Abstract
Conjugated linoleic acids (CLAs) affect body fat gain, carcinogenesis, insulin resistance, and lipid peroxidation in mammals. Several isomers of CLA exist, of which the (9Z, 11E) and (10E, 12Z) isomers have beneficial effects on human metabolism but are scarce in foods. Bacterial polyunsaturated fatty acid isomerases are promising biotechnological catalysts for CLA production. We describe six crystal structures of the Propionibacterium acnes polyunsaturated fatty acid isomerase PAI in apo- and product-bound forms. The three-domain flavoprotein has previously undescribed folds outside the FAD-binding site. Conformational changes in a hydrophobic channel toward the active site reveal a unique gating mechanism for substrate specificity. The geometry of the substrate-binding site explains the length preferences for C18 fatty acids. A catalytic mechanism for double-bond isomerization is formulated that may be altered to change substrate specificity for syntheses of rare CLAs from easily accessible precursors.
Keywords: conjugated linoleic acid, flavoprotein, polyenoic fatty acid isomerase, structure-based mechanism
Conjugated linoleic acid (CLA) has been reported to regulate body fat gain, inhibit carcinogenesis, and modulate the immune response and insulin tolerance in animals and humans (1, 2). These effects are mediated mainly by the CLA isomers (9Z, 11E) and (10E, 12Z), further called 9,11-CLA and 10,12-CLA. The main sources of CLA in the human diet are meat and milk from ruminants (3). CLA and other conjugated fatty acids are produced in vivo by double-bond isomerization of polyunsaturated fatty acid (PUFA) precursors (4–6). Isomerase activity was found in bacteria (4) and algae (5, 6). The beneficial effects of CLA in treatment of cancer and multifunctional diseases such as obesity and diabetes have raised interest in enrichment of the human diet with these fatty acids by biotechnological means. Because the currently available sources for CLA are ill-defined mixtures of several isomers, a detailed analysis of CLA effects requires synthesis of the pure isomers (1, 2). Chemical methods for pure CLA isomer production are expensive, and CLA production by recombinant bacterial enzymes offers an alternative. However, CLA production by this avenue requires optimization of the existing enzymes, which accept only free fatty acids instead of lipids (7). The fatty acid isomerase from Propionibacterium acnes (PAI), which catalyzes the isomerization of linoleic acid (LA) to 10,12-CLA, is a promising candidate for this approach (Fig. 1a).
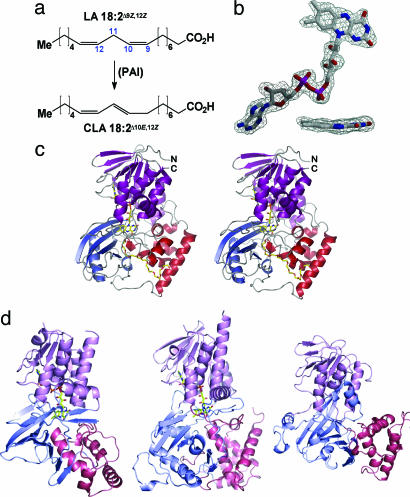
Fig. 1.
The PAI reaction and structure. (a) Reaction catalyzed by PAI. (b) σA-weighted mFo − DFc omit electron density maps contoured at 2σ of FAD and a side-on view of the isoalloxazine ring (Inset). (c) Architecture of PAI with the FAD-binding domain colored in magenta, domain 2 in red, and domain 3 in blue. FAD and PEG 400 are shown as stick models in this stereo figure. (d) PAI structural analogs: polyaminooxidase (PDB ID code 1RSG) (Left), UDP-galactopyranose mutase (1I8T) (Center), and guanine nucleotide dissociation inhibitor (1GND) (Right). The domains are colored as for PAI but in lighter hue.
The isomerization reaction of CLA is conceptually simple, requiring abstraction of a hydrogen radical or anion from C11, followed by a double-bond shift and rehydrogenation at either C13 or C9 (Fig. 1a). However, the underlying mechanism of the conversion of the pentadienyl moiety into a conjugated diene is poorly understood. To date, biochemical studies were reported for PUFA isomerases from Butyrivibrio fibrisolvens (BFI) (8, 9) and the red alga Ptilota filicina (PFI) (6, 10). PFI presumably contains a flavin adenine dinucleotide (FAD) cofactor (6), and sequence analysis of PAI reveals homology to dinucleotide-binding domains present in FAD-containing aminooxidases. However, no structure of any PUFA isomerase is currently available to put the available biochemical data on a structural basis and to determine how FAD can catalyze the nonredox PUFA isomerization.
We determined the crystal structure of PAI to define the active site and extract a structure-based mechanism for polyenoic fatty acid isomerization. PAI is an FAD-containing monomer consisting of three intricately connected domains. The N-terminal domain shares similarity with modules found in other FAD-binding proteins, and the overall fold is in part similar to yeast polyamine oxidase (11). The geometry of the substrate-binding pocket determined for the PAI–LA complex reveals that fatty acid is bound with the methyl end inside and delineates the residues that are involved in the stabilization of reaction intermediates. Because the activity of heterologously produced isomerases in bacteria and plants on fatty acids is still unsatisfactory (6, 7, 10, 12), the PAI structure can serve as a framework for engineering of the substrate specificity such that not free fatty acids but plant oils are directly converted to PUFAs.
Results
PUFA isomerase from P. acnes, a commensal of human skin, is a yellow-colored monomeric enzyme of 424 residues. Absorption spectroscopy and HPLC analysis (data not shown) revealed the presence of a noncovalently bound oxidized FAD, which is retained during crystallization (Fig. 1b). The structure of PAI was determined in-house by the single isomorphous replacement with anomalous scattering method (see Materials and Methods) using iodide as the heavy atom and refined to a resolution of 1.95 Å. Five more crystal structures of PAI in space groups I213 and C2, and either in the apoform or bound to products or polyethylene glycol (PEG) 400, were subsequently determined by molecular replacement and refined to resolutions of 2.25 Å or higher (Table 1). The maximum rms deviation within these structures is 0.3 Å, independent of crystal lattice or complex state, thus eliminating crystal-packing artifacts on the conformation of PAI.
Table 1.
Data collection, phasing, and refinement statistics
| Data set | 2B9W–native 1 (PEG 400) | 2B9X–KI soak (PEG 400) | 2BA9–native 2 (PEG 400) | 2B9Y–apo | 2BAB–CLA complex | 2BAC–LnA complex |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Resolution range,* Å | 50–1.86 (1.92–1.86) | 50–2.21 (2.29–2.21) | 50–1.95 (2.02–1.95) | 50–2.2 (2.28–2.20) | 50–2.0 (2.07–2.00) | 40–2.25 (2.33–2.25) |
| Space group | I213 | I213 | C2 | I213 | I213 | C2 |
| Cell dimensions, Å; ° | 160.4 | 160.3 | a = 133.7, b = 60.8, c = 72.2, β = 115.8 | 160.4 | 161.8 | a = 133.7, b = 60.8, c = 71.8, β = 117.7 |
| Unique reflections* | 49,143 (2282) | 58,536 (1322) | 37,527 (3804) | 32,918 (1695) | 46,145 (3465) | 20,816 (356) |
| Multiplicity* | 5.3 (1.9) | 5.9 (1.2) | 4.3 (4.3) | 5.0 (1.9) | 5.2 (1.9) | 3.2 (1.7) |
| Completeness,* % | 98.4 (26.9) | 87.9 (19.7) | 100 (98.1) | 94.3 (49.2) | 97.3 (73.8) | 85.3 (14.7) |
| Mosaicity, ° | 0.49 | 0.85 | 0.65 | 0.96 | 0.57 | 1.05 |
| Rsym,* %† | 9.9 (75.6) | 12.9 (55.5) | 12.3 (61.2) | 8.7 (49.1) | 10.4 (53.3) | 9.0 (24.4) |
| Average I/σ(I)* | 15.2 (1.0) | 12.1 (1.0) | 11.4 (2.5) | 16.5 (1.5) | 13.3 (1.5) | 12.8 (3.0) |
| SIRAS phasing | ||||||
| Resolution, Å | 20–4 | |||||
| No. of iodide sites | 13 | |||||
| FOM/CCmap | 0.56/0.80 | |||||
| Refinement | ||||||
| Resolution range,* Å | 35.9–1.95 (2.00–1.95) | 22.7–2.22 (2.28–2.22) | 46.4–1.95 (2.00–1.95) | 42.9–2.20 (2.27–2.20) | 27.0–2.00 (2.05–2.00) | 40.0–2.30 (2.36–2.30) |
| Rcryst,*‡ % | 16.4 (28.0) | 17.7 (27.1) | 21.3 (27.3) | 15.6 (25.2) | 16.3 (26.9) | 15.8 (18.6) |
| Rfree,*‡ % | 19.0 (32.5) | 21.5 (37.2) | 25.9 (30.5) | 20.4 (29.6) | 20.0 (31.6) | 22.8 (23.2) |
| No. of residues/waters | 424/465 | 423/354 | 423/432 | 423/511 | 426/559 | 424/467 |
| Coordinate error,§ Å | 0.081 | 0.127 | 0.182 | 0.118 | 0.088 | 0.158 |
| rmsd Bonds/angles, Å; ° | 0.015/1.44 | 0.010/1.21 | 0.010/1.22 | 0.013/1.34 | 0.011/1.24 | 0.011/1.31 |
| Ramachandran plot,¶ % | 89.2/10.5/0.3 | 87.3/12.2/0.5 | 89.5/10.0/0.5 | 87.9/11.9/0.3 | 87.9/11.9/0.2 | 89.2/10.5/0.3 |
| Average B values, Å2 | 38.3 ± 8.5 | 48.6 ± 7.2 | 24.3 ± 4.9 | 35.5 ± 8.0 | 40.9 ± 8.5 | 27.3 ± 6.3 |
SIRAS, single isomorphous replacement with anomalous scattering; rmsd, rms deviation.
*Values in parentheses correspond to the highest-resolution shell.
†Rsym = 100·ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h and 〈I(h)〉 is the average value of the reflection intensity.
‡Rcryst = Σ|Fo| − |Fc|/Σ|Fo|, where Fo and Fc are the structure factor amplitudes from the data and the m odel, respectively. Rfree is Rcryst with 5% of test set structure factors.
§Based on maximum likelihood.
¶Calculated using procheck. Numbers reflect the percentage amino acid residues in the core, allowed, and generous allowed regions, respectively.
Structure Description.
PAI comprises three intricately connected domains. Domain 1 (residues 1–77, 196–275, and 371–424) is a mixed α/β FAD-binding domain, domain 2 (residues 78–195) is predominantly α-helical, and domain 3 (residues 276–370) comprises a four-stranded β-sheet interspersed with two α-helices (Fig. 1c). Domain 1 closely resembles a variant of the Rossmann fold for dinucleotide binding found in the glutathione reductase family GR1 (13), consisting of a central four-stranded parallel β-sheet surrounded by two α-helices. The conserved GxGxxG(x)18E motif of the GR1 family, which contains the P-loop and a glutamate residue that bind to the phosphate and ribose moieties of FAD, respectively, is located within the first 37 N-terminal residues of PAI. This region is highly conserved among GR1-flavoproteins and also contains several residues that engage in hydrophobic contacts with FAD (see Fig. 5, which is published as supporting information on the PNAS web site). Accordingly, the closest structural homologues of PAI domain 1 identified by the dali program (www.ebi.ac.uk/dali) are FAD-containing oxidoreductases and isomerases such as yeast polyamine oxidase (11) and UDP-galactopyranose mutase (12), with Z-scores of 24.9 and 15.5, respectively (Fig. 1d). The closest non-flavin-containing structural homologue is the guanine nucleotide dissociation inhibitor (GDI) (Fig. 1d) with a dali Z-score of 13.1 (14). Because GDI does not bind FAD, the reason for its close structural homology to flavoproteins is unclear.
PAI domains 2 and 3 have no structural homologues in the dali database but display a roughly similar organization as in UDP-galactopyranose mutase and polyamine oxidase [Protein Data Bank (PDB) ID codes 1I8T and 1B37] (Fig. 1d) with high rms deviation values of 12 Å after superposition of all Cα atoms. A narrow, C-shaped channel of ≈30 Å length is formed by domains 2 and 3 and stretches from the surface of PAI toward the isoalloxazine ring of FAD. Thus, whereas domain 1 binds to FAD, domains 2 and 3 are responsible for substrate binding and specificity, as well as completion of the active site (see below).
FAD Cofactor.
Electron density maps calculated with unbiased single isomorphous replacement with anomalous scattering phases unambiguously revealed the FAD cofactor (Fig. 1b), which adopts an extended conformation and orients its re-face toward the active site. As in other flavin-containing enzymes, the catalytically active isoalloxazine ring is buried in a hydrophobic pocket and inaccessible to bulk solvent. The ring is bent such that the pyridoxine and xylene moieties adopt a 23° angle (Fig. 1b). A similar deviation of isoalloxazine from planarity was described for cholesterol oxidase (PDB ID code 1MXT), polyamine oxidase, and trimethylamine dehydrogenase (PDB ID code 1DJN). Although the flavin ring of FAD entertains only three direct hydrogen bonds (H-bonds) with PAI, it is contacted by 9 hydrophobic residues with a total of 62 van der Waals contacts, underscoring the hydrophobic nature of the FAD-binding pocket. In flavin-containing oxidases, the FAD N5 atom is usually H-bonded to a protein main- or side-chain atom (15). This feature is absent in PAI, and the space in the vicinity of N5 is part of the substrate-binding site.
Fatty Acid Binding Mode.
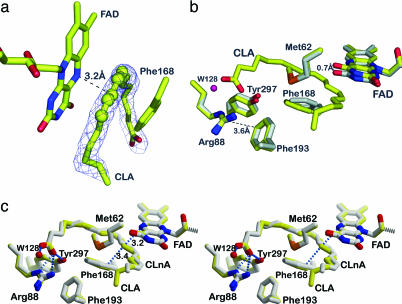
To reveal the exact binding mode of PUFAs, PAI was cocrystallized with LA (PDB ID code 2BAB; Table 1) and α-linolenic acid (LnA) (PDB ID code 2BAC). The identity of the fatty acid (for instance, the substrate LA or the product 10,12-CLA) cannot be judged from electron density because both LA and CLA can adopt a planar conformation of C9 to C13 carbons (Fig. 2a). Thus, GC-MS analysis was used to confirm the nature of the fatty acid in the crystals (see Fig. 6, which is published as supporting information on the PNAS web site). In case of cocrystallization with LA, the crystals contained exclusively the product 10,12-CLA, indicating turnover of the substrate before crystallization. The structure of the PAI-product complex is an important hallmark of the catalytic cycle and also illuminates the principles of substrate binding.
Fig. 2.
Substrate binding to PAI. (a) The σA-weighted mFo − DFc omit electron density map of 10,12-CLA is contoured at 2σ. CLA adopts a U-shaped form. Atoms C9–C13 are shown as spheres to visualize the planarity of the double bond system. (b) Conformational changes in the PAI–CLA complex. Superposition of the active sites of apo-PAI (gray) and bound to CLA (yellow). Phe-168, FAD, and Tyr-270 undergo small conformational change upon substrate binding. The side chain of Phe-168 becomes coplanar with the conjugated double bonds of CLA. The FAD isoalloxazine ring is shifted backwards to accommodate the fatty acid. The OH-group of Tyr-270 moves toward the carboxylate of CLA. (c) Structure of PAI in complex with (11E, 13E, 15Z)–CLnA (gray). The PAI–CLA complex (yellow) is shown for reference in this stereo figure. The conjugated triene bond system is planar (atoms C10–C16), and the fatty acid molecule is more strongly bent compared with CLA.
CLA adopts a strongly bent U-shape when bound to PAI such that C1 and C18 are only 9.4 Å apart (Fig. 2a). This bent conformation is imposed by Met-62, which functions as a rigid pole around which the fatty acid hydrocarbon chain wraps (Fig. 2b). Only small conformational changes of PAI side chains and FAD accompany fatty acid binding. The hydrocarbon chain of the fatty acid displaces the isoalloxazine ring of FAD by up to 0.7 Å relative to its position in the apostructure. Likewise, Phe-168 rotates around χ2 by ≈25° and moves away from the fatty acid by up to 1.5Å compared with the apostructure (Fig. 2b). The result is a coplanar orientation of the pentadienyl moiety of the fatty acid with FAD and Phe-168, which puts the site of hydrogen abstraction, C11, in 3.2-Å distance to the electrophilic FAD atom N5. The carboxylate of CLA is H-bonded to Arg-88 and Tyr-297, which shifts closer toward the fatty acid by 0.4 Å in the complex structure. The entrance of the substrate channel is blocked by parallel π-stacking of Arg-88 and Phe-193, excluding water from the active site (Fig. 2 b and c).
Up to four water molecules are displaced from the vicinity of the isoalloxazine moiety upon substrate binding. Exclusion of water by substrate binding is essential to prevent side-reactions with electrophilic intermediates that are proposed in the catalytic cycle (see below). A similar water-exclusion mechanism by substrate binding has been described for polyamine oxidase (16) and acyl-CoA dehydrogenases (17).
We also determined the structure of PAI in complex with conjugated LnA (CLnA), a C18 fatty acid containing three double bonds (Fig. 2c). The structure of the PAI–CLnA complex comprises all salient features of the PAI–CLA complex with the exception of the conformation of the methyl end of CLnA, which apparently is mobile as judged from increased B-values in this region (Fig. 2c). Structural plasticity is also apparent for the conjugated triene moiety of CLnA, which is displaced by 0.6 Å relative to CLA. Such shifts of the fatty acid may be required for efficient H-atom transfer during catalysis. Importantly, a 418-Å3 cavity is located at the methyl end of the fatty acid, which could accommodate two more carbon atoms, and extends the substrate palette for PAI toward C20 and C22 PUFAs (7).
Gating Mechanism for Substrate Entry.
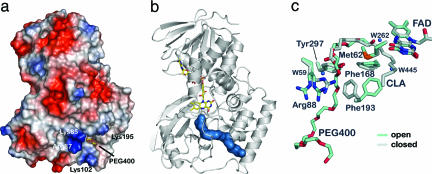
A hallmark of FAD-containing proteins is a buried cofactor, which requires substrate to enter through a channel that can be as long as 30 Å. The electrostatic potential of PAI reveals a positively charged patch generated by residues Lys-85, Arg-87, Lys-102, and Arg-195 near the entrance of such a channel that leads to the FAD cofactor (Fig. 3a). This positive patch may serve as an initial recognition site for the negatively charged carboxylate group of fatty acid substrates. Three structures of PAI (PDB ID codes 2B9W, 2B9X, and 2BA9) were determined from crystals of space groups I213 and C2 (Table 1), all of which include a PEG 400 molecule. PEG 400 is located inside the channel between domains 2 and 3 that stretches from the surface of PAI to the FAD cofactor (Fig. 3b). The channel is lined by hydrophobic residues, congruent with the hydrophobic nature of PAI substrates. The absence of hydrophilic contacts to PEG 400 and of alternative paths from the surface to the PAI active site shows that PEG 400 mimics a fatty acid hydrocarbon chain and, therefore, precisely delineates the path to the substrate-binding site. However, PEG 400 is an inhibitor rather than a substrate or product analogue because, unlike LA or LnA, it does not completely enter the active site (Fig. 3c).
Fig. 3.
Substrate entry channel and gating mechanism in PAI. (a) The surface potential of PAI shows an electropositive area localized at the entrance of the channel that is created by Lys-85, Arg-87, Lys-102, and Lys-195. The PEG 400 molecule marks the entry of the channel. (b) The molecular surface (blue) of part of the PEG 400 molecule bound to PAI in the absence of substrate/product shows the 30-Å path from the surface to the active site FAD (drawn as sticks). (c) Conformational changes in active site associated with PEG 400 binding reveal the gating mechanism. PEG 400, Phe-193, and Arg-88 are in the open conformation when PEG 400 is bound (blue) compared with the apoenzyme (gray). Arg-88 displays two conformations in the open form of PAI, both of which point away from the entering substrate.
Flavoproteins can exclude water from the substrate-binding and active site by various gating mechanisms that include domain movements, mobile loops, and also side-chain switches (15). Comparison of the PEG 400-bound structures with the apostructure (PDB ID code 2B9Y) revealed that the latter option is realized in PAI. Superposition of these structures shows that only Arg-88 and Phe-193 change their side-chain conformations when PEG 400 enters the channel, and, thus, these residues function as a gate. In the presence of PEG 400 the side chains of Arg-88 and Phe-193 are oriented away from the channel and in the “open” position. However, in the apoform or after fatty-acid binding, these residues swing back into the “closed” position and block the entry channel to the active site (Fig. 3c). The two positions are characterized by a rotation of the Phe-193 side chain by 78° around χ1 and a reorientation of the Arg-88 side chain, which leads to distances of 11.1 and 3.3 Å, respectively, between the tips of each side chain in the open and closed states.
Closure of the gate is driven by the substrate itself. Arg-88 can H-bond to the carboxylate group of the substrate only in the closed state. Phe-193 switches back to the closed state and stacks onto the guanidinium group of Arg-88 in a π-cation interaction. In addition, the hydrocarbon chain of the bound substrate displaces Phe-193 from the open position. This mechanism of gate closure may help to select fatty acid with a maximum chain length: If a fatty acid with a long hydrocarbon chain displaces Phe-193 before the carboxylate is bound by Arg-88, it is either no or only a poor PAI substrate. Interestingly, PAI also catalyzes conjugated triene formation from LnA but not from γ-linolenic acid (γ-LnA) (18:3Δ6Z,Δ9Z,Δ12Z). The double bonds in γ-LnA are shifted by three carbon atoms toward the C terminus compared with LnA (7). Thus, the distance between the lock and FAD can explain positional specificity of PAI: ω-3 PUFAs yield a mixture of diconjugated and triconjugated products because the hydrogen atom can be abstracted from either C11 or C14. However, in ω-6 fatty acids, C11 and C8 would have to be attacked, but C8 is located too far from FAD, and conversion of ω-6 fatty acids leads to sole formation of the diconjugated fatty acid derivative.
Structure-Based Mechanism of Fatty Acid Isomerization.
Biochemical studies on PFI, which presumably contains a FAD cofactor, have suggested that this enzyme operates by means of transfer of the 11-pro-S hydrogen of γ-LnA to the C13 position along with the removal of the 8-pro-R hydrogen and protonation at C11, resulting in isomerization of two double bonds (6, 10). Given the space restriction of the fatty acids when bound to PAI (Fig. 2), any movements of substrate and products should be on a small scale of ≈2 Å. Therefore, the C11 pro-R hydrogen of LA would point directly toward FAD atom N5 in the initial PAI–substrate complex, ready for abstraction as the first step of the isomerization reaction.
To date, several examples of flavoenzymes catalyzing reactions with no net redox change via one- or two-electron transfer are known (18). A main issue regarding isomerization of PUFAs by PAI is whether the catalytic mechanism is radical or ionic. Both the radical and anionic intermediate of the isoalloxazine ring are stabilized by π-conjugation, which makes distinction between these alternative reaction routes difficult (15). However, in PAI a more efficient stabilization of a cationic intermediate is provided by the edge-on sandwiching between FAD and Phe-168. Likewise, the negative charge on FAD can be efficiently stabilized by PAI. It has been noted before that the N1–C2 O2 locus of the isoalloxazine ring in flavoproteins is ≈3.5 Å to a positively charged residue or the N terminus of an α-helix dipole, which can stabilize a negative charge on FAD during catalysis (15, 18). In PAI, such a charge is partially neutralized by the dipole of the C-terminal α-helix (residues F408–F422), which has counterparts in UDP-mutase and polyamine oxidase (Fig. 1d). In conclusion, the possibilities of π-cation and helix dipole stabilization of a substrate carbocation render an ionic reaction mechanism extremely likely.
O2 locus of the isoalloxazine ring in flavoproteins is ≈3.5 Å to a positively charged residue or the N terminus of an α-helix dipole, which can stabilize a negative charge on FAD during catalysis (15, 18). In PAI, such a charge is partially neutralized by the dipole of the C-terminal α-helix (residues F408–F422), which has counterparts in UDP-mutase and polyamine oxidase (Fig. 1d). In conclusion, the possibilities of π-cation and helix dipole stabilization of a substrate carbocation render an ionic reaction mechanism extremely likely.
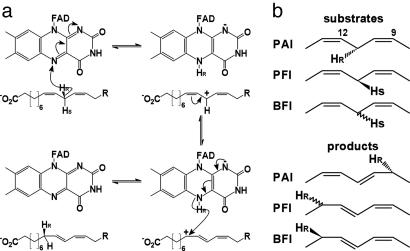
Readdition of a hydride at C9 then fixes one of three possible mesomeric structures as the product (Fig. 4a). For this hydride addition, either the substrate or FAD must shift by 2.6 Å from the central C-atom (C11 in case of LA) to C9. Because FAD is tightly bound to PAI by numerous interactions, the intermediate is likely to move after hydride abstraction. Structural plasticity of the bound fatty acid is already apparent from comparison of the two product complexes (Fig. 2c), and the intermediate probably shifts in the plane of the pentadienyl moiety during catalysis. Independent proof that the same hydrogen atom abstracted by FAD is transferred back to LA at C9 will come from analyses with C11-dideuterated LA as the substrate. It is very likely that PAI follows a similar catalytic mechanism to that described for PFI (5).
Fig. 4.
Stereochemistry of PAI reaction. (a) Structure-based isomerization mechanism of LA to 10,12-CLA. Pro-R Hydride abstraction by FAD atom N5 yields an intermediate LA carbocation, which is stabilized by π-conjugation. Hydride transfer to the Re-side of LA atom C9 yields 10,12-CLA and regenerates the oxidized form of FAD. (b) Stereochemistry of hydrogen transfer in PUFA isomerases. In PAI the hydride is abstracted from and transferred back to the fatty acid from the Re-side. For PFI, abstraction from the Si-side was reported, but no information is available for the back-transfer of the H-atom. The situation is reversed in BFI, where the abstraction face is unknown, but the H-atom is transferred back from the Re-side.
Discussion
PAI represents the third structure of a carbon–carbon double bond isomerase, but its reaction mechanism is entirely different from that of the other two enzymes: Isopentenylpyrophosphate–dimethylallylpyrophosphate isomerase is a flavoprotein with bound flavin mononucleotide (FMN) but also requires NADPH. Δ3-Δ2-enoyl-CoA isomerase uses a glutamate residue as the catalytic base for proton abstraction from a carbon atom next to an activating carbonyl group of the fatty acid. There is no structural similarity between these isomerases and PAI. Rather, the domain arrangement in PAI resembles that of UDP-galactopyranose mutase, which probably operates by means of a sugar radical (12, 19), and therefore its mechanism will not bear any resemblance to the mechanism used by PAI. Thus, the PAI structure is a prototype for PUFA isomerases and can serve as a framework for PUFA isomerization in general.
Hitherto, the only crystal structure of an enzyme bound to free PUFA is prostaglandine endoperoxide H synthase-1 (PDB ID codes 1IGX, 1IGZ, and 1FE2) (20), but the overall shape of the fatty acid and the mode of carboxylate recognition is different from PAI. In PAI, residues Arg-88 and Phe-193 act as a lock, fixing the carboxylate of the fatty acid near the entrance of the active site. The positive potential on the PAI surface generated by Lys-85, Arg-87, Lys-102, and Lys-195 reveals a site for initial recognition of substrate carboxylate groups. Similar to PAI, BFI and PFI prefer a carboxylic group over methyl esters as substrates (5, 7, 9), and PFI also contains FAD (5). Thus, the Arg–Phe lock is likely a general feature of FAD-containing PUFA isomerases.
An important aspect of the reaction mechanism is how the hydrogen transfer occurs during catalysis. The close distance between C11 and N5 of FAD in the 10,12-CLA–PAI complex strongly suggests that FAD acts as a hydrogen-abstracting electrophile. Because no cysteine, tryptophan, or acidic residues are present in the active site, non-FAD redox and proton abstraction processes can be excluded. Both in PFI and BFI, atom C11 was shown to be the hydrogen abstraction site (5, 9). The chain length preferences for different isomerases could be explained by a fixed distance between FAD and the substrate carboxylate, which is 11 carbon atoms in the case of LA, LnA, and γ-LnA and 10 carbon atoms in the case of arachidonic or eicosapentadienoic acid (7).
Given these biochemically derived length criteria and the coplanar positioning of the polyene moiety with FAD, we suggest that the first step in the catalytic sequence for PAI, in analogy to BFI and PFI, is abstraction of the pro-R hydrogen atom from C11 by the FAD N5 atom (Fig. 4b Upper). C11 of CLA and FAD N5 are 3.2 Å apart and assuming a similar spatial arrangement for LA (Fig. 2a), the C–H bond would be aligned with the lowest unoccupied molecular orbital of FAD enabling hydride transfer to N5. The intermediate carbocation is then stabilized by stacking with the Phe-168 and flavin aromatic systems, and the isomerization of the C9–C10 double bond occurs by regiospecific hydride transfer to the pro-R position, which completes the catalytic cycle. The preferred direction of the allylic shift is explained by the relative position of Phe-168 and the carbocation. Because the phenyl ring is closer to the C10–C9 bond, the mesomer with a partial positive charge on C9 will be more stable, and the (10E, 12Z)-isomer will be fixed by hydride transfer from FAD. Indeed, GC-MS data with (11S)-deutero γ-LnA as PFI substrate showed that deuterium transfer occurs from C11 to C13 (5). The question as to the stereochemistry of this transfer can be answered by the mode of fixing of the fatty acid in the PAI active site. Because a complete reorientation of the fatty acid other than a shift along the direction of the carbon chain during catalysis is impossible, the pro-R positions at both sites must be involved. For conjugated triene formation as in case of LnA, a second hydrogen abstraction is required at C14. Inspection of the active site in the PAI–CLnA complex does not reveal significant differences compared with the apoenzyme, suggesting that substrate shifting occurs during the catalytic cycle to bring C14 close to FAD N5. Protonation at C10 then would complete the reaction for the triene isomerization.
The mechanism described here allows comparison of PUFA isomerases in terms of regiospecificity and stereospecificity. First, for all three enzymes C11 in case of C18-PUFAs is the site of hydrogen abstraction. This hydrogen is then transferred back to C9 (PAI) or C13 (BFI, PFI). Assuming that the active sites in BFI and PFI have similar architectures to PAI, including FAD and an aromatic residue for stabilization of the intermediate carbocation, the regiospecificity of isomerization could be determined by positioning of the aromatic residue (Phe-168 in PAI) relative to C9 and C13. Furthermore, the abstraction and readdition of the bis-allylic hydrogen is regiospecific, i.e., from the same side of the pentadienyl system. The 11-pro-S hydrogen is abstracted from γ-LnA and transferred to C13 for PFI, but the stereochemistry of the product was not determined (5). For BFI, hydrogen transfer from C11 to the pro-R position at C13 was demonstrated, but no data on the stereospecificity are available (9). Although structural data on PFI and BFI are lacking, these data strongly indicate that the orientation of fatty acid in PFI and BFI is opposite to that in PAI. This goal can be achieved either by reverse head-to-tail orientation or by positioning of the fatty acid with the si-face toward FAD.
In summary, the structures of PAI and the PAI–CLA and PAI–LnA complexes reveal three determinants for regiospecificity and stereospecificity of PUFA isomerases: (i) the distance between the Arg–Phe lock and the N5 atom of FAD, (ii) the orientation of the fatty acid in the binding pocket of the enzyme, and (iii) the position of a stabilizing aromatic residue relative to the intermediate carbocation. As a long-term goal, site-directed mutagenesis of Arg-88 and Phe-193 in PAI to small, hydrophobic residues should therefore increase the enzyme affinity toward lipids rather than fatty acids and create lipid-specific isomerases with desired specificity.
Materials and Methods
Protein Production and Purification.
PAI was cloned in pGEX 6-1, overproduced in Escherichia coli BL21(DE3), and purified at 4°C as a GST-fusion containing a PreScission protease site as described in ref. 7. Briefly, cells were disrupted by sonication in 10 mM Tris·HCl (pH 8.0)/150 mM NaCl/1 mM EDTA/2 mM DTT/1.5% N-laurylsarcosine. The supernatant was adjusted to 2% Triton X-100 and GST–PAI was bound to glutathione (GSH) Sepharose, washed with 100 mM Tris·HCl (pH 7.5)/150 mM NaCl and eluted with 100 mM Tris·HCl (pH 7.5)/150 mM NaCl/1 mM EDTA/2 mM DTT/0.01% Triton X-100/20 mM GSH. The fusion protein was cleaved with PreScission protease during dialysis against 50 mM Tris·HCl (pH 7.5)/30 mM NaCl/1 mM EDTA/2 mM DTT. Anion exchange chromatography on Source 30Q (Amersham Pharmacia) equilibrated in the above buffer was used to separate PAI from GST. PAI was concentrated to 10 mg/ml in 20 mM Hepes/NaOH (pH 7.5) by ultrafiltration (Amicon). Protein concentration was estimated by absorption using a calculated ε280 of 74,500 M−1·cm−1.
Crystallization, Data Collection, Structure Determination, and Refinement.
Cubic-shaped crystals of PAI were grown at 10°C by sitting-drop vapor diffusion from Tris·HCl (pH 8.5)/2 M Li2SO4 and either 2% PEG 400 or 2% 1,4-butanediol. Plate-like crystals grew from 0.1 M Mes/NaOH (pH 6.0)/2 M (NH4)2SO4/5% PEG 400. The cubic crystals were directly cryocooled in liquid nitrogen while plates were cryoprotected in mother liquor supplemented with 12.5% PEG 400. Crystals of the PAI–CLA complex were obtained by microseeding using cubic seeds obtained from mother liquor containing 1,4-butanediol to avoid competition with PEG 400 and crystallization solutions containing 1 mM LA. Crystals of the PAI–CLnA complex were grown from 0.1 M Hepes·NaOH (pH 7.5)/2 M (NH4)2SO4/0.8% 2-methyl-2,4-pentanediol. The plate-like crystals were used for seeding in presence of 1 mM LnA. The fatty acid content of PAI–CLA and PAI–CLnA crystals was analyzed by GC and GC-MS as described in ref. 7. Crystals were washed five times with 3 μl of mother liquor, resuspended in 50 μl of H2O, and extracted with 100 μl of CHCl3:CH3OH (1:1). The lower phase was dried and used for analysis. Data were collected in-house at 100 K on a MAR345 image plate detector-mounted on a MicroMax 007 generator and reduced with the hkl programs (HKL Research, Charlottesville, VA) (Table 1). The space group of the cubic crystals is I213 with one molecule per asymmetric unit, Matthews coefficient VM = 3.7, and solvent content 65%. The plate-like crystals are of space group C2 with one molecule in the asymmetric unit, Matthews coefficient VM = 2.8, and solvent content 55%. The structure was determined by single isomorphous replacement with anomalous scattering of an iodide-soaked (mother liquor plus 0.25 M KI and 12.5% PEG 400) cubic crystal against data set native 1 (Table 1). A 13-atom substructure was found by shelxd (21) at 3-Å resolution. Phase extension to 1.86-Å resolution and density modification was carried out in shelxe (22). Automated model building with arp/warp (23) placed 417 of 426 residues. coot (24) was used for manual model building. Refinement was performed with refmac5 (25) with the same set of 5% of reflections reserved for Rfree cross-validation (26) against data truncated at 1.95-Å resolution. All other PAI structures were determined by molecular replacement using the first PAI structure as the starting model. The quality of the models was assessed with procheck (27) and whatcheck (28). Data collection and refinement statistics are summarized in Table 1. Possible H-bonds, salt bridges, and van der Waals contacts were detected with hbplus (29) and contacsym (30) using default parameters. Secondary structure was assigned with stride (31). Structure figures were created with bobscript (32) and rendered with raster3d (33) or pymol (www.pymol.org).
Supplementary Material
Acknowledgments
We thank the staff at the Deutsches Elektronen Synchrotron and Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung for beamtime and guidance. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to M.G.R.). A.L. is supported by International Max Planck Research School Molecular Biology (Göttingen).
Abbreviations
- LA
linoleic acid
- CLA
conjugated LA
- LnA
α-linolenic acid
- CLnA
conjugated LnA
- PUFA
polyunsaturated fatty acid
- BFI
Butyrivibrio fibrisolvens polyenoic fatty acid isomerase
- PAI
Propionibacterium acnes PUFA isomerase
- PFI
Ptilota filicina polyenoic fatty acid isomerase
- PDB
Protein Data Bank
- PEG
polyethylene glycol
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2B9W, 2B9X, 2BA9, 2B9Y, 2BAB, and 2BAC).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wahle K. W., Heys S. D., Rotondo D. Prog. Lipid Res. 2004;43:553–587. doi: 10.1016/j.plipres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Pariza M. W., Park Y., Cook M. E. Proc. Soc. Exp. Biol. Med; 2000. pp. 8–13. [DOI] [PubMed] [Google Scholar]
- 3.Fritsche J., Rickert R., Steinhart H., Yurawecz M. P., Mossoba M. M., Sehat N., Roach J. A. G., Kramer J. K. G., Ku Y. Fett Lipid. 1999;101:272–276. [Google Scholar]
- 4.Kepler C. R., Tove S. B. J. Biol. Chem. 1967;242:5686–5692. [PubMed] [Google Scholar]
- 5.Wise M. L., Hamberg M., Gerwick W. H. Biochemistry. 1994;33:15223–15232. doi: 10.1021/bi00255a002. [DOI] [PubMed] [Google Scholar]
- 6.Conacher H. B. S., Gunstone F. D. Chem. Phys. Lipids. 1969;3:203–220. doi: 10.1016/0009-3084(69)90013-9. [DOI] [PubMed] [Google Scholar]
- 7.Hornung E., Krueger C., Pernstich C., Gipmans M., Porzel A., Feussner I. Biochem. Biophys. Acta. 2005;1738:105–114. doi: 10.1016/j.bbalip.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Kepler C. R., Tucker W. P., Tove S. B. J. Biol. Chem. 1970;245:3612–3620. [PubMed] [Google Scholar]
- 9.Kepler C. R., Tucker W. P., Tove S. B. J. Biol. Chem. 1971;246:2765–2771. [PubMed] [Google Scholar]
- 10.Zheng W., Wise M. L., Wyrick A., Metz J. G., Yuan L., Gerwick W. H. Arch. Biochem. Biophys. 2002;401:11–20. doi: 10.1016/S0003-9861(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 11.Binda C., Angelini R., Federico R., Ascenzi P., Mattevi A. Biochemistry. 2001;40:2766–2776. doi: 10.1021/bi002751j. [DOI] [PubMed] [Google Scholar]
- 12.Sanders D. A., Staines A. G., McMahon S. A., McNeil M. R., Whitfield C., Naismith J. H. Nat. Struct. Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 13.Dym O., Eisenberg D. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schalk I., Zeng K., Wu S. K., Stura E. A., Matteson J., Huang M., Tandon A., Wilson I. A., Balch W. E. Nature. 1996;381:42–48. doi: 10.1038/381042a0. [DOI] [PubMed] [Google Scholar]
- 15.Fraaije M. W., Mattevi A. Trends Biochem. Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 16.Binda C., Coda A., Angelini R., Federico R., Ascenzi P., Mattevi A. Struct. Fold Design. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim J. J., Wang M., Paschke R. Proc. Natl. Acad. Sci. USA. 1993;90:7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghisla S., Massey V. Eur. J. Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang Z., Zhang Q., Liu H. W. Bioorg. Chem. 2003;31:494–502. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Malkowski M. G., Thuresson E. D., Lakkides K. M., Rieke C. J., Micielli R., Smith W. L., Garavito R. M. J. Biol. Chem. 2001;276:37547–37555. doi: 10.1074/jbc.M105982200. [DOI] [PubMed] [Google Scholar]
- 21.Schneider T. R., Sheldrick G. M. Acta Crystallogr. D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 22.Sheldrick G. M. Z. Kristallogr. 2002;217:644–650. [Google Scholar]
- 23.Lamzin V. S., Wilson K. S. Acta Crystallogr. D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.CCP4 Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Brünger A. T. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 28.Hooft R. W., Vriend G., Sander C., Abola E. E. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 29.McDonald I. K., Thornton J. M. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 30.Sheriff S., Hendrickson W. A., Smith J. L. J. Mol. Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 31.Frishman D., Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 32.Esnouf R. M. J. Mol. Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 33.Merritt E. A., Murphy M. E. P. Acta Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.