Abstract
The herpes simplex virus 1 ORF UL41 encodes a protein (virion host shutoff or vhs) associated with selective degradation of mRNA early in infection. Some mRNAs, exemplified by GAPDH or β-actin mRNAs, are degraded rapidly. Others, for example IEX-1 mRNA, are degraded in two stages: whereas the 3′ domain disappears rapidly, a large 5′ domain fragment of the mRNA lingers for several hours. Still a third, exemplified by tristetraprolin mRNA, is not degraded, allowing its protein product to accumulate in infected cells. Here we report the following: (i) a GST-vhs protein produced in Escherichia coli, solubilized and purified to homogeneity acts as bona fide endoribonuclease when tested on in vitro transcribed IEX-1 probes. A GST-vhs protein in which three key vhs amino acids were replaced with alanines, solubilized and purified by the same protocol, had no enzymatic activity. (ii) The number of fragments generated by cleavage of a truncated IEX-1 RNA by vhs appears to be small; the cleavage sites are centered at or near the AU-rich elements located at the 3′ untranslated region of the mRNA. A truncated RNA containing only the IEX-1 coding domain was cleaved numerous times. (iii) In cells infected at high multiplicity and exposed to a large number of particles per cell, the vhs protein accumulated within 3 h after infection, in small uniform cytoplasmic granules raising the possibility that vhs colocalizes with tristerapolin, a protein induced after infection, in structures involved in degradation of RNA.
Keywords: endoribonuclease, mRNA, virion host shutoff
One of the key early events in the replication of herpes simplex virus 1 (HSV-1) is the shutoff of host macromolecular metabolism (1). The shutoff of protein synthesis was mapped to a gene designated UL41, and the protein product was designated virion host shutoff or vhs (2, 3). During productive infection, copies of vhs protein, which enter the cell as components of the virion tegument, cause the degradation of preexisting and newly transcribed mRNAs during the first few hours after infection (2–4). After the onset of viral transcription, vhs also accelerates the turnover of viral mRNAs, facilitating the sequential expression of different classes of viral genes (3, 5, 6). At later times after infection, newly made vhs is sequestered and rendered inactive by another viral protein, VP16 also known as α-trans inducing factor, the product of UL48 (7). The vast literature of the past decade, reviewed in ref. 8, speaks eloquently of the importance of vhs in the biology of HSV. Some of the key studies, relevant to this report, are as follows: (i) vhs degrades mRNA in the absence of other viral proteins, as shown by inhibition of reporter gene expression in mammalian cells transiently cotransfected with a vhs expression vector (9, 10). (ii) vhs, along with homologues from other alphaherpesviruses, shares amino acid sequence similarity with a larger family of human, yeast, bacterial and phage nucleases and mutations of highly conserved residues, known to be essential for the catalytic activity of cellular nucleases, totally abolish the ability of vhs to inhibit the expression of a reporter gene (11, 12). (iii) Extracts of detergent-solubilized virions contain a vhs-dependent nuclease activity which is found in wild-type but not in vhs-mutant virions and can be specifically blocked by anti-vhs antisera (13). (iv) vhs induces endonucleolytic cleavage of a number of RNA substrates when it is expressed as the only viral protein in the rabbit reticulocyte lysate in vitro translation system (14, 15). (v) Although vhs does not discriminate between cellular and viral mRNAs, it exhibits a strong preference for mRNAs, as opposed to rRNA and tRNA either in vivo (6) or in in vitro decay reactions containing cytoplasmic extracts from infected cells (16) and/or in vitro translated vhs (13). (vi) It has been reported that the cleavage of the mRNA does not occur at random sites but it appears to initiate near regions of translation initiation; sequences at the 5′ termini are degraded before those near the 3′ ends of the transcripts (4, 17). (vii) vhs interacts with the cellular translation initiation factor eIF4H, and the related factor eIF4B, in the yeast two-hybrid system and in mammalian cells. This interaction has been proposed as a mechanism for targeting vhs to the regions of translation initiation of mRNAs (12, 18).
Although we do not contest the conclusion that the vhs-dependent degradation of some mRNAs initiates at or near the 5′ terminus, studies carried out in our laboratory do not support the concept that vhs mediates indiscriminate degradation of mRNAs or that the degradation invariably proceeds 5′ to 3′ (19). Examination of the relative rates of degradation of several cellular mRNAs upon HSV-1 infection led to the conclusion that cellular RNAs form at least three classes based on their fate after infection. There are (i) constitutively expressed mRNAs (i.e., GAPDH, β-actin) that were rapidly degraded (20); (ii) some inducible mRNAs containing AU-rich elements (AREs) in their 3′ UTR (i.e., stress-inducible immediate-early responsive gene or IEX-1, IκBα, c-fos) that were deadenylated, subjected to endonucleolytic cleavage near the 3′ end and to 3′-to-5′ degradation, with the 5′-truncated mRNA fragments persisting in wild-type virus-infected cells for many hours (20, 21), and (iii) some inducible mRNAs, exemplified by GADD45β and tristetraprolin, which showed no evidence of degradation and whose proteins readily accumulated in infected cells (19, 22). All of these processes require vhs because none of them was detected in cells infected with the ΔUL41-mutant virus.
Taken together, the foregoing results suggest that vhs is an endoribonuclease or a component of a ribonuclease complex. However, the evidence that vhs interacts with cellular proteins involved with translation of mRNA coupled with the absence of studies based on vhs purified to homogeneity precludes a definitive conclusion as to the function of this protein. Here we report that a soluble GST-tagged full-length vhs that was purified to homogeneity exhibits endoribonuclease activity in the absence of any other cellular or viral proteins.
Results
Expression of a Full-Length Soluble GST-vhs Fusion Protein and Generation of Anti-vhs Antiserum.
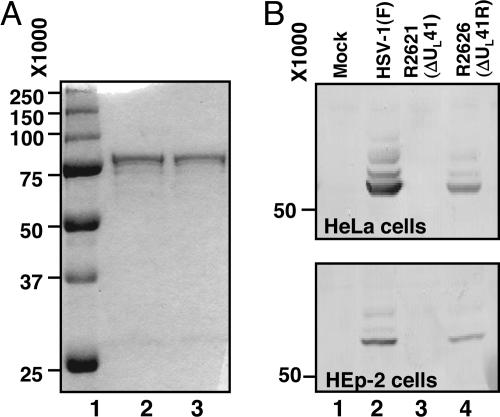
Vhs is an insoluble or, at best, poorly soluble protein. We were able to successfully purify a GST-vhs fusion protein from E. coli by customizing a general purification protocol published elsewhere (23). The procedure described in Materials and Methods was derived by testing different Sarkosyl concentrations for solubilization of fusion proteins, different amounts of the glutathione Sepharose (GS) beads and Triton X-100 for efficient binding, and different elution buffers for the best protein recovery. The fusion protein eluted from GS beads with a buffer containing a low concentration of SDS appeared as a single band of the expected size (≈75 kDa) in the comassie-blue stained gel (Fig. 1A, lane 2). The protein remained soluble even after dialysis against a buffer lacking SDS (Fig. 1A, lane 3). The purified GST-vhs fusion protein was then used to raise an antiserum in rabbits. The antiserum was tested in Western blots for specificity. As expected and shown in Fig. 1B, the antiserum cross-reacted with a single polypeptide of the expected size in HeLa or HEp-2 cells infected with either wild-type [HSV-1(F)] or ΔUL41R repaired viruses (R2626) (lanes 2 and 4, respectively), but not in mock-infected cells or cells infected with a ΔUL41 mutant virus (R2621) (lanes 1 and 3, respectively).
Fig. 1.
Expression of recombinant GST-vhs fusion protein and specificity of anti-vhs antiserum. (A) Coomassie blue-stained gel of GST-vhs fusion protein bound to GS and eluted with buffer containing 75 mM Hepes (pH 7.4), 150 mM NaCl, 5 mM DTT, and 0.08% SDS before (lane 2) and after (lane 3) dialysis against buffer lacking SDS. The molecular weight marker is also shown (lane 1). (B) HeLa and Hep2 cells were either mock infected (lane 1) or infected with 10 PFU/cell of HSV-1(F) (lane 2), ΔUL41 mutant virus (lane 3) or ΔUL41R repaired virus (lane 4). Cells were harvested 18 h after infection and subjected to electrophoretic separation in denaturing gels and immunoblotting with the anti-vhs rabbit antiserum.
Purified GST-vhs Fusion Protein Exhibits RNase Activity.
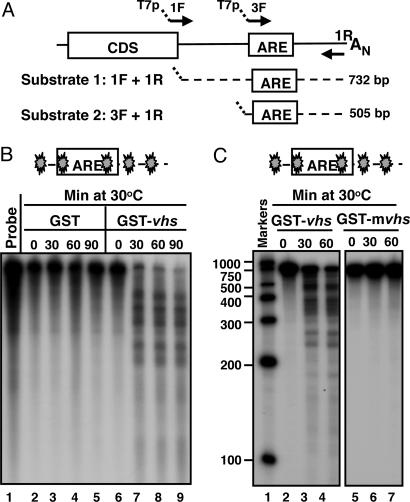
The objective of the experiments described in this section was to test whether the purified GST-vhs fusion protein exhibited RNase activity in an in vitro assay. The entire 3′UTR of human IEX-1 mRNA was selected at first as RNA substrate (substrate 1; Fig. 2A). In the experiments described here, the 32P-internally labeled substrate 1 was generated by in vitro transcription as described in Materials and Methods. Equal amounts of GST-vhs fusion protein or GST alone were incubated at 30°C with the reporter RNA in the presence of Mg2+. Samples were withdrawn at various times and analyzed by denaturing polyacrylamide gel electrophoresis followed by autoradiography. The results, shown in Fig. 2B, were as follows: the RNA substrate mixed with GST protein alone remained stable through the 90 min incubation time (lanes 2–5), whereas the RNA reacted with the GST-vhs fusion protein was cleaved into several fragments of discrete size (lanes 7–9). The fragments appeared at the earliest time point tested (lane 7) and were not further degraded during the reaction time (lanes 8 and 9), suggesting that the degradation products were generated by endonucleolytic cleavage. To verify that the observed RNase activity was specifically driven by vhs and not by a contaminating bacterial nuclease co-purified with GST, a GST-vhs mutant protein (GST-mvhs), bearing substitutions of three amino acid residues (E192, D194, D195), previously reported to play a critical role in vhs nuclease activity (12), was generated and purified by the same procedure as the GST-vhs protein. As shown in Fig. 2C, no degradation products were detected when the RNA substrate was incubated with the mutant vhs (lane 5–7). In addition, the GST-vhs nuclease activity could be totally blocked by addition of an RNase inhibitor (RNasin Plus) to the reaction mixture (data not shown). These results indicate that a biologically active form of GST-vhs protein has been successfully purified from bacteria and that vhs functions as a ribonuclease in the absence of other viral or cellular proteins. Furthermore, the pattern of RNA degradation indicates that vhs is indeed an endoribonuclease.
Fig. 2.
GST-vhs fusion protein is biologically active. (A) Schematic representation of the full-length IEX-1 mRNA and location of the primers used to generate the DNA templates for in vitro transcription of substrates 1 and 2. (B) Five micrograms of either GST (lanes 2–5) or GST-vhs fusion protein (lanes 6–9) were incubated at 30°C with 32P internally labeled IEX-1 3′ UTR RNA (substrate 1). Samples were removed at the indicated times, purified, and resolved on a sequencing gel, followed by autoradiography as described in Materials and Methods. Free probe (lane 1) was extracted and analyzed as well. (C) Five micrograms of either GST-vhs wild-type protein (lanes 2–4) or GST-mvhs mutant protein, bearing alanine substitutions of E192, D194, and D195 (lanes 5–7) were incubated at 30°C with 32P internally labeled substrate 1 for time intervals shown and RNA samples were analyzed as in A. Internally labeled RNA marker was loaded in lane 1, and the length of the fragments is reported on the side. The filled symbol marks the site of labeling of the RNA.
The Locations of vhs Cleavage Sites in Truncated Transcripts Containing AREs.
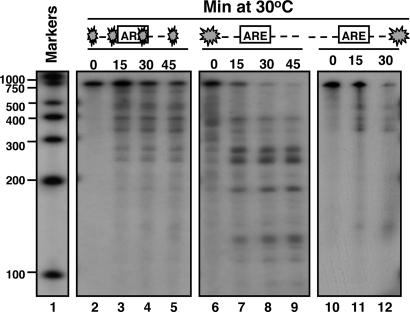
The experiments described above were carried out with IEX-1 3′UTR substrate internally labeled with 32P. Even though the size of the fragments could provide some hints about the location of the cleavage sites, results of analyses based on terminally labeled mRNA would be more persuasive. To this end, IEX-1 3′UTR substrates labeled at the 5′ or 3′ end were generated as described in Materials and Methods. The substrates were then incubated at 30°C with 5 μg of purified GST-vhs fusion protein for 0, 15, 30, and 45 min. After incubation, the RNA samples were purified, loaded on a sequencing gel, and exposed to x-ray film. The results shown in Fig. 3 were as follows: whereas the internally labeled IEX-1 3′UTR substrate (lanes 3–5) was cleaved into several fragments ranging from 500 to 200 nt, the majority of the fragments produced by digestion of the 5′ end-labeled vhs substrate were of shorter size, ranging from 300 to 100 nt (lanes 7–9). This pattern of degradation indicates that the cleavage sites were most likely located closer to the 5′ end of the IEX-1 3′ UTR substrate. If the prediction were correct, it would be expected that incubation of GST-vhs with the same substrate, but labeled at the 3′ end, should result in the accumulation of only longer fragments, ranging from 700 to 400 nt. This was indeed the case (lanes 11 and 12).
Fig. 3.
vhs cleavage sites in truncated transcripts containing AREs. Substrate 1 was either 32P internally labeled (lanes 2–5), or labeled at the 5′end (lanes 6–9) or the 3′end (lanes 10–12). The substrates were incubated at 30°C with 5 μg of purified GST-vhs for the indicated times. Samples were purified and resolved on a sequencing gel, followed by autoradiography as described in Materials and Methods. Internally labeled RNA marker was loaded in lane 1, and the length of the fragments is reported on the side.
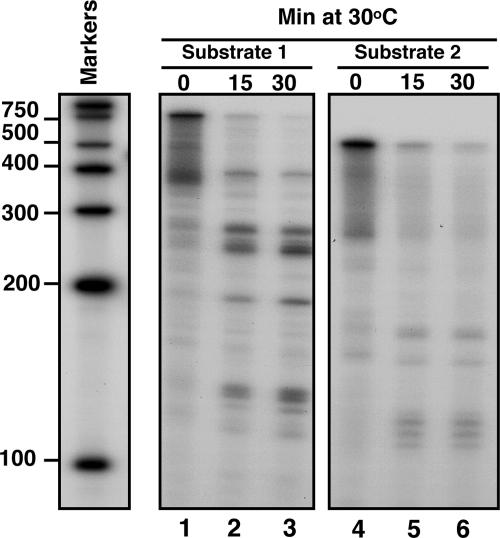
To verify our findings, we generated another substrate (substrate 2), lacking the region upstream of the AREs of IEX-1 3′-UTR (Fig. 2A). The 5′ end-labeled substrate 2 was then incubated with the purified GST-vhs and analyzed as before along with the 5′end-labeled substrate 1. The prediction was that removing the region upstream of the AREs would result in the disappearance of the longer fragments (ranging from 300 to 200 nt) generated by the cleavages occurring inside the AREs. As predicted and shown in Fig. 4, only few short fragments appeared after digestion of substrate 2 with GST-vhs (lanes 5 and 6).
Fig. 4.
vhs cleaves truncated transcripts containing AREs at preferential sites. Substrates 1 (lanes 1–3) and 2 (lanes 4–6) were labeled with γ32P at the 5′end and incubated with 5 μg of purified GST-vhs protein at 30°C for the indicated times. Samples were purified and resolved on a sequencing gel, followed by autoradiography as described in Materials and Methods. Internally labeled RNA marker was also loaded, and the length of the fragments is reported on the side.
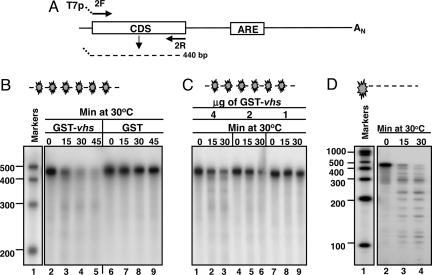
The IEX-1 Coding Domain RNA Substrate Is Cleaved by GST-vhs at Multiple Sites.
The results described above were in agreement with the pattern of degradation of IEX-1 mRNA observed in HSV-1-infected cells and specifically with the lingering of truncated 5′ domains, generated by vhs-dependent endonucleolytic cleavage within the AREs (19, 21). The objective of the experiments described in this section was to test whether a substrate, consisting of only IEX-1 coding sequence domain (CDS, substrate 3), was cleaved by the purified GST-vhs fusion protein. A schematic diagram of substrate 3 is shown in Fig. 5A. The internally labeled substrate 3 was incubated with 5 μg of either purified GST-vhs fusion protein or GST protein alone and analyzed by denaturing polyacrylamide gel electrophoresis followed by autoradiography. The results, shown in Fig. 5B were as follows: as expected, the internally labeled substrate 3 mixed with GST protein alone was not degraded (lanes 6 to 9), whereas the sample incubated with GST-vhs fusion protein was largely degraded as early as 15 min after incubation at 30°C (lanes 3–5). In addition, the GST-vhs nuclease activity was dose-dependent (Fig. 5C) inasmuch as it decreased with the progressive reduction of GST-vhs protein concentration in the reaction mixture. The unexpected total degradation of IEX-1 coding domain substrate suggested that vhs-dependent cleavage occurred at multiple sites. Indeed, incubation of 5′ end-labeled substrate 3 with GST-vhs protein generated several fragments ranging in size between 400 and 100 nt (Fig. 5D, lanes 3 and 4), validating our prediction.
Fig. 5.
vhs cleaves truncated transcripts containing the IEX-1 coding domains at multiple sites. (A) Schematic representation of the full-length IEX-1 mRNA and location of the primers used to generate the DNA template for in vitro transcription of IEX-1 coding domain (substrate 3). (B) Five micrograms of either GST-vhs fusion protein (lanes 2–5) or GST (lanes 6–9) were incubated at 30°C with internally labeled substrate 3. Samples were removed at the indicated times, purified, and resolved on a sequencing gel, followed by autoradiography as described in Materials and Methods. Internally labeled RNA marker was loaded in lane 1, and the length of the fragments is reported on the side. (C) Internally labeled substrate 3 was incubated with different amounts of GST-vhs fusion protein: 4 μg (lanes 1–3), 2 μg (lanes 4–6), and 1 μg (lanes 7–9). Samples were analyzed as in B. (D) 5′ end-labeled substrate 3 was incubated with 5 μg of purified GST-vhs fusion protein at 30°C for the indicated times. Samples were analyzed as in B and C.
Localization of vhs in Cellular Compartments During Infection.
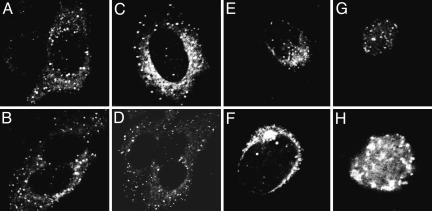
Studies on localization of vhs during infection yielded mixed results. One study reported that vhs accumulates in nuclei after infection (24). Another reported that vhs accumulates in the cytoplasm but at times when newly made vhs would be expected to be made (25). To resolve the conflicting results, two series of experiments were carried out. In the first, HSV-1(F) virions purified in connection with other studies (26) were used to expose SK-N-SH cells grown in four-well slides at a ratio of 100 plaque-forming units (PFU) per cell. At 3 h after infection, the cells were reacted with primary (rabbit polyclonal anti-vhs) and secondary (anti-rabbit Ig conjugated with fluorescein isothiocyanate) antibodies and examined with a Zeiss confocal microscope as described in Materials and Methods. As shown in Fig. 6A–D, vhs carried into cells with the virus accumulated in small punctuate granules dispersed in the cytoplasm.
Fig. 6.
vhs introduced into cells during infection and newly made vhs accumulate in the cytoplasm and perinuclear region of infected cells. (A–D) Representative SK-N-SH cells exposed to 100 PFU of purified HSV-1(F) and fixed and stained with primary anti-vhs and secondary anti-rabbit conjugated antibody at 3 h after infection. (E–H) HEp-2 cells fixed 16 h after exposure to 10 PFU of HSV-1(F) and reacted with either anti-vhs (E and F) or anti-VP22 antibodies (G and H). The cultures were reacted with secondary antibody conjugated with fluorescein isothiocyanate as described in Materials and Methods.
In the second series of experiments, HEp-2 cells were exposed to 10 PFU of HSV-1 per cell. The cultures were fixed at 5, 9, or 16 h after infection and reacted with either rabbit anti- vhs or rabbit anti-UL49 (or VP22) antibodies. At all times vhs was found exclusively in the cytoplasm and perinuclear region. In contrast, VP22 was found in nuclei. Fig. 6 E–H shows representative cells fixed at 16 h and stained with either anti-vhs (Fig. 6 E and F) or anti-VP22 (Fig. 6 G and H) antibodies.
Discussion
We were able to successfully purify a GST-tagged full-length vhs in a soluble form from bacteria by determining the conditions required for solubilization, binding to GS beads and elution of the chimeric protein. The purified GST-vhs protein exhibits RNase activity in an in vitro assay. The nuclease activity was specifically caused by vhs inasmuch as neither GST nor the fusion protein GST-mvhs bearing substitutions of three residues in vhs (12) and purified by the same protocol exhibited exhibit RNase activity when tested in parallel. The specific pattern of degradation of the RNA substrates into fragments of discrete sizes is consistent with the hypothesis that vhs acts as an endoribonuclease. Moreover, the vhs nuclease activity is dose-dependent and blocked by RNase inhibitor. Our results thus provide unambiguous evidence that vhs is itself a nuclease in the absence of any other cellular or viral proteins. This conclusion does not obviate the possibility that vhs forms complexes with either viral or cellular proteins to express additional or alternative functions. One interesting property of the proteins encoded by HSV-1 is that they tend to perform multiple, sometimes unrelated functions (27).
The activity of purified GST-vhs protein was tested in vitro on a single substrate, IEX-1 mRNA. The choice of this specific substrate was based on earlier studies showing that, in infected cells, IEX-1 mRNA was transcribed but that the protein was not made (21). Extensive analyses showed that IEX-1 mRNA was degraded in a unique, vhs-dependent manner (19, 21). Specifically, the mRNA was deadenylated and subjected to 3′-to-5′ degradation and the truncated 5′ mRNA fragments tended to persist in the cytoplasm of wild-type virus infected cells for many hours. A more accurate analysis (19) revealed that the truncated 5′ mRNA fragments were generated by endonucleolytic cleavage within or near the AREs located in the 3′ UTR of IEX-1 mRNA.
In our in vitro assays, we generated three different substrates, representing the entire 3′ UTR (substrate 1), the region downstream of the AREs (substrate 2), and the coding domain (substrate 3) of IEX-1 mRNA. The results obtained with substrates 1 and 2 are consistent with the analyses of the fate of the IEX-1 mRNA in infected cells. Specifically, incubation of 5′end-labeled substrates with GST-vhs generates a relatively small number of fragments whose sizes point to a location of the cleavage sites upstream or within the AREs. Preliminary analyses of the sequences surrounding the potential cleavage sites did not provide a clear idea whether vhs recognizes a specific sequence or secondary structure. Analyses of the digests of GST-vhs protein with substrate 3, representing only the IEX-1 coding domain indicated that vhs cleaved this transcript at multiple locations without exhibiting preferences for specific sites. One hypothesis that remains to be tested is that vhs recognizes specific secondary structures that are formed by full-length IEX-1 mRNA inside the cells or by the 3′UTR fragment in vitro, but not by the coding domain transcribed in vitro.
Lastly, mRNA is degraded in two known structures, in exosomes and stress granules (SGs). SGs are cytoplasmic microdomains to which poly(A)+ mRNAs are recruited by TIA-1 (T cell internal antigen 1) or TIA-1 related protein (TIAR) and which serve as a site of triage for poly(A)+ RNA during stress responses. The SG-associated RNAs are in a dynamic equilibrium with polyribosomes (28, 29). In earlier studies, we showed that TIA-1/TIAR are activated and translocated to the cytoplasm of HSV-1 infected cells but we could not demonstrate the formation of stress granules (22). Exosomes consist of complexes of ribonucleases to which ARE mRNAs are recruited by tristetraprolin (TTP) for degradation and earlier studies have shown that TTP is activated and accumulates in the cytoplasm in infected cells (19, 22). The nature of the granules to which vhs localizes early in infection are not known. They could be modified exosome structures or SGs to which TTP could also be recruited.
Materials and Methods
Expression and Purification of GST-vhs Fusion Proteins.
The full-length UL41 gene was amplified by PCR and cloned into pGEX4T-1 vector (Amersham Pharmacia), creating a translational fusion between residue 1 of vhs and the C terminus of GST. To generate the GST-mvhs mutant construct, the glutamic acid residue 192 and the aspartic acid residues 194 and 195 in the wild-type vhs were replaced with alanines with the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The plasmids encoding the vhs mutant was sequenced to verify the nucleotide changes. The plasmids encoding the GST-vhs fusion proteins or GST alone were transformed in E. coli BL21(DE3), and the expression of the proteins was induced in a log phase culture by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.2 mM. The GST-vhs fusion proteins were successful purified in a soluble form by customizing a general purification protocol previously reported (23). Briefly, the pellets of fusion protein-expressing bacteria were resuspended in ice-cold STE buffer (10 mM Tris·HCl, pH 8.0/150 mM NaCl/1 mM EDTA/100 μg/ml PMSF) and treated with lysozyme (100 μg/ml), followed by the addition of 5 mM DTT. To solubilize the fusion proteins, Sarkosyl was added to the lysozyme-treated bacteria to a final concentration of 0.35% and the mixture was sonicated at low setting in ice. The samples were then centrifuged at 10,000 × g for 25 min at 4°C and the supernatant fluids were transferred to fresh tubes. Triton X-100 was added to a final concentration of 1% along with an appropriate volume of 50% (vol/vol) slurry of GS-4B beads (Amersham Pharmacia) in PBS. The samples were then reacted at 4°C for at least 1 h with continuous agitation. The supernatant fluids were removed, and the GS bead pellets were washed five times with ice-cold PBS. Finally, the GST-vhs fusion proteins were eluted at room temperature in a buffer containing 75 mM Hepes (pH 7.4), 150 mM NaCl, 5 mM DTT, and 0.08% SDS, dialyzed overnight against a buffer containing 20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM DTT, and 10% glycerol and stored at −80°C in small aliquots.
Production of an Anti-vhs Rabbit Serum.
Anti-vhs polyclonal rabbit serum was prepared by Jossman (Napa, CA).
Cells and Viruses.
SK-N-SH, HEp-2, and HeLa cells obtained from the American Type Culture Collection were propagated in DMEM supplemented with 5% newborn calf serum. HSV-1(F) is the prototype HSV-1 strain used in our laboratory (30). The ΔUL41 mutant virus R2621 and the virus in which the gene was restored (R2626) were reported elsewhere (31). HSV-1(F) was purified as described (26).
Immunoblots.
HeLa or HEp-2 cells were mock infected or infected with 10 PFU of the wild-type or mutant viruses per cell. The procedures for harvesting, solubilization, protein quantification, SDS/PAGE, transfer to nitrocellulose membranes and reaction with antibody to specific protein were as reported (21). The anti-vhs polyclonal rabbit serum was used at a 1:1,000 dilution.
Indirect Immunofluorescence Staining.
The studies were done in cells grown in four-well slides according to procedures reported elsewhere (22). The rabbit anti-UL49 (VP22) polyclonal antibody has been described (32).
Preparation of DNA Templates for in Vitro Transcription.
To generate the DNA templates used for the production of IEX-1 RNA substrates, the entire IEX-1–3′UTR region (substrate 1) was amplified by RT-PCR of total RNA purified from HSV-1-infected HeLa cells with Superscript III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. A specific primer complementary to the IEX-1 poly(A) site (1R: 5′-ATTTTATTTGTGTTCACAGAACATACTAGGCG-3′) was used to prime the RT reaction. The RT mixture was amplified with 1R primer along with the forward primer (1F: 5′-TAATACGACTCACTATAGGGCGCCTTCTAACTGTGACTCC-3′) encompassing the IEX-1 stop codon and containing the T7 promoter sequence embedded at the 5′ end. The 3′ end portion of the IEX-1–3′ UTR (substrate 2) was generated by PCR amplification of the substrate 1 using the following primers, 3F: 5′-TAATACGACTCACTATAGGGCGGTGGGAAGGAGAGCGTC-3′, containing the T7 promoter sequence at the 5′ end; and the 1R reverse primer. The coding domain sequence (CDS) of IEX-1 mRNA (substrate 3) was amplified from the previously described plasmid IEX-1-pRB5850 (21) with the following primers: 2F: 5′-TAATA CGACTCACTATAGGGATGACCATCCTGCAGGCCC-3′, encompassing the IEX-1 start codon and containing the T7 promoter sequence embedded at the 5′ end, in pair with IEX-1-reverse (21) or 2R. The resulting fragments of 732, 505, and 440 bp, respectively, were purified from agarose gels by using the QIAquick Gel extraction kit (Qiagen) and quantified by OD reading.
Preparation of Unlabeled and Labeled RNA Substrates.
Unlabeled and [α-32P]UTP internally labeled RNA substrates were generated by in vitro transcription of PCR-amplified DNA templates with the aid of the MAXIscript kit (Ambion, Austin, TX), according to the manufacturer’s instructions. 5′ end-labeled reporter RNAs were generated by incubation of unlabeled transcripts with T4 polynucleotide kinase in presence of [γ-32P]ATP according to the protocol supplied with the KinaseMax 5′ end-labeling kit (Ambion). 3′ end-labeled reporter RNAs were generated by reacting unlabeled transcripts with T4 RNA ligase (Ambion) in presence of [32P]pCp (cytidine-3′,5′-bis-phosphate) and ATP at 4°C for 72 h. All labeled RNA substrates were purified with the aid of NucAway spin columns (Ambion), according to the company’s instructions, and counted in a Beckman LS8000 scintillation counter. For the generation of [α-32P]UTP internally labeled RNA markers, RNA Century marker plus template (Ambion) was transcribed in vitro as suggested by the manufacturer.
vhs in Vitro Assays.
Five micrograms (10 μl) of either GST-vhs or GST protein preparations were incubated at 30°C with 5 × 105 cpm of the specific substrate in a 50-μl reaction mixture containing 25 mM Tris·HCl (pH 8), 80 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM DTT, and 0.1 mM EDTA. Samples (10 μl), removed at various times, were immediately added to 400 μl of HSCB buffer (25 mM Tris·HCl, pH 7.6/400 mM NaCl/0.1% SDS) to stop the reaction. As control, samples were also collected immediately after the reaction mixture was added to the purified proteins (time 0) to monitor the degradation status of the substrates before the incubation at 30°C. The samples were extracted once with phenol-chloroform-isoamylic alcohol (pH 7.9; Ambion) and precipitated with three volumes of ethanol plus 1/100 volume of GlycoBlue (Ambion) as coprecipitant. The RNA pellets were resuspended in 10 μl of Gel loading buffer II (Ambion) and loaded onto a denaturing polyacrylamide gel (5% polyacrylamide/8 M urea/1× TBE). The gel were transferred to filter paper and exposed to x-ray film for signal detection.
Acknowledgments
These studies were aided by National Cancer Institute Grants CA87661, CA83939, CA71933, CA78766, and CA88860.
Abbreviations
- HSV-1
herpes simplex virus 1
- ARE
AU-rich element
- GS
glutathione Sepharose
- PFU
plaque-forming units.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Roizman B., Borman G. S., Kamali-Rousta M. Nature. 1965;206:1374–1375. doi: 10.1038/2061374a0. [DOI] [PubMed] [Google Scholar]
- 2.Kwong A. D., Frenkel N. Proc. Natl. Acad. Sci. USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strom T., Frenkel N. J. Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karr B. M., Read G. S. Virology. 1999;264:195–204. doi: 10.1006/viro.1999.9986. [DOI] [PubMed] [Google Scholar]
- 5.Kwong A. D., Kruper J. A., Frenkel N. J. Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oroskar A. A., Read G. S. J. Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smibert C. A., Popova B., Xiao P., Capone J. P., Smiley J. R. J. Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smiley J. R. J. Virol. 2004;78:1063–1068. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones F. E., Smibert C. A., Smiley J. R. J. Virol. 1995;69:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak A. S., Everly D. N., Knight K., Read G. S. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 11.Everly D. N., Jr., Read G. S. J. Virol. 1999;73:9117–9129. doi: 10.1128/jvi.73.11.9117-9129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everly D. N., Jr., Feng P., Mian I. S., Read G. S. J. Virol. 2002;76:8560–8571. doi: 10.1128/JVI.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zelus B. D., Stewart R. S., Ross J. J. Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elgadi M. M., Hayes C. E., Smiley J. R. J. Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elgadi M. M., Smiley J .R. J. Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krikorian C. R., Read G. S. J. Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Parada J., Saffran H. A., Smiley J. R. J. Virol. 2004;78:13391–13394. doi: 10.1128/JVI.78.23.13391-13394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doepker R. C., Hsu W. L., Saffran H. A., Smiley J. R. J. Virol. 2004;78:4684–4699. doi: 10.1128/JVI.78.9.4684-4699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esclatine A., Taddeo B., Evans L., Roizman B. Proc. Natl. Acad. Sci. USA. 2004;101:3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esclatine A., Taddeo B., Roizman B. Proc. Natl. Acad. Sci. USA. 2004;101:18165–18170. doi: 10.1073/pnas.0408272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taddeo B., Esclatine A., Zhang W., Roizman B. J. Virol. 2003;77:6178–6187. doi: 10.1128/JVI.77.11.6178-6187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esclatine A., Taddeo B., Roizman B. J. Virol. 2004;78:8582–8592. doi: 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercado-Pimentel M. E., Jordan N. C., Aisemberg G. Protein Expr. Purif. 2002;26:260–265. doi: 10.1016/s1046-5928(02)00524-7. [DOI] [PubMed] [Google Scholar]
- 24.Yedowitz J. C., Kotsakis A., Schlegel E. F., Blaho J. A. J. Virol. 2005;79:4730–4743. doi: 10.1128/JVI.79.8.4730-4743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata T., Goshima F., Daikoku T., Inagaki-Ohara K., Takakuwa H., Kato K., Nishiyama Y. J. Gen. Virol. 2000;81:401–406. doi: 10.1099/0022-1317-81-2-401. [DOI] [PubMed] [Google Scholar]
- 26.Sciortino M. T., Suzuki M., Taddeo B., Roizman B. J. Virol. 2001;75:8105–8116. doi: 10.1128/JVI.75.17.8105-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B., Knipe D. M. In: Virology. Knipe D. M., Howley P. M., editors. Philadelphia: Lippincott; 2001. pp. 2399–2459. [Google Scholar]
- 28.Anderson P., Kedersha N. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 29.Kedersha N., Anderson P. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 30.Ejercito P. M., Kieff E. D., Roizman B. J. Gen. Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 31.Poon A. P., Roizman B. Virology. 1997;229:98–105. doi: 10.1006/viro.1996.8425. [DOI] [PubMed] [Google Scholar]
- 32.Blaho J. A., Mitchell C., Roizman B. J. Biol. Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]