 M−1·cm−1 (27). All protein concentrations are reported in monomer equivalents.
Biophysical Measurements.
Far-UV CD spectra and GdmCl-induced denaturation curves (Figs. 4 and 5) were recorded on a PiStar 180 CDF Spectrometer system from Applied Photophysics (Surrey, U.K.) that was also equipped with an automatic titration system. All of the denaturation curves were recorded in buffer containing 20 mM Tris·HCl (pH 7.5), 100 mM KCl, and 0.2 mM EDTA at 25°C in a thermostated cell by using the far-UV CD signal at 222 nm, except in the cases of the (OV41)Arc* and (OL36)CopG* analogues where the far-UV CD signal at 230 nm was used. The chemical denaturation curves were analyzed by assuming a two-state (folded dimer and unfolded monomer) model for each folding reaction to extract ΔGf and m values, as described for the wild-type proteins (24, 28).
Chemical denaturation curves for (OV41)Arc* and (OL36)CopG* could not be acquired with only GdmCl. These analogues were only partially folded at 0 M GdmCl, thus the pretransition baseline, which is required for quantitative analysis of the unfolding transition, could not be determined. Therefore, the ΔGf values reported for these analogues were extracted from GdmCl denaturation curves recorded in the presence of the stabilizing osmolyte (TMAO), an approach that has been previously reported (19, 20). In these experiments GdmCl-induced denaturation curves were acquired by using different TMAO concentrations that ranged from 1.50 to 2.25 M for (OV41)Arc* and from 1.75 to 2.50 M for (OL36)CopG* (Fig. 5). In separate experiments it was determined that the (OV41)Arc* and (OL36)CopG* analogues were completely folded in 0 M GdmCl at these TMAO concentrations (data not shown).
Biophysical Calculations.
Solvent accessible surface area calculations were performed by using x-ray crystallographic data that were available for (PL8)Arc (the Arc* construct in this work) with Protein Data Bank (PDB) code 1MYK (17). The PDB code used for calculations on the CopG system was 2CPG (16). The data were obtained to calculate the degree to which each mutation site was buried upon folding (29, 30) by using the program what if and taking a probe radius of 1.4 Å2.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM61680 (to M.C.F.).
Abbreviations
- Gdm
guanidinium
- TMAO
trimethylamine N-oxide.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
All Arc* analogues are identical to wild-type Arc repressor except that they contain a Pro-to-Leu mutation at position 8, the effects of which are described in the text, and the Arc** analogue contains the Pro-to-Leu mutation at position 8 and an Asp-to-Ala mutation at position 20. All CopG* analogues are identical to wild-type CopG except that they contain Glu-to-Ala and amide-to-ester bond mutations at position 11, the effects of which are described in the text. The CopG** analogue includes the CopG* mutations noted above as well as a Glu-to-Ala mutation at position 15. In all parenthetical notations, the O refers to the ester bond, the second letter refers to the one-letter code of the amino acid (e.g., A, alanine; L, leucine; I, isoleucine; V, valine) that is C-terminal to the ester bond, and the number refers to the position of this C-terminal amino acid in the protein’s polypeptide chain. For example, (OL12)Arc* means that there is an ester-bond mutation N-terminal to the Leu at position 12 in the 53-aa polypeptide chain of Arc*.
Two additional CopG analogues were also chemically synthesized and subject to thermodynamic analysis. The analogues included (KOA3)CopG* that contained both Lys-to-Ala and ester-bond mutations at the third amino acid position in CopG*’s polypeptide chain (see the β-b position in Fig. 2D), and (KA3)CopG* that contained just the Lys-to-Ala mutation. The Ala mutation facilitated the incorporation of the ester bond and had minimal impact on the protein’s thermodynamic stability (as determined in GdmCl-induced equilibrium unfolding experiments) or on the protein’s three-dimensional structure (as determined by far-UV CD spectroscopy) (see Figs. 3 and 4). The ester bond effectively eliminated the outermost two backbone–backbone hydrogen bonds in the intersubunit β-sheet region of CopG* and generated a subunit interface with the identical number of hydrogen bonds as an ester bond-containing Arc analogue (POL8)Arc [or (OL8)Arc*] on which we have previously reported (7).
References
- 1.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant A., Lee D., Orengo C. Genome Biol. 2004;5:107. doi: 10.1186/gb-2004-5-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman E., Thorson J. S., Schultz P. G. J. Am. Chem. Soc. 1997;119:7151–7152. [Google Scholar]
- 4.Koh J. T., Cornish V. W., Schultz P. G. Biochemistry. 1997;36:11314–11322. doi: 10.1021/bi9707685. [DOI] [PubMed] [Google Scholar]
- 5.Shin I., Ting A. Y., Schultz P. G. J. Am. Chem. Soc. 1997;119:12667–12668. [Google Scholar]
- 6.Haque T. S., Little J. C., Gellman S. H. J. Am. Chem. Soc. 1996;118:6975–6985. [Google Scholar]
- 7.Wales T. E., Fitzgerald M. C. J. Am. Chem. Soc. 2001;123:7709–7710. doi: 10.1021/ja010775n. [DOI] [PubMed] [Google Scholar]
- 8.Silinski P., Fitzgerald M. C. Biochemistry. 2003;42:6620–6630. doi: 10.1021/bi020700b. [DOI] [PubMed] [Google Scholar]
- 9.Beligere G. S., Dawson P. E. J. Am. Chem. Soc. 2000;122:12079–12082. [Google Scholar]
- 10.Blankenship J. W., Balambika R., Dawson P. E. Biochemistry. 2002;41:15676–15684. doi: 10.1021/bi026862p. [DOI] [PubMed] [Google Scholar]
- 11.Deechongkit S., Nguyen H., Powers E. T., Dawson P. E., Gruebele M., Kelly J. W. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 12.Deechongkit S., Dawson P. E., Kelly J. W. J. Am. Chem. Soc. 2004;126:16762–16771. doi: 10.1021/ja045934s. [DOI] [PubMed] [Google Scholar]
- 13.Dawson P. E., Kent S. B. H. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 14.Mendel D., Cornish V. W., Schultz P. G. Annu. Rev. Biophys. Biomol. Struct. 1995;24:435–462. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- 15.Raumann B. E., Rould M. A., Pabo C. O., Sauer R. T. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 16.Gomis-Rüth F. X., Solà M., Acebo P., Párraga A., Guasch A., Eritja R., González A., Espinosa M., del Solar G., Coll M. EMBO J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schildbach J. F., Milla M. E., Jeffrey P. D., Raumann B. E., Sauer R. T. Biochemistry. 1995;34:1405–1412. doi: 10.1021/bi00004a035. [DOI] [PubMed] [Google Scholar]
- 18.Arnett E. M., Mitchell E. J., Murty T. S. S. R. J. Am. Chem. Soc. 1974;96:3875–3891. [Google Scholar]
- 19.Wang A., Bolen D. W. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 20.Mello C. C., Barrick D. Protein Sci. 2003;12:1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pace N. C., Horn G., Hebert E. J., Bechert J., Shaw K., Urbanikova L., Scholtz J. M., Sevcik J. J. Mol. Biol. 2001;312:393–404. doi: 10.1006/jmbi.2001.4956. [DOI] [PubMed] [Google Scholar]
- 22.Hebert E. J., Giletto A., Sevcik J., Urbanikova L., Wilson K. S., Dauter Z., Pace C. N. Biochemistry. 1998;37:16192–16200. doi: 10.1021/bi9815243. [DOI] [PubMed] [Google Scholar]
- 23.Takano K., Scholtz J. M., Sacchettini J. C., Pace C. N. J. Biol. Chem. 2003;278:31790–31795. doi: 10.1074/jbc.M304177200. [DOI] [PubMed] [Google Scholar]
- 24.Wales T. E., Richardson J. S., Fitzgerald M. C. Protein Sci. 2004;13:1918–1926. doi: 10.1110/ps.04671804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu W., Qasim M. A., Laskowski M., Jr., Kent S. B. H. Biochemistry. 1997;36:673–679. doi: 10.1021/bi9625612. [DOI] [PubMed] [Google Scholar]
- 26.Waddell W. J. J. Lab. Clin. Med. 1956;48:311–314. [PubMed] [Google Scholar]
- 27.Brown B. M., Bowie J. U., Sauer R. T. Biochemistry. 1990;29:11189–11195. doi: 10.1021/bi00503a006. [DOI] [PubMed] [Google Scholar]
- 28.Bowie J. U., Sauer R. T. Biochemistry. 1989;28:7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 29.Lee B., Richards F. M. J. Mol. Biol. 1971;55:379–380. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 30.Myers J. K., Pace C. N., Scholtz J. M. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.