Abstract
p8 is a small-stress protein involved in several cellular functions including apoptosis. To identify its putative partners, we screened a HeLa cDNA library by using the two-hybrid technique and found that p8 binds the antiapoptotic protein prothymosin α (ProTα). Fluorescence spectroscopy, circular dichroism, and NMR spectroscopy showed that p8 and ProTα formed a complex. Binding resulted in important changes in the secondary and tertiary structures of the proteins. Because p8 and ProTα form a complex, they could act in concert to regulate the apoptotic cascade. We induced apoptosis in HeLa cells by staurosporine treatment and monitored the effects of knocking down p8 and/or ProTα or overexpressing p8 and/or ProTα on caspase 3/7 and 9 activities and on cell death. Transfecting ProTα or p8 small interfering RNAs increased the activities of both caspases and the number of apoptotic nuclei. However, transfecting both small interfering RNAs resulted in no further increase. Overexpressing p8 or ProTα did not alter caspase activities, whereas overexpressing both resulted in a significant reduction of caspase activities. These results strongly suggest that the antiapoptotic response of HeLa cells upon staurosporine treatment requires expression of both p8 and ProTα.
Keywords: caspases, unfolded proteins, CytoTrap, stress proteins
The p8 gene, first described as overexpressed in the pancreas during pancreatitis (1), encodes a ubiquitous stress protein (2). p8 is a highly basic 80-aa polypeptide containing a canonical bipartite domain of positively charged amino acids typical of nuclear-targeting signals (1). A nuclear/cytoplasmic location was demonstrated in Cos7 cells (3), in pancreatic β cells (4), and in several cancer cell lines. The p8 protein shares biochemical and biophysical features with the high mobility group HMG-I/Y family (5). Sequence identity of p8 and HMG-I/Y is only ≈35%, but molecular mass, isoelectric points, hydrophilicity plots, and charge distributions along the polypeptides are very similar (5). An architectural role in transcription was proposed for p8 by analogy with HMG-I/Y, and recent reports are consistent with this hypothesis (6–8).
Several apparently unrelated functions have been ascribed to p8. It was involved in cell cycle regulation as a growth promoter (1, 3, 4) or inhibitor (9, 10), its overexpression was also associated with a low apoptotic index in pancreatic cancer cells (11), and p8 could control tumor progression (12, 13). Also, p8 activation stops cell growth upon nutriment deprivation in Drosophila melanogaster (14). Moreover, p8 is an important component of the defense program against lipopolysaccharide challenge (15), and it improves liver response to CCl4 challenge (16) and pancreatic response to acute pancreatitis by enhancing the expression of the antiinflammatory protein PAP I (8). To account for these various functions, it is suggested that the small size of the protein, its lack of specific tridimensional structure, and its nuclear/cytoplasmic localization (5) allow its interaction with several partners to target different signaling pathways.
Prothymosin α (ProTα) is a small, 12.4-kDa protein, highly acidic and lacking secondary structure, with a wide tissue distribution and a highly conserved among mammals (17). ProTα was associated with cellular proliferation and carcinogenesis (18, 19), cellular and viral transcription (19, 20), and remodeling of chromatin (21). More recently, ProTα was shown to inhibit caspase 9 activation by blocking apoptosome formation, which prevents the activation of effector caspases (22)
p8 and ProTα are two small proteins without stable secondary structure in solution, showing opposite electrostatic charges at neutral pH. They could therefore interact and promote mutual stabilization of their structures in a particular conformation, the resulting p8/ProTα complex becoming able to exert a specific function. In this work, we show that p8 and ProTα can indeed form a complex involved in the regulation of staurosporine-induced apoptosis.
Results
ProTα Is a Partner of p8.
We identified proteins interacting with p8 by two-hybrid screening of a cDNA library. The p8 cDNA subcloned into pSos provided the bait to screen a HeLa cDNA library constructed in pMyr. After cotransfection into Saccharomyces cerevisiae, strain cdc25H, 2 × 106 clones were screened, and 51 positives were identified. All clones were PCR-amplified. Their sequences were identified by comparison with the GenBank repertoire (Table 1, which is published as supporting information on the PNAS web site).
The interaction between p8 and ProTα was confirmed by transforming S. cerevisiae with both pMyr-ProTα and pSos-p8 constructs and allowing the transformants grow on synthetic dropout (SD) glucose and galactose agar plates lacking leucine and uracil [SD/glu(-LU) and SD/gal(-LU)] at the stringent temperature of 37°C. Clones growing on SD/gal(-LU) plates but not on SD/glu(-LU) plates at 37°C are interaction-positive clones. Growth was observed when pSos-p8 and pMyr-ProTα were both present but not when pSos-p8 or pMyr-ProTα was used separately. Negative and positive controls were grown as suggested by the manufacturer with expected results. These data show that p8 interacts with ProTα and that the interaction is specific.
The pMyr-ProTα-1–54 and pMyr-ProTα-42–110 constructs that encode the N-terminal and C-terminal parts of the human ProTα were used separately in two-hybrid experiments to identify which part of the ProTα molecule is involved in p8 binding. The constructs were cotransfected with the p-Sos-p8 plasmid. Surprisingly, neither the N-terminal nor the C-terminal parts of ProTα showed interaction with p8, indicating that binding requires most, if not all, of the ProTα sequence.
Interaction of p8 and ProTα in Cells.
Interaction between p8 and ProTα was controlled by coimmunoprecipitation assays. 293T cells were transfected with ProTα-V5 and p8-GFP, alone or in combination. ProTα-V5 and p8-GFP were immunoprecipitated from cell extracts with anti-ProTα or anti-p8 antibodies, respectively, and analyzed by Western blot. Anti-V5 antibody was used to detect tagged ProTα and anti-GFP was used to detect tagged p8. ProTα-V5 was detected in the complex containing p8, whereas p8-GFP was detected in the complex containing ProTα-V5. Tags were not detected in the negative control (Fig. 1A). These results confirm that ProTα interacts with p8 in 293T cells.
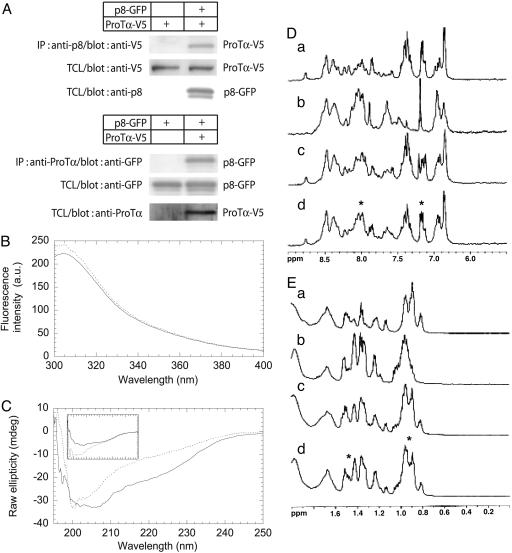
Fig. 1.
Interaction of p8 with ProTα. (A) 293T cells were cotransfected with 2 μg of pcDNA4-ProTα-V5 and pEGFP-C2p8 as indicated. Immunoprecipitation was performed by using an anti-p8 rabbit antibody or an anti-ProTα mouse monoclonal antibody. Cell lysates prepared with a buffer containing 1% Triton X-100 were used for precipitations with Sepharose beads. The precipitated material was immunoblotted with anti-GFP or anti-V5 antibodies. TCL, total cell lysate. (B) Fluorescence spectrum of the p8/ProTα complex. Emission fluorescence spectra were obtained in 25 mM phosphate buffer (pH 7)/150 mM NaCl at 298 K with 20 μM p8 (continuous line) and a mixture of p8 and ProTα (20 μM each) (dotted line). (C) Far-UV CD of the p8/ProTα complex. The spectrum obtained by the sum of the individual spectra of p8 and ProTα (30 μM each, continuous line) differs from that of the mixture of p8 and ProTα (30 μM each, dotted line). (C Inset) p8 isolated in solution (dotted line); ProTα isolated in solution (continuous line). Solution conditions were the same as in B. (D and E) 1D-1H-NMR spectra of p8 and ProTα were obtained in the amide and aromatic region (D) and methyl region (E) for p8 (150 μM) (a), ProTα (150 μM) (b), p8-ProTα mixture (150 μM each) (c), and spectrum resulting from the sum of p8 and ProTα individual spectra (d). Asterisks indicate the regions where the most important changes were detected (see text for details). Solution conditions were the same as in B.
Structural Analysis of p8 Binding to ProTα.
Fluorescence spectroscopy.
p8 has two tyrosine residues (3), Tyr-30 and Tyr-36. The emission fluorescence spectrum of p8 showed a maximum at 305 nm (Fig. 1B), as expected for a protein containing only tyrosine residues (23). Addition of an equal concentration of ProTα induced an increment in the intensity of the emission spectrum (Fig. 1B), showing that the structural environment of a fluorescent residue had changed. Because ProTα does not contain any fluorescent residue, this increase must be due to conformational rearrangements around at least one of the two tyrosines of p8 upon interaction with ProTα.
Far-UV CD.
The far-UV CD spectrum of p8 shows typical features of a random-coil protein (5), as does the ProTα spectrum (24). The far-UV CD spectrum of an equimolar mixture of the two proteins (Fig. 1C) differed from the spectrum expected for the same mixture in the absence of binding, which would be the mere addition of the individual spectra of the two proteins. The major difference was a decrease in raw ellipticity between 200 and 230 nm, indicating major modifications of secondary structure. These findings suggest that the secondary structure of p8, ProTα, or both changed upon binding.
NMR spectroscopy.
The spectra of p8 and ProTα showed typical features of random-coil chains (5, 24, 25): All of the amide protons appeared clustered between 8.0 and 8.7 ppm (Fig. 1D), with complete lack of dispersion in the methyl region (Fig. 1E). The amide protons of both proteins exchanged within 5 min in the presence of deuterated water, suggesting weak hydrogen bonding, if any. The spectrum of the mixture was different from that obtained by superposing the individual spectra acquired for both proteins. For instance, in the amide region, the positions of the aromatic protons and of some of the amide protons around 8.0 ppm were different. Because there is no aromatic side chain in ProTα, changes in the aromatic region must be attributed to changes in the environment of Tyr-30 and/or Tyr-36 in p8, in agreement with results from fluorescence experiments. Small changes were also observed in signals corresponding to protons of the methyl groups of Leu, Ile, and Val side chains in p8 and ProTα sequences, and also near 1.7 ppm, corresponding to the protons of the long side chain residues Ile, Leu, Lys, and Arg (Fig. 1E). It is interesting to note that three Leu residues are present in the vicinity of Tyr-30 and Tyr-36 (Leu-29, Leu-32, and Leu-37).
To further confirm the binding, we compared the height of the strongest methyl resonance (SMR) signal, at 0.8 ppm in the spectrum of the protein mixture (Fig. 1E), with the same peak obtained from the superposition of the two individual spectra (26). The SMR signal was 10% smaller in the spectrum of the mixture than in the superposed spectra. This change in the intensity revealed protein binding, as described in ref. 26.
Influence of p8 and ProTα Knockdown on Caspase 3/7 and Caspase 9 Activities.
In response to apoptogenic stimuli, Apaf1 oligomerizes to form the apoptosome that recruits and activates caspase 9, that, in turn, activates effector caspases 3, 6, and 7. ProTα impedes apoptosome formation, which prevents caspase 9 activation and blocks the apoptotic cascade (22). Because p8 and ProTα form a complex, the two proteins could act in concert to regulate the apoptotic cascade. To check this hypothesis, we monitored the effects of knocking down p8 and/or ProTα or overexpressing p8 and/or ProTα on caspase activities after inducing apoptosis with staurosporine.
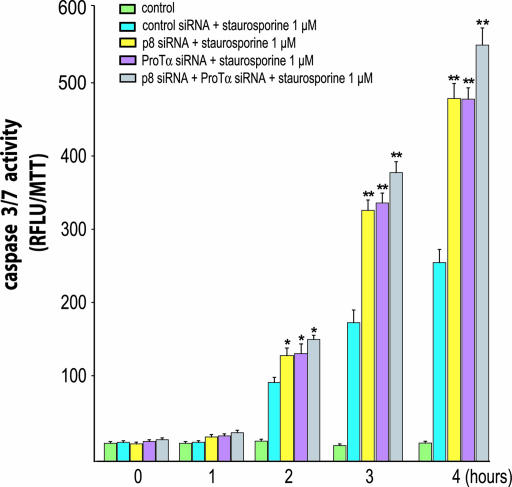
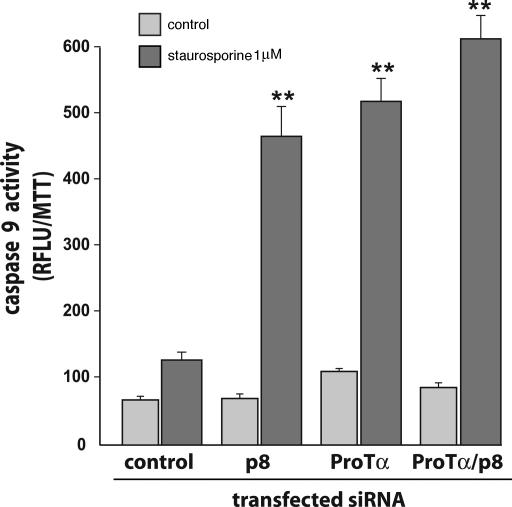
Expression of p8 and/or ProTα was knocked down in HeLa cells (Fig. 6, which is published as supporting information on the PNAS web site), and cells were treated with staurosporine for 4 h. Caspase 3/7 activity was measured after 1, 2, 3, and 4 h. Staurosporine treatment induced caspase 3/7 activity (Fig. 2) in cells transfected with control small interfering RNA (siRNA) with the same intensity as in nontransfected HeLa cells (data not shown). In cells in which ProTα or p8 had been knocked down with appropriate siRNAs, staurosporine treatment resulted in a larger increase in caspase 3/7 activation, ≈2-fold the level observed with control siRNA. However, when p8 and ProTα were concomitantly knocked down, caspase 3/7 activity was ≈2.4-fold higher than in cells treated with control siRNA, only slightly more than after ProTα or p8 knock down, showing that the effects of inhibiting ProTα and p8 expressions were not cumulative. Similar results were obtained when monitoring caspase 9 activity (Fig. 3). As control, these experiments were repeated by using other siRNAs against p8 and ProTα, with comparable results (data not shown). Finally, similar data were obtained when the pancreatic cancer cell line MiaPaca2 was used in the same experimental setup (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
Kinetics of caspase 3/7 activity in p8 and ProTα knocked-down cells. HeLa cells transfected with control siRNA or siRNAs for p8 and/or ProTα, were plated at 2.5 × 104 cells per well in 48-well plates. After 24 h, cells were incubated with staurosporine (1 μM) for the indicated time, and caspase 3/7 activity was quantified. Activity was measured by cleavage of the fluorescent caspase 3/7 substrate Z-DEVD-R110. Results are expressed as relative fluorescence units (RFLU) per cell viability as measured by the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrasodium bromide assay. Error bars represent the SEM from six experiments. Statistically significant differences from cells treated with control siRNA plus staurosporine are indicated as ∗, P < 0.05 and ∗∗, P < 0.01. Inhibition of p8 or ProTα expression enhanced caspase 3/7 activation.
Fig. 3.
p8 and ProTα are required for caspase 9 activation in staurosporine-treated cells. Cells transfected with control siRNA or with siRNAs for p8 and ProTα, alone or together were plated at 2.5 × 104 cells per well in 48-well plates. After 24 h, cells were incubated 4 h with 1 μM staurosporine. Caspase 9 activity was measured as described in Methods. Results are expressed as in Fig. 2. Caspase 9 activity was enhanced upon p8 or ProTα knockdown.
Caspase 3/7 and Caspase 9 Activities in Cells Overexpressing p8 and ProTα.
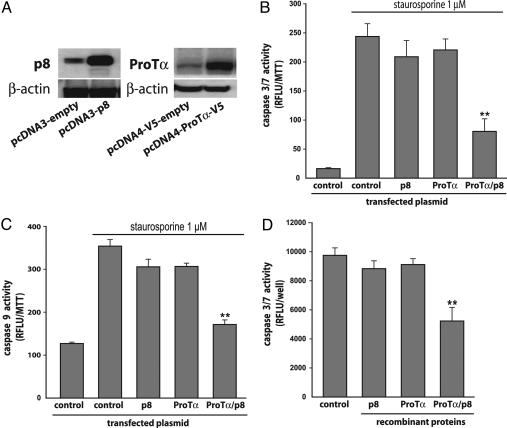
HeLa cells were transiently transfected with p8 and/or ProTα expression plasmids, then treated with staurosporine for 4 h before caspase 3/7 and caspase 9 activities were measured (Fig. 4). Overexpression of ProTα or p8 resulted in a moderate decrease in the induction of caspase 3/7 by staurosporine (15 and 20%, respectively). By contrast, induction of caspase 3/7 activity was dramatically inhibited (≈70%) when p8 and ProTα were concomitantly overexpressed. Similar results were found when induction of caspase 9 activity by staurosporine was monitored (Fig. 4).
Fig. 4.
p8 and ProTα inhibit caspase3/7 and 9 activations. HeLa cells were transfected with pcDNA3-p8 and/or with pcDNA4-ProTα-V5. (A) Immunoblot analyses by using the anti-p8 and anti-ProTα antibodies were performed with whole protein extracts. β-actin was used as control. (B and C) Caspase 3/7 (B) and caspase 9 (C) activities were monitored after incubating cells 4 h with 1 μM staurosporine. Results are expressed as in Fig. 2. Cotransfection of p8 and ProTα expression vectors inhibited the activations of both caspases. (D) Fourteen micrograms of HeLa S-100 cytosolic fraction was incubated in the presence of a 150 nM concentration of recombinant p8, ProTα, or both and the fluorescent caspase 3/7 substrate Z-DEVD-R110 in a final volume of 80 μl. Fluorescence was measured, and results are expressed as in Fig. 2.
Caspase 3/7 Activity in HeLa S-100 Cytosolic Fraction Is Inhibited by p8 and ProTα.
As shown in Fig. 4, basal caspase 3/7 activity is found in the S-100 fraction of HeLa cell extracts as published in ref. 22. Interestingly, the presence of a 150 nM concentration of p8 or ProTα recombinant proteins didn’t induced inhibition of caspase 3/7 activity. On the contrary, a significant inhibition was found when p8 and ProTα were added together to the reaction mix.
Effect of p8 and ProTα Knockdown on Staurosporine-Induced Cell Death.
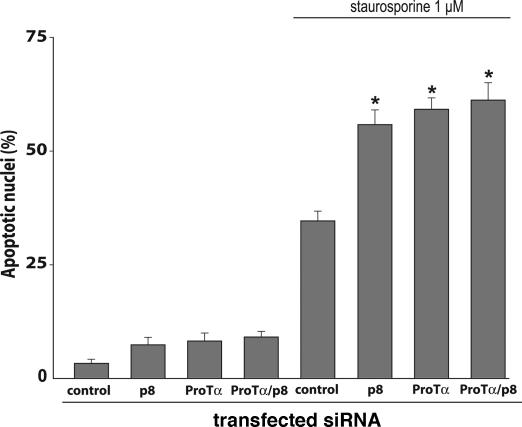
We used specific siRNAs to assess the effects of p8 and/or ProTα on staurosporine-induced cell death (Fig. 5). Transfection of HeLa cells with p8 siRNA, ProTα siRNA, or both led after 24 h to the appearance of condensed and fragmented chromatin typical of apoptosis in 6–9% of nuclei, compared with <2% in cells transfected with control siRNA. After staurosporine treatment, apoptotic nuclei were strikingly more abundant in cells with reduced expression of p8 (57%), ProTα (60%), or both (61%) than in cells transfected with control siRNA (33%). These experiments were repeated by using other siRNAs against p8 and ProTα with comparable results (data not shown).
Fig. 5.
Correlation between caspase activation and apoptosis. Cell viability was assessed by morphological observation of nuclei counterstained with Hoechst 33258. Staurosporine (1 μM) strongly induced apoptosis, this effect being enhanced in the absence of p8 and ProTα. Results are expressed as in Fig. 2.
Discussion
p8 and ProTα are stress proteins involved in limiting apoptosis. It is surprising that proteins so small in size (≈8–10 kDa), with no stable tridimensional structure (5), can play such an important role. Yet, an increasing number of functional proteins with unordered backbone structure have been described over the last years because of increased performances of biophysical techniques, helped by the development of algorithms allowing accurate identification of extended regions of disordered structure (27). Several cases have been reported of unordered proteins that acquire a stable structure after binding to the correct partner. Some eukaryotic transcription factors presenting with a basic region largely unstructured can fold into a stable helical structure upon binding to DNA (28). This structural stabilization is also the case for p8 upon interaction with DNA (5). Finally, many activation domains fold only upon binding to their protein target. For example, the cyclin-dependent kinase inhibitor p21 is unfolded in the absence of its ligand CDK2. Two-hybrid screening actually shows that p8 binds a large number of potential biological partners (Table 1), which might account for its various functions.
ProTα, one of p8 potential ligands revealed by our screening, was particularly interesting because, like p8, it is involved in several regulatory pathways, and the two proteins share strong implication into apoptosis. ProTα is a small ubiquitous protein, generally abundant, and present both in nucleus and cytoplasm. It is involved in the proliferation of mammalian cells (19), in transcription (20), in chromatin remodeling, and in protection against apoptosis (22). The molecular mechanisms are unknown, except for the antiapoptotic activity of ProTα, which occurs through inhibition of apoptosome formation (22). We wished to test the hypothesis that the two proteins require binding to each other to acquire their antiapoptotic properties.
We first confirmed the results of the two-hybrid screening, even if the CytoTrap technique used in this study is known to generate very few false positives and ProTα represented >10% of the positive clones. Coimmunoprecipitation of tagged proteins from 293T cell extracts showed that p8 and ProTα actually formed a complex (Fig. 1). Biophysical techniques were used to further characterize the complex (Fig. 1). Fluorescence and NMR spectroscopies showed that the binding involved the regions around the tyrosine residues in p8. Far-UV CD revealed that, upon binding, at least one of the proteins acquires secondary structure.
Coimmunoprecipitation experiments indicated that the p8/ProTα complex was stable enough to be functional, and we could assess the contribution of each member of the complex to the control of staurosporine-induced apoptosis. We first knocked down each of the two proteins, then both of them, and compared the extent of apoptosis in the cells, keeping in mind that inhibition of expression by siRNAs is never complete. Suppressing the expression of p8 or ProTα led to the same increase in caspase activity (Figs. 2 and 3) and the number of apoptotic nuclei (Fig. 5), suggesting that the two proteins are equally important to the antiapoptotic function. Suppressing both expressions had no further effect. This result suggests that each protein is indispensable to the mechanism by which apoptosis is inhibited. Results from experiments in which p8, ProTα, or both were overexpressed led to the same conclusion: Overexpressing one of the two proteins had no effect on caspase activities (Fig. 4), whereas overexpressing both led to a significant decrease. In addition, in vitro experiments also support the fact that both proteins are necessary to inhibit the caspase activation. Altogether, these findings are in agreement with the hypothesis that p8 and ProTα require forming a complex to exert their antiapoptotic activity.
In conclusion, p8 and ProTα are two small proteins with unordered backbones which, when associated in a heterodimer, inhibit staurosporine-induced apoptosis. Demonstration that p8 can gain a specific function when associated with a specific partner suggests that other potential partners of p8 identified by two-hybrid screen might, upon binding to p8, generate complexes with important functions presently attributed to p8 alone.
Methods
Yeast Two-Hybrid Screen.
We used the CytoTrap two-hybrid system (Stratagene). The complete coding sequence of human p8 (3) was subcloned into the Mlu1 restriction site of the pSos vector to generate the fusion protein pSos-p8. That construct was used as bait to screen a human HeLa cDNA library (Stratagene) constructed into the pMyr vector; pMyr-ProTα-1–54 and pMyr-ProTα-42–110, encoding ProTα fragments from amino acids 1 to 54 and 42 to 110, respectively, were obtained by subcloning specific RT-PCR-amplified DNA fragments into the pMyr vector. Screening was conducted as recommended by the manufacturer.
Cell Viability.
HeLa, 293T, and the human pancreatic cancer-derived MiaPaca2 cell lines were cultivated and treated as recommended by American Type Culture Collection. Their viability was determined by using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrasodium bromide viability assay (Promega) according to the manufacturer’s recommendations.
Coprecipitation Assays.
The full-length sequence of ProTα was cloned into pcDNA4 to generate the ProTα-V5 recombinant protein. p8-GFP was generated by in-frame ligation of the full-length sequence of human p8 into the pEGFP-C2 vector (Clontech). Plasmids were transfected into 293T cells by using Lipofectamine 2000 and following the manufacturer’s recommendations (Invitrogen). These cells are easily transfected, with an efficiency close to 100%. They produce high amounts of recombinant proteins. After cell lysis in 50 mM Tris·HCl, pH 8.0/0.5% Nonidet P-40 with protease inhibitors, p8 and ProTα were immunoprecipitated with rabbit antibodies against the ERKLVTKLQNSERKKRGARR p8 epitope (3) or with an anti-ProTα monoclonal antibody (Axxora), respectively. After a 2-h incubation with rocking, 20 μl of protein A-Sepharose or protein G-Sepharose conjugate (Zymed Laboratories) was added, and incubation was continued 2 h at 4°C. After three washes, Sepharose beads were treated for SDS/PAGE and Western blotting with anti-V5 (Invitrogen) or anti-GFP (Roche Diagnostics) antibodies.
Detection of Morphological Changes in Nuclei.
Morphological changes in chromatin structure typical of apoptotic cell death were detected by Hoechst 33258 staining. Briefly, attached cells were washed with PBS and fixed in PBS/paraformaldehyde 4% for 20 min at room temperature. Cells were permeabilized in PBS/0.2% Triton X-100 for 10 min and stained with 2 g/liter Hoechst 33258 in the dark for 10 min. Cell suspensions were mounted on glass slides and examined by fluorescence microscopy. Apoptotic cells were identified by their morphology and their staining.
siRNA Transfection.
p8–4 siRNA [sense 5′-GGAGGACCCAGGACAGGAUd(TT)-3′] and ProTα-1 siRNA [sense 5′-AUCACCACCAAGGACUUAAd(TT)-3′] were the most efficient of the eight siRNA obtained from Qiagen (Valencia, CA). Silencing by these siRNAs was maximal after 72 h. Cells were plated in 6-well plates to give 60–80% confluence. The next day, they were washed once with serum-free medium and transfected by using a mix composed of 6 μl of XtremeGENE (Roche Diagnostics) and 10 μl of p8 siRNA, ProTα siRNA, or control siRNA diluted in 240 μl of serum-free medium. After incubating 4 h at 37°C, the transfection medium was replaced by fresh medium. Twenty-four hours later, 2.5 × 104 cells were plated in 48-well plates for 24 h and then treated with 1 μM staurosporine (Sigma).
Transfection of p8 and ProTα Expression Plasmids.
The full-length cDNA encoding human ProTα was subcloned into the pcDNA4-V5 expression vector; pcDNA3-p8 (3) and pcDNA4-ProTα-V5 expression plasmids were transfected by using the FuGENE reagent by following the manufacturer’s recommendations (Roche Diagnostics) and allowed to express for 24 h. Empty vectors were used as control or to complete to identical amounts the mass of transfected DNA.
Semiquantitative RT-PCR Analysis.
Expressions of p8, ProTα, and TATA box-binding protein (TBP) mRNAs were determined by semiquantitative RT-PCR on RNA extracted from HeLa cells by using the forward (5′-TAGAGACGGGACTGCG-3′) and reverse (5′-GCGTGTCTATTTATTGTTGC-3′) primers for p8 amplification (32 cycles), the forward (5′-TCAGACGCAGCCGTAGACAC-3′) and reverse (5′-TCTAGTCATCCTCGTCGGTCT-3′) primers for ProTα amplification (25 cycles), and the forward (5′-TGCACAGGAGCCAAGAGTGAA-3′) and reverse (5′-CACATCACAGCTCCCCACCA-3′) primers for TBP amplification (22 cycles). Amplification products were submitted to electrophoresis on a 1.5% agarose gel and stained with ethidium bromide.
Quantitative RT-PCR Analysis.
First-strand cDNA was synthesized in a 20-μl reaction mix with 1 μg of total RNA by using Expand Reverse Transcriptase (Roche Diagnostics) for 10 min at 30°C, then 45 min at 42°C. Quantitative PCR was performed with a LightCycler (Roche Diagnostics) and Takara reagents. Five microliters of 10-fold diluted cDNA was mixed with 10 μl of SYBR Premix Ex Taq (including Taq polymerase, reaction buffer, MgCl2, SYBR green I dye, and dNTP mix) and 4 nmol of forward and reverse primers in a volume of 20 μl. After an initial Taq activation for 10 sec at 95°C, LightCycler PCR was performed at 45–55 cycles with the following cycling conditions: 95°C for 5 sec, 58°C for 6 sec, and 72°C for 12 sec. Each sample was analyzed in duplicate, and the experiment was repeated two to three times. Results were analyzed by using realquant (Roche Diagnostics) and expressed as a percent of control values.
Measurement of Caspase 3/7 and Caspase 9 Activities.
Caspase 3/7 and caspase 9 activities were measured by using the Apo-ONE Homogeneous Caspase 3/7 Assay Fluorometric Kit and Caspase-Glo 9 Assay, respectively (Promega), and performed as recommended by the manufacturer. Results are expressed as relative fluorescence units per cell viability as measured by the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrasodium bromide assay.
Structural Studies.
Protein expression and purification.
The p8 protein was expressed and purified as described in ref. 5 and stored in 25 mM sodium phosphate buffer, pH 6.0. ProTα was cloned into the pQE30 vector, expressed as a His-tagged protein and purified as described in ref. 5. Purity of the samples (>95%) was assessed by SDS/PAGE and confirmed by MALDI mass spectrometry (data not shown). Protein concentration was calculated by using the extinction coefficients of amino acids for p8 (29) and the molar extinction coefficient at 215 nm for ProTα (24).
Fluorescence measurements.
Fluorescence spectra were collected in a Cary Eclipse spectrofluorometer (Varian) interfaced with a Peltier cell. A 1-cm path-length quartz cell (Hellma, Forest Hills, NY) was used. Experiments were acquired at 298 K, with a 20 μM concentration of each molecule in PBS, pH 7. Samples were excited at 280 nm, and the slit widths were 5 nm for excitation and emission. The signal was acquired for 1 sec, and the increment of wavelength was set to 1 nm. Blank corrections were made in all spectra. Experiments were repeated four times with independent samples and good reproducibility.
Far-UV CD measurements.
Protein concentrations were 30 μM for both p8 and ProTα in phosphate buffer, pH 7/100 mM NaCl. CD spectra were collected on a Jasco (Easton, MD) J810 spectropolarimeter fitted with a thermostated cell holder and interfaced with a Neslab RTE-111 water bath, set at 298 K. Spectra were acquired at a scan speed of 50 nm/min with a response time of 2 sec and averaged over six scans. Reproducibility was assessed by repeating experiments four times with fresh samples.
NMR Spectroscopy.
Spectra were recorded in a Bruker DRX-500 spectrometer at 298 K and processed by using the bruker-xwinnmr software on a personal computer workstation. An exponential window function and polynomial base line corrections were applied in all cases. 1H chemical shifts were quoted relative to external reference 3-(trimethylsilyl)propionic-(2,2,3,3- 2H4) acid sodium salt. The 1D-1H-NMR spectra were acquired with 32 K data points by using the watergate sequence (30). Typically, 2 K scans were acquired, and the spectral width was 7,000 Hz. p8 and ProTα concentrations were 150 μM at pH 7 in 25 mM phosphate buffer in 90% H2O/10% 2H2O (150 μM each for experiments with both proteins). The final 1D-1H-NMR file contained 64 K data points.
Caspase 3/7 Activity in HeLa S-100 Cytosolic Fraction.
The S-100 fraction of HeLa cell extracts was prepared as described by Jiang et al. (22). Fourteen micrograms of S-100 were incubated 30 min in the presence of the fluorescent caspase 3/7 substrate Z-DEVD-R110 (Promega) with a 150 nM concentration of recombinant p8, ProTα, or both in a final volume of 80 μl. Fluorescence was measured by using a Wallac (Gaithersburg, MD) Victor 2 Multilabel counter 1420 with excitation at 495 nm and emission at 525 nm. Results are expressed as relative fluorescence units per well.
Statistics.
Statistical analysis was performed by ANOVA with post hoc analysis Student–Neuman–Keuls test. Results shown represent mean ± SE.
Supplementary Material
Acknowledgments
This work was supported by grants from Institut National de la Santé et de la Recherche Médicale, Ligue Contre le Cancer, and Canceropole PACA (to J.L.I.), Ministerio de Educación y Ciencia Grant CTQ2004-04474, Generalitat Valenciana Grant GV04B-402, and an institutional grant from Urbasa to the Instituto de Biología Molecular y Celular (to J.L.N.). C.M. was supported by Association pour la Recherche sur le Cancer.
Abbreviation
- ProTα
prothymosin α
- SD
synthetic dropout
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mallo G. V., Fiedler F., Calvo E. L., Ortiz E. M., Vasseur S., Keim V., Morisset J., Iovanna J. L. J. Biol. Chem. 1997;272:32360–32369. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Montero A., Vasseur S., Mallo G. V., Soubeyran P., Dagorn J. C., Iovanna J. L. Eur J. Cell Biol. 2001;80:720–725. doi: 10.1078/0171-9335-00209. [DOI] [PubMed] [Google Scholar]
- 3.Vasseur S., Vidal Mallo G., Fiedler F., Bodeker H., Canepa E., Moreno S., Iovanna J. L. Eur. J. Biochem. 1999;259:670–675. doi: 10.1046/j.1432-1327.1999.00092.x. [DOI] [PubMed] [Google Scholar]
- 4.>Path G., Opel A., Knoll A., Seufert J. Diabetes. 2004;53(Suppl. 1):S82–S85. doi: 10.2337/diabetes.53.2007.s82. [DOI] [PubMed] [Google Scholar]
- 5.Encinar J. A., Mallo G. V., Mizyrycki C., Giono L., Gonzalez-Ros J. M., Rico M., Canepa E., Moreno S., Neira J. L., Iovanna J. L. J. Biol. Chem. 2001;276:2742–2751. doi: 10.1074/jbc.M008594200. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmeister A., Ropolo A., Vasseur S., Mallo G. V., Bodeker H., Ritz-Laser B., Dressler G. R., Vaccaro M. I., Dagorn J. C., Moreno S., et al. J. Biol. Chem. 2002;277:22314–22319. doi: 10.1074/jbc.M201657200. [DOI] [PubMed] [Google Scholar]
- 7.Quirk C. C., Seachrist D. D., Nilson J. H. J. Biol. Chem. 2003;278:1680–1685. doi: 10.1074/jbc.M209906200. [DOI] [PubMed] [Google Scholar]
- 8.Vasseur S., Folch-Puy E., Hlouschek V., Garcia S., Fiedler F., Lerch M. M., Dagorn J. C., Closa D., Iovanna J. L. J. Biol. Chem. 2004;279:7199–7207. doi: 10.1074/jbc.M309152200. [DOI] [PubMed] [Google Scholar]
- 9.Vasseur S., Hoffmeister A., Garcia-Montero A. C., Mallo G. V., Feil R., Kuhbandner S., Dagorn J. C., Iovanna J. L. Oncogene. 2002;21:1685–1694. doi: 10.1038/sj.onc.1205222. [DOI] [PubMed] [Google Scholar]
- 10.Malicet C., Lesavre N., Vasseur S., Iovanna J. L. Mol. Cancer. 2003;2:37. doi: 10.1186/1476-4598-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su S. B., Motoo Y., Iovanna J. L., Berthezene P., Xie M. J., Mouri H., Ohtsubo K., Matsubara F., Sawabu N. Clin. Cancer Res. 2001;7:1320–1324. [PubMed] [Google Scholar]
- 12.Ree A. H., Tvermyr M., Engebraaten O., Rooman M., Rosok O., Hovig E., Meza-Zepeda L. A., Bruland O. S., Fodstad O. Cancer Res. 1999;59:4675–4680. [PubMed] [Google Scholar]
- 13.Vasseur S., Hoffmeister A., Garcia S., Bagnis C., Dagorn J. C., Iovanna J. L. EMBO Rep. 2002;3:165–170. doi: 10.1093/embo-reports/kvf023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinke I., Schutz C. S., Katzenberger J. D., Bauer M., Pankratz M. J. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasseur S., Hoffmeister A., Garcia-Montero A. C., Barthet M., Saint-Michel L., Berthezene P., Fiedler F., Closa D., Dagorn J. C., Iovanna J. L. BMC Gastroenterol. 2003;3:25. doi: 10.1186/1471-230X-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taieb D., Malicet C., Garcia S., Rocchi P., Arnaud C., Dagorn J. C., Iovanna J. L., Vasseur S. Hepatology. 2005;42:176–182. doi: 10.1002/hep.20759. [DOI] [PubMed] [Google Scholar]
- 17.Hannappel E., Huff T. Vitam. Horm. (San Francisco) 2003;6:257–296. doi: 10.1016/s0083-6729(03)01007-0. [DOI] [PubMed] [Google Scholar]
- 18.Eschenfeldt W. H., Berger S. L. Proc. Natl. Acad. Sci. USA. 1986;83:9403–9407. doi: 10.1073/pnas.83.24.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Marquez J., Segade F., Dosil M., Pichel J. G., Bustelo X. R., Freire M. J. Biol. Chem. 1989;264:8451–8454. [PubMed] [Google Scholar]
- 20.Cotter M. A., Robertson E. S. Mol. Cell. Biol. 2000;20:5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karetsou Z., Sandaltzopoulos R., Frangou-Lazaridis M., Lai C. Y., Tsolas O., Becker P. B., Papamarcaki T. Nucleic Acids Res. 1998;26:3111–3118. doi: 10.1093/nar/26.13.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., et al. Science. 2003;299:223–226. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 23.Pace C. N., Scholtz J. M. In: Protein Structure. 2nd Ed. Creighton T E., editor. Oxford: Oxford Univ. Press; pp. 253–259. [Google Scholar]
- 24.Gast K., Damaschum H., Eckert K., Schulze-Forster K., Maure H. R., Müller-Frohne M., Zirwer D., Czarnecki J., Damachun G. Biochemistry. 1995;34:13211–13218. doi: 10.1021/bi00040a037. [DOI] [PubMed] [Google Scholar]
- 25.Watts J. D., Cary P. D., Sautiere P., Crane-Robinson C. Eur. J. Biochem. 1990;192:643–651. doi: 10.1111/j.1432-1033.1990.tb19271.x. [DOI] [PubMed] [Google Scholar]
- 26.Araç D., Murphy T., Rizo J. Biochemistry. 2003;42:2774–2780. doi: 10.1021/bi0272050. [DOI] [PubMed] [Google Scholar]
- 27.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 29.Lakowicz J. R. Principles of Fluorescence Spectroscopy. 2nd Ed. New York: Plenum; [Google Scholar]
- 30.Piotto M., Saudek V., Sklenar V. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.