Abstract
Cell-type-selective expression of the TFIID subunit TAFII105 (renamed TAF4b) in the ovary is essential for proper follicle development. Although a multitude of signaling pathways required for folliculogenesis have been identified, downstream transcriptional integrators of these signals remain largely unknown. Here, we show that TAF4b controls the granulosa-cell-specific expression of the proto-oncogene c-jun, and together they regulate transcription of ovary-selective promoters. Instead of using cell-type-specific activators, our findings suggest that the coactivator TAF4b regulates the expression of tissue-specific genes, at least in part, through the cell-type-specific induction of c-jun, a ubiquitous activator. Importantly, the loss of TAF4b in ovarian granulosa cells disrupts cellular morphologies and interactions during follicle growth that likely contribute to the infertility observed in TAF4b-null female mice. These data highlight a mechanism for potentiating tissue-selective functions of the basal transcription machinery and reveal intricate networks of gene expression that orchestrate ovarian-specific functions and cell morphology.
Keywords: ovary, TFIID, short interfering RNA, chromatin immunoprecipitation, granulosa cells
Specific targeting of RNA polymerase (Pol) II to heteromorphic promoters dispersed throughout the genome and packaged into chromatin requires the function of a multitude of individual proteins and multiprotein complexes that collectively direct the modulation of RNA synthesis. At the core of this machinery are the general transcription factors that were first posited to be global in nature and not directly implicated in control of cell-type-specific gene expression. Rather, cell-type-specific expression patterns were largely attributed to DNA-binding activator and repressor proteins that were thought to be expressed in a highly cell-type-specific fashion (1). The general transcription factor IID (TFIID) is a large multiprotein transcription factor that lies at the core of the basal transcription machinery. TFIID is composed of the TATA-box-binding protein (TBP) and a common subset of TBP-associated factors (TAFs) that together perform promoter-selective and coactivator functions during transcription initiation (2). Together, genetic and biochemical studies suggest a conserved and formidable role of TFIID in the execution of global programs of gene expression.
The identification of specialized, tissue-restricted components of TFIID revealed that the basal machinery has evolved additional critical roles in the execution of cell-type-specific patterns of gene expression, especially during germ-line development (3). In Drosophila, multiple testis-specific TAFs have been shown to be precisely expressed in the male germ line and absolutely required for spermatogenesis (4). In mammals, the TBP homologue TRF 2 (also called TLF) has been shown to be required for spermiogenesis during male gametogenesis (5). To examine the cell-type-specific role of TFIID in mammals, we have characterized the function of the TAF4b subunit of TFIID, which was the first identified tissue-specific TAF (6). In mice, the elevated expression level of Taf4b mRNA in the ovary and testis indicates that TAF4b may play a selective role in the regulation of gonadal-specific programs of gene expression (7). The presence of cell-type-specific components of the general transcription machinery in metazoan organisms suggests that, in addition to performing global functions in gene expression, selective components of the basal machinery may be intimately involved in promoting cell-type-specific patterns of gene expression.
Postnatal ovarian follicles are composed of intimately associated cell types including somatic granulosa cells (GCs) and germ cells that collectively contribute to the production of a functional oocyte. Folliculogenesis is a cyclic process in which cohorts of meiotically arrested oocytes resume growth while their accompanying GCs proliferate and then further differentiate in response to pituitary gonadotropins (8). Intriguingly, in the wild-type ovary, Taf4b mRNA is expressed highly in the layers of GCs that support the growing oocyte through ovulation. To elucidate such a selective role in the gonads, we have previously targeted disruption of Taf4b in mice, revealing essential functions of TAF4b in ovarian follicle development in addition to the maintenance of spermatogenesis in males (7, 9). Female TAF4b-null mice are infertile owing to severe abnormalities in folliculogenesis. Because defective follicle development in TAF4b-null females is accompanied by detrimental changes in tissue-specific gene expression, the promoters of these genes may be direct targets of TAF4b transcriptional regulation in GCs. By using mammalian folliculogenesis as a model system of coordinated cell-type specification, we have begun to characterize how complex differential programs of gene expression are achieved in mammals and how such programs function during reproduction and ovarian follicle development.
Results
Establishment of TAF4b-Overexpressing GCs.
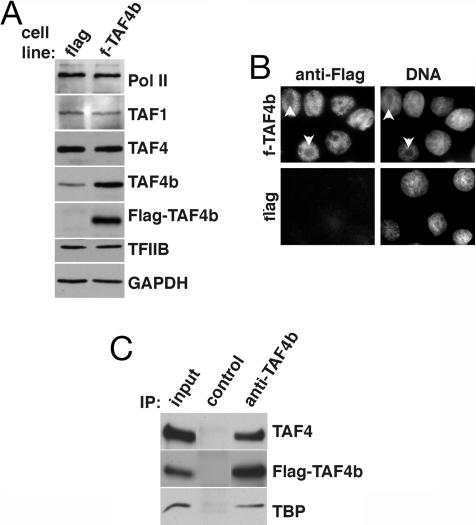
To dissect the molecular mechanisms underlying female infertility in the Taf4b−/− mice, a rat spontaneously immortalized GC (SIGC) line was used as a model system to identify TAF4b-dependent promoters and to probe additional networks of gene expression programs that are regulated by TAF4b (10). Because transcriptional activation by numerous activators can be potentiated by overexpression of individual TFIID subunits in mammalian cells (11), we established several stable SIGC lines that constitutively express the Flag-tagged TAF4b (f-TAF4b) or express the control Flag-tag alone (flag). From these stable cell lines, we further tested and selected one of each, a control flag and f-TAF4b cell line, for further analysis. Expression of Flag-TAF4b and endogenous TAF4b was monitored by Western blot analysis using anti-Flag and anti-TAF4b antibodies, respectively. SIGCs containing Flag-TAF4b express approximately three to five times more TAF4b than the control flag cell line, which had equivalent levels of the GAPDH control (Fig. 1A). Overexpression of TAF4b did not affect the levels of the 220-kDa subunit of RNA Pol II, TAF1 (formerly TAFII250), TAF4 (formerly TAFII130), or TFIIB (Fig. 1A).
Fig. 1.
Overexpression of TAF4b in GCs. (A) Immunoblotting of total cell extracts from stable f-TAF4b and flag control cell lines was performed to determine levels of TFIID subunits and general transcription factors by using anti-Pol II, anti-TAF1, anti-TAF4, anti-TFIIB, anti-Flag, and an anti-TAF4b monoclonal antibody. Anti-GAPDH antibodies were used to confirm equal loading. (B) Nuclear localization of Flag-TAF4b in SIGCs was detected by indirect immunofluorescence using an anti-Flag antibody. Nuclei were counterstained with Hoechst dye to label DNA. Arrows indicate nucleoli. (C) Partially purified nuclear extracts from f-TAF4b cells were precipitated with a control and anti-TAF4b polyclonal antibodies, followed by immunoblotting with anti-TAF4, anti-Flag, and anti-TBP antibodies.
Data from indirect immunofluorescence microscopy of SIGCs demonstrated that Flag-TAF4b was predominately detected in the nucleus, with the nucleoli (arrows) being devoid of Flag-TAF4b staining, which is expected for a protein involved in RNA Pol II transcription (Fig. 1B). The constitutive nuclear localization of TAF4b is consistent with an essential transcriptional role in regulating GC gene expression.
The composition of TFIID in the stable f-TAF4b SIGCs was investigated by specific immunoprecipitation (IP) of TAF4b from nuclear extract fractions that were partially purified over a phosphocellulose column. The presence of Flag-TAF4b, TAF4, and TBP were clearly detected in coprecipitates from the anti-TAF4b IP, but not from a control IP by Western blot analysis (Fig. 1C). Conversely, we immunoprecipitated TAF4-containing complexes from total cell extracts of stable SIGC lines and subsequently detected Flag-TAF4b in the precipitates and further confirmed the association of TAF4b with TBP and TAF1 in these cells (see Fig. 6, which is published as supporting information on the PNAS web site). The ability to coimmunoprecipitate TAF4 and TAF4b suggests that GCs contain a population of TFIID complexes or partial TAF subcomplexes containing both subunits, thus paralleling our previous findings (6).
Activation of Ovary-Specific Promoters by TAF4b.
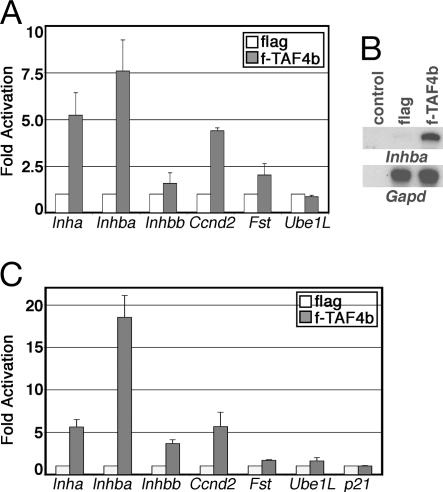
If loss of TAF4b activity contributes to the reduction of gene expression observed in the TAF4b-null mice, then overexpression of TAF4b in GCs is predicted to stimulate transcription of potential TAF4b-target genes (7). To this end, reporter constructs containing native ovary-selective promoters driving the expression of the luciferase reporter gene were transfected into the f-TAF4b and control flag cells. As shown in Fig. 2A, the activities of the inhibin-α (Inha) and -βA (Inhba) subunit promoters were significantly induced compared with the βB subunit (Inhbb) promoter. In addition, the cyclin D2 (Ccnd2) and follistatin (Fst) promoters were also induced in f-TAF4b cells compared with a control TAF4b-independent promoter, the ubiquitin-activating enzyme E1-like (Ube1L) promoter (Fig. 2A). Because these promoters did not display equivalent increases in activity, overexpression of TAF4b in GCs does not appear to induce a global increase in general transcription activity, but, instead, this tissue-specific component of TFIID mediated the transcriptional activation of a select subset of promoters. These results strongly suggest a TAF4b-selective role in activating the differential expression of ovarian-specific genes previously determined to be reduced in TAF4b-null ovaries.
Fig. 2.
Activation of tissue-selective promoters by TAF4b. (A) Luciferase assays demonstrate the activation of GC-specific promoters in Flag-TAF4b overexpressing SIGCs (gray bars) compared with flag control cells (white bars). Fold activation was obtained by comparison of relative luciferase activity measured from f-TAF4b stable cells versus control flag cells. Error bars represent SDs from at least three independent experiments. (B) The autoradiography of RPAs using RNA isolated from f-TAF4b and flag stable SIGCs and detection of the endogenous Inhba mRNA or an internal control Gapd mRNA. Control, yeast RNA. (C) Luciferase assays demonstrate a specific activation of GC-selective promoters in NIH/3T3 cells that overexpress Flag-TAF4b (gray bars) compared with flag control cells (white bars). Fold induction was measured as described in A.
We next asked whether overexpression of TAF4b could, indeed, activate the levels of an endogenous Inh gene within the context of chromosomal DNA. The levels of Inhba mRNA measured by RNase protection assays (RPAs) revealed an ≈5-fold higher level of Inhba in f-TAF4b cells compared with the control flag cell line, whereas the levels of an internal control Gapd gene were unchanged (Fig. 2B). Importantly, our data suggest that TAF4b regulates folliculogenesis, at least in part, through the transcriptional coactivation of genes that are primarily expressed in GCs and essential for follicle growth.
We also tested whether TAF4b induction of ovary-selective promoters required GCs and their corresponding putative cell-type-specific transcriptional activators, as might be expected. Nonovarian NIH/3T3 cells were stably transfected with Flag-TAF4b or the control Flag vector. The overall levels of Flag-TAF4b overexpression in NIH/3T3 cells were considerably lower than in the SIGCs (see Fig. 7, which is published as supporting information on the PNAS web site). Surprisingly, despite the apparent low levels of TAF4b expression in these nonovarian cells, TAF4b nevertheless potently induced the expression of multiple GC-specific promoters to various levels, with Inhba promoter again displaying the highest levels of activity (Fig. 2C). In contrast, control non-cell-type-specific promoters such as p21 were not induced, whereas the Fst and Ube1L promoters had barely above-background levels of activity in the f-TAF4b cell line (Fig. 2C). Taken together, these results suggest that an adaptation of TFIID by incorporation of TAF4b may be sufficient, at least in part, to reprogram the expression of a subset of cell-type-specific genes independent of cell-type-specific activators.
TAF4b Stimulates the Expression of c-jun.
In an attempt to identify additional TAF4b-target genes expressed primarily in GCs, DNA microarray analysis was used to monitor changes in the gene expression profiles of SIGC stable cell lines. Approximately 82 genes were consistently and specifically up-regulated at least 2-fold in f-TAF4b stable cells compared with the control flag cells (Table 1 and data not shown). Interestingly, the data indicated that c-jun (7.9-fold) and fra-1 (2.5-fold) are two of the potential TAF4b-responsive genes in GCs. Consistent with this result, a number of known c-Jun/activator protein 1 (AP-1) target genes were also highly up-regulated in f-TAF4b stable cells, some of which are growth factors and cytoskeletal proteins (Table 1). Real-time RT-PCR was performed to confirm a subset of the microarray results and to independently quantitate increases in mRNA levels (see Fig. 8, which is published as supporting information on the PNAS web site). This result suggests that TAF4b is involved in the transcription of a subset of GC-specific as well as non-cell-type-selective genes.
Table 1.
Microarray analysis of genes up-regulated in f-TAF4b SIGCs
| Gene title | Average fold induction |
|---|---|
| Osteopontin | 38.25 |
| Vimentin | 22.63 |
| Insulin-like growth-factor-binding protein 3 | 13.22 |
| Fibronectin | 11.08 |
| Matrix metalloproteinase-3 | 9.25 |
| c-Jun | 7.88 |
| α-Tubulin | 7.31 |
| Vascular endothelial growth factor-B | 6.38 |
| Latent transforming growth-factor-β binding protein 1 | 4.35 |
| Caldesmon-1 | 4.27 |
| Disabled-2 | 3.79 |
| Interleukin 1α | 3.28 |
| Brain-derived neurotrophic factor | 3.14 |
| Amphiregulin | 2.78 |
| RhoB | 2.76 |
| Kruppel-like factor 6 | 2.54 |
| Fos-related antigen 1 | 2.52 |
| Follistatin | 2.52 |
| Transforming growth factor-α | 2.38 |
TAF4b Stimulation of c-jun Expression Is Cell-Type-Specific.
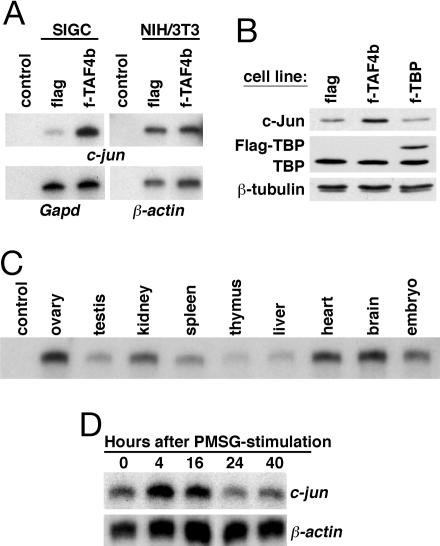
The transcription factor c-Jun is an immediate early gene product that can be rapidly induced by a myriad of extracellular stimuli independent of cell type. Therefore, we next asked whether Flag-TAF4b-dependent induction of c-jun expression was cell-type-specific. In SIGCs expressing elevated levels of Flag-TAF4b, we detect 5-fold more c-jun mRNA than in the control flag cell line, consistent with our previous DNA microarray analysis (Fig. 3A Left). Surprisingly, however, in NIH/3T3 cells, both f-TAF4b-expressing and flag-control stable cell lines exhibited equivalent mRNA levels of c-jun, indicating that TAF4b-dependent c-jun induction may be cell-type-specific (Fig. 3A Right). To confirm that constitutive overexpression of a TFIID subunit does not indirectly induce c-Jun levels in the SIGCs, we established a Flag-tagged TBP (f-TBP) cell line and measured the levels of c-Jun protein. SIGCs expressing either the control Flag vector or Flag-TBP possessed equivalently low levels of c-Jun, whereas f-TAF4b cells had higher levels, as expected (Fig. 3B). Thus, although NIH/3T3 cells can be stimulated to activate the promoters of ovarian-specific reporter constructs (e.g., Inhba) by overexpression of TAF4b, a key ubiquitous activator like c-Jun is induced only in a cell-type-specific manner.
Fig. 3.
Activation of gene expression in stable f-TAF4b cells and mouse tissues. (A) RPAs of c-jun expression were performed on RNA isolated from the SIGC and NIH/3T3 stable cell lines. Gapd and β-actin mRNAs were used as internal controls. Control, yeast RNA. (B) Immunoblotting of total cell extracts from stable control flag, f-TAF4b, and f-TBP SIGCs was performed to determine levels of c-Jun, TBP, and Flag-TBP. Anti-β-tubulin was used to confirm equal loading. (C) RPA of c-jun expression in adult mouse tissues and embryos. Control, yeast RNA. (D) Time course demonstrating the rapid induction of c-jun expression after PMSG-stimulation in adult mouse ovaries in comparison to β-actin as determined by RPA. Autoradiographic images are shown in A, C, and D.
Elevated c-jun mRNA and Rapid Pregnant Mare Serum Gonadotropin (PMSG)-Responsiveness in the Mouse Ovary.
We were surprised to find that overexpression of TAF4b resulted in elevated levels of c-jun in the SIGCs, because we did not observe detectable changes in the levels of c-jun in our initial microarray analysis of TAF4b-null ovaries. In wild-type mice, c-jun mRNA levels are most elevated in the ovary, heart, and brain compared with several other mouse tissues as determined by RPA (Fig. 3C). The control 18S rRNA had similar expression levels (data not shown), previously reported for these same tissue samples (7).
To correlate c-jun expression levels with folliculogenesis, we stimulated wild-type mice with PMSG, which mimics the in vivo activity of the follicle-stimulating hormone (FSH) by activating follicle growth, and measured c-jun levels by RPA. As compared with a β-actin control mRNA that was relatively unchanged, the c-jun mRNA levels accumulated at 4 and 16 h after PMSG injection to levels between 2- and 4-fold above uninduced levels (Fig. 3D). Our data suggest that, whereas c-jun mRNA is expressed at some basal level in many different cell types, it is highly expressed in the ovary, where it specifically responds to the action of gonadotropins. Accordingly, we postulate that c-Jun performs a critical function in the execution of the transcriptional response to gonadotropin signaling and GC proliferation in concert with TAF4b.
TAF4b Knockdown in SIGCs Decreases c-jun Expression.
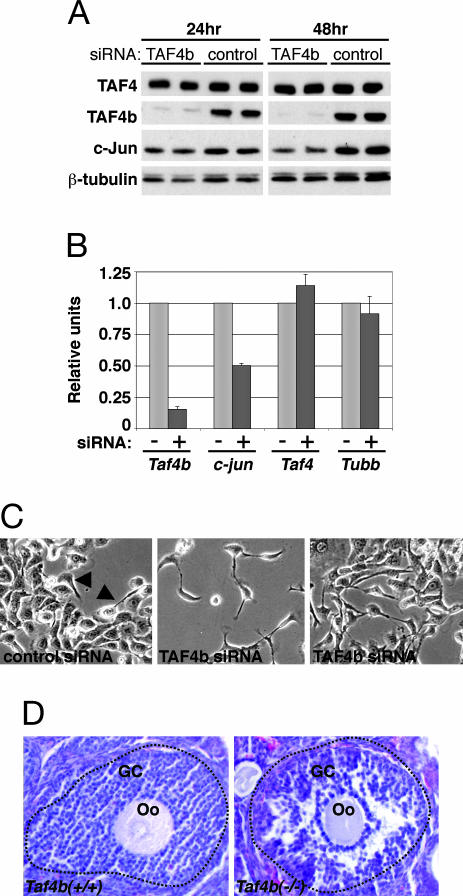
Short interfering (si)RNA-mediated knockdown of TAF4b activity in parental SIGCs was used to determine its effect on c-jun expression and GC function. An ≈90% or greater reduction in TAF4b protein levels was achieved at 24 and 48 h after TAF4b siRNA treatment as compared with the control siRNA without any significant effect on the levels of TAF4 or β-tubulin (Fig. 4A). In contrast, the loss of TAF4b activity was accompanied by a modest decrease in c-Jun at 24 h and a significant 1.5- to 2.5-times reduction in c-Jun protein levels at 48 h after TAF4b siRNA treatment (Fig. 4A). Quantitative real-time RT-PCR analysis of mRNA isolated from these cells confirmed that c-jun levels were significantly reduced in SIGCs that had an 85% loss in Taf4b mRNA at 48 h after siRNA treatment, whereas Taf4 and β-Tubulin (Tubb) mRNAs were essentially unchanged (Fig. 4B). These results establish a selective dependence of c-jun expression on TAF4b in GCs.
Fig. 4.
Phenotype of SIGCs and ovaries that lack TAF4b expression. (A) SIGCs were treated with TAF4b and control siRNAs for the indicated time points, followed by immunoblot analysis using anti-TAF4, anti-TAF4b, anti-c-Jun, and anti-β-tubulin antibodies as a loading control. (B) Treatment of SIGCs with TAF4b (+) in comparison with control (–) siRNAs reduces c-jun expression. Gene expression was determined by real-time RT-PCR analysis of RNAs from TAF4b siRNA-treated cells after standardization to control siRNAs and using Gapd as an internal control. Error bars represent SDs from three independent experiments. (C) Live-cell images of siRNA-treated SIGCs at ×10 magnification. An occasional fibroblastic cell can be identified in control siRNA-treated cells (arrows). (D) Hematoxylin-and-eosin-stained ovary sections are shown. Disorganized GCs surrounding an oocyte (Oo) are evident in Taf4b−/− follicles compared with wild-type Taf4b+/+ follicles, which are demarcated with a dotted line.
Unexpectedly, siRNA knockdown of TAF4b in SIGCs resulted in significant alterations in their cellular morphology. Normally, SIGCs grow as a homogeneous monolayer of cells with round, epithelial morphologies and extensive cell–cell contacts replete with gap junctions and desmosomes (10). Control siRNA treated SIGCs retained epithelial cell characteristics, with an occasional cell having a fibroblastic appearance (Fig. 4C Left, arrows). Strikingly, TAF4b siRNA-treated cells predominantly appeared to have a fibroblastic morphology, develop long processes, and possess fewer cell–cell attachments (Fig. 4C Center and Right). To further investigate this phenotype, we next examined TAF4b-null ovaries for similar defects in GC organization. Remarkably, TAF4b-null ovaries contain follicles that possess abnormally arranged layers of GCs that appear to have detached GCs compared with age-matched wild-type controls (Fig. 4D). Therefore, TAF4b activity appears to play a role in the preservation of essential GC-cell contacts and morphology in cultured SIGCs and in vivo.
TAF4b and c-Jun Promoter Cooccupancy in GCs.
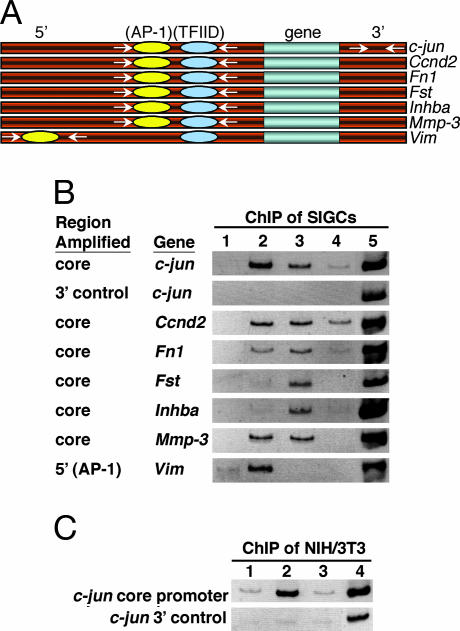
One potential mechanism explaining the selective activation of gene expression in GCs by TAF4b is the direct recruitment or binding of TAF4b-containing TFIID complexes to a specific subset of core promoters. In fact, it has recently been suggested that TAF4b possesses intrinsic DNA-binding activity (12). The expression of several genes up-regulated in the f-TAF4b SIGCs are known to be activated by c-Jun/AP-1 (Table 1). The core promoter regions of the c-jun (13), Ccnd2 (14), Fn1 (15), Fst (16), Inhba (17), Mmp-3 (18), and a 5′ distal enhancer site within the Vim (19) promoters all possess c-Jun/AP-1 DNA-binding elements (Fig. 5A).
Fig. 5.
Direct association of TFIID and c-Jun with target gene promoters. (A) Schematic representation of genomic loci analyzed by ChIP. At core promoter regions, only AP-1- and TFIID-binding sites are shown. For promoters (c-jun, Ccnd2, and Fst) that lack canonical TATA-box elements, TFIID-binding sites are depicted upstream of transcriptional start sites (not shown). For promoters (c-jun, Ccnd2, Fst, Inhba, and Vim) that have multiple predicted c-Jun/AP-1-binding sites, only one site is represented. Primer pairs (arrows) used to amplify a 5′ distal enhancer, core promoters, and a 3′ control region are shown. A box is used to represent each gene. (B) ChIP assays using stable f-TAF4b SIGC cells from genomic regions illustrated in A. Lane 1, control antibody; lane 2, anti-c-Jun; lane 3, anti-Flag; lane 4, anti-TAF4; lane 5, input. (C) ChIP assays from a stable f-TAF4b NIH/3T3 cell line. Lane 1, control antibody; lane 2, anti-c-Jun; lane 3; anti-Flag; lane 4, input. Inverted images of ethidium-bromide-stained PCR products are shown.
To determine whether TAF4b stimulation of c-jun transcription was directly because of its recruitment at the c-jun promoter, chromatin IP (ChIP) assays were performed on stable f-TAF4b SIGCs by using anti-c-Jun and anti-TAF antibodies. Our results indicated that the TFIID subunits TAF4b and TAF4 and one of its activators, c-Jun itself, are bound to the c-jun core promoter in vivo (Fig. 5B, lanes 2–4). In contrast, no specific c-jun core promoter fragments were recovered by using a control antibody (Fig. 5B, lane 1). TFIID and c-Jun binding was specific to the promoter region, because no DNA fragments were precipitated from the 3′ noncoding region of the gene (Fig. 5B, 3′ control). Importantly, we also detected the binding of TAF4b to the core promoter regions of the Ccnd2, Fn1, Fst, Inhba, and Mmp-3 genes but not to the 5′ distal AP-1 enhancer region of the Vim promoter. In contrast, c-Jun was bound at a 5′ distal AP-1 site within the Vim promoter and at the core promoter regions of the Ccnd2, Fn1, and Mmp-3 genes. TAF4 was readily detected only at the core promoters of c-jun and Ccnd2 and, to a lesser extent, at the Inhba and Fn1 promoters, unlike TAF4b, which was detected at each core promoter tested (Fig. 5B).
Because the ability of Flag-TAF4b to activate expression of c-jun was cell-type-specific, we reasoned that Flag-TAF4b may not be recruited to the c-jun promoter in NIH/3T3 cells. Indeed, we failed to detect the association of Flag-TAF4b above control levels in NIH/3T3 cells by ChIP assays (Fig. 5C, lane 3). However, as expected, c-Jun was bound to its own promoter in the NIH/3T3 cells (Fig. 5C, lane 2). These results suggest that TAF4b regulates the levels of c-jun expression in a cell-type-specific manner by directly associating with the core promoter.
Discussion
TAF4b-Dependent Regulation of Ovarian Gene Expression.
Our initial analysis of putative TAF4b-target genes was performed with intact mouse ovaries (7). Identification of TAF4b-dependent genes whose expression is not cell-type-specific but essential for GC function may have been masked by their abundant expression in additional ovarian cells that do not express or require TAF4b for transcription initiation. Herein, we have uncovered additional TAF4b-dependent genes by analyzing the function of TAF4b in a pure population of GCs grown in culture. Importantly, we have identified key ovarian programs of gene expression that are directly depend on the levels of TAF4b. Specifically, we have determined that TAF4b binds to and activates the native promoters of the Inh-subunit, Ccnd2, and Fst genes when constitutively overexpressed in cultured GCs. These results suggest that the loss of TAF4b transcriptional activity in the knockout mice contributes to the down-regulated levels of their expression in TAF4b-null females, which ultimately leads to an inadequate supply of healthy follicles expressing these transcripts and, thus, the observed female fertility defect (7).
In addition to inducing GC-specific gene expression, we show that TAF4b also induced the expression of AP-1 family members and directly regulates the c-jun promoter in a cell-type-selective context. Consequently, only a subset of promoters that contain putative AP-1-binding sites or genes known to be responsive to c-Jun stimulation were up-regulated in these cells. Our results strongly support the notion that TAF4b can function as a specific transcriptional coactivator for only a limited subset of genes, some of which are also c-Jun/AP-1 targets, and whose coordinated expression is essential for proper folliculogenesis.
TAF4b Coactivation in the Absence of Ovary-Specific Activators.
Surprisingly, we found that TAF4b was able to stimulate selective ovarian promoters in a dose-dependent manner without a complement of GC-specific activators as induction was also observed in NIH/3T3 cells that express elevated levels of TAF4b (Fig. 2C). Our results indicate that the restrictive expression of TAF4b in a cell-type-specific fashion may be sufficient to alter TFIID’s activity or specificity for a subset of core promoter elements that is independent of cell-type-specific activators. Consistent with this notion, overexpression of TAF4b in Taf4-null embryonic fibroblasts is also capable of inducing transcription, especially that of osteopontin expression (20), which was highly induced in GCs that overexpress TAF4b (Table 1). Because TAF4 does not compensate for the loss of TAF4b in the knockout mice, it is likely that TAF4b may use ubiquitous transcription factors that differ from those used by TAF4.
Remarkably, Taf4-null embryonic fibroblasts that possess TAF4b also have increased expression of Mmp-3 and Bdnf, two transcripts that were also induced in SIGCs that overexpress TAF4b (Table 1), indicating that TAF4b may regulate their expression in a non-cell-type-specific manner (20). However, c-jun was not induced in Taf4-null embryonic fibroblasts that express TAF4b, indicating that c-jun’s regulation by TAF4b is strictly cell-type-specific. Thus, in GCs, TAF4b-containing TFIID complexes may be constitutively bound to specific core promoter elements within the c-jun promoter to maintain basal levels of c-jun expression and to be poised for immediate induction. In this regard, c-jun expression in GCs can be simultaneously responsive to a greater spectrum of distinct stimuli by using dedicated TAF4b–TFIID promoter-recognition complexes.
TAF4b Is Essential for Ovarian Folliculogenesis.
In response to gonadotropin-induced transcriptional signaling networks, GCs rapidly proliferate and continue to differentiate into more specialized GC subtypes as ovarian follicular development ensues. Importantly, hormonally primed TAF4b-null ovaries fail to activate the expression of a subset of FSH-responsive genes to the same levels as wild-type controls, suggesting an important role for TAF4b in a FSH signaling pathway (7). Therefore, we predict that FSH (an inducer of cAMP production) potentiates the transcriptional activity of TAF4b-containing TFIID complexes at target promoters such as c-jun, which can be partially bypassed by directly overexpressing TAF4b in GCs (see Fig. 9, which is published as supporting information on the PNAS web site). Indeed, forskolin/cAMP stimulation (used to mimic the effects of FSH) of a human GC line results in the transient phosphorylation of TAF4b, with the potential to affect its function (21).
In contrast, the loss of TAF4b activity results in the aberrant transcription of critical GC-specific genes that likely impact GC morphology and function. Specifically, intercellular communications and their requisite cellular adhesions are essential linkages for GC–cell and GC–oocyte interactions during normal folliculogenesis (22). Consistent with this necessity, we specifically demonstrate that TAF4b-null ovaries possess vastly disorganized GCs that appear to be deficient in intimate cell–cell attachments, which may contribute to the abnormal development of follicles in the TAF4b-null ovaries (Fig. 4D). Furthermore, knockdown of TAF4b in cultured GCs, which still express normal levels of TAF4, develop abnormal cell morphologies and subsequently display significantly disrupted cell–cell attachments. Similarly, Taf4-null embryonic fibroblasts also display irregular and elongated morphologies despite the presence of TAF4b in these cells, suggesting that the paralogous TAF4b and TAF4 proteins share a common nonredundant function in maintaining normal cellular morphologies in distinct cell types (20). Thus, loss of Taf4b and TAF4b-dependent target gene expression results in substantial defects in GC transcription, follicle development, and cell morphology in both cultured GCs and in vivo.
Taken together, our data suggest that TAF4b is, indeed, a key cell-type-specific transcription coactivator component essential for target gene expression such as that of c-jun, as we have determined by both TAF4b overexpression and knockdown in GCs. TAF4b represents a unique class of cell-type-specific transcription factors that, surprisingly, can control c-jun expression independent of its well documented regulation as a rapidly induced immediate early gene. The dynamic composition of AP-1 transcription factors during GC proliferation and differentiation likely reflects the necessity to direct specific programs of gene expression unique to each stage of follicle development in response to multiple signaling pathways. Therefore, the specific expression of TAF4b in GCs provides a potentially unique mechanism to initiate a cascade of gene-expression changes by modulating the expression and activity of specific transcription activators and growth regulatory factors involved in cell proliferation, differentiation, and morphology.
Materials and Methods
Western Blot and IP.
Total protein extracts were prepared from cells in log phase of growth that were lysed in 1× SDS-gel-loading buffer. Western blot analysis was performed with anti-Pol II (H-224; Santa Cruz Biotechnology), anti-TAF4, anti-TFIIB (BD Transduction Laboratories), anti-Flag M2 (Sigma), anti-GAPDH (Abcam), anti-TAF1, and anti-TAF4b antibodies. Partial purification of f-TAF4b nuclear extracts by phosphocellulose chromatography was performed as described in ref. 23. IP of TAF4b was performed with anti-TAF4b polyclonal antibodies or a control antibody (mouse preimmune serum), followed by Western blot analysis with anti-TAF4, anti-Flag M2, and anti-TBP (Biodesign, Saco, ME) antibodies.
Indirect Immunofluorescence.
SIGC stable cell lines were grown on chamber slides (Nalge Nunc International), fixed in 2% paraformaldehyde, and permeabilized with 0.5% Triton X-100. Cells were immunostained with anti-Flag M2 antibody, detected with anti-mouse Alexa Fluor 488 (Molecular Probes) containing Hoechst 33342 (Sigma), and mounted with Vectashield (Vector Laboratories). Cells were observed under a Zeiss Axioplan microscope and imaged with a Leica LEI-750TD digital video camera (Optronics, Goleta, CA).
Luciferase Assays.
Stable cell lines were transiently transfected by using Effectene (Qiagen, Valencia, CA). The activities of promoter-luciferase reporters were normalized to the expression of HSV TK-Renilla control vector (Promega) for SIGC and SV40-Renilla control vector for NIH/3T3. A dual-luciferase reporter assay (Promega) was used to measure luciferase activities from three independent experiments that were performed in triplicate. The ratio of average luciferase values and SDs in the f-TAF4b cells as compared with control flag cells (set at 1) are shown.
RPA.
Total RNA was isolated from cells and ovaries by using TRIzol (Invitrogen). RPA probes were synthesized with Maxiscript (Ambion). The RPAIII kit (Ambion) was used to perform RPA. RPA of mouse tissue RNAs is described in ref. 7. Mice were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. For hormone treatments, 7- to 8-week-old wild-type C57BL/6 female mice (The Jackson Laboratory) were i.p. primed with 5 IU of PMSG (Sigma). Ovariectomy was performed on one control-uninjected female at the start of the time course and subsequently on one PMSG-injected animal at indicated time points.
DNA Microarray Analysis.
f-TAF4b and flag SIGC RNAs were used to probe microarray RG-U34A according to standard protocols and manufacturer’s instructions (Affymetrix). Details of microarray analysis can be found in Supporting Materials and Methods and Tables 2 and 3, which are published as supporting information on the PNAS web site.
Quantitative RT-PCR Analysis.
RNAs used for DNA microarray analysis and 48 h after siRNA treatments were digested with DNase I (Ambion) and reverse transcribed with Transcriptor (Roche) or iScript cDNA synthesis kit (Bio-Rad), respectively. Real-time PCR analysis was performed with 1× SYBR green PCR master mix (Bio-Rad) on a DNA Engine Opticon 2 (MJ Research). PCRs were performed three times in triplicate. Gapd was used for normalization. Fold-induction is defined as the ratio of relative average values for f-TAF4b cells versus flag control amplifications, or TAF4b siRNA versus nontargeting control siRNA. Primer sequences are available upon request.
TAF4b Knockdown in SIGCs.
Parental SIGCs were transfected with rat TAF4b and nontargeting control siRNA SMART pools by using DharmaFECT4 according to manufacturer’s suggestions (Dharmacon Research, Layfayette, CO). After 24 h, complexes were removed, and cells were harvested or cultured for an additional 24 h. Cells were lysed in either 1× SDS-gel-loading buffer or TRI reagent (Sigma). Live-cell images were acquired 48 h after siRNA treatment on a Zeiss Axiovert35 microscope equipped with a Zeiss ZVS-3C75DE video camera. Ovary sections from 4-week-old hormonally synchronized Taf4b+/+ and Taf4b−/− mice were prepared as described in ref. 7.
ChIP.
ChIP assays were performed essentially as described in ref. 24, but f-TAF4b cells were cross-linked by 1% formaldehyde for 12 min. Specific details of chromatin processing and IP can be found in Supporting Materials and Methods.
Plasmids, Cell Lines, and Antibodies.
Details of plasmids, antibody production, and cell lines used in this study can be found in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Coleman, M. Haggart, P. Hu, Y. Isogai, M. Marr, and O. Puig for assistance and helpful suggestions; B. Meyer for use of microscopy equipment; R. Coleman, Y. Fong, S. Martin, and O. Puig for critical comments on the manuscript; and members of the Tjian laboratory for useful ideas and technical assistance. K.G.G. was supported by Postdoctoral Fellowship #PF-02-113-01-GMC from the American Cancer Society. R.N.F. is supported, in part, by Center of Biomedical Research Excellence Grant P20 RR-15578 from the National Institutes of Health (NIH)/National Center for Research Resources. R.T. was supported, in part, by a grant from NIH.
Abbreviations
- AP-1
activator protein 1
- ChIP
chromatin IP
- GC
granulosa cell
- FSH
follicle-stimulating hormone
- IP
immunoprecipitation
- PMSG
pregnant mare serum gonadotropin
- Pol
polymerase
- RPA
RNase protection assay
- SIGC
spontaneously immortalized GC
- siRNA
short interfering RNA
- TBP
TATA-box protein
- TAF
TBP-associated factor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lemon B., Tjian R. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 2.Albright S. R., Tjian R. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 3.Hochheimer A., Tjian R. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 4.Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T. Y., Marino S., Fuller M. T. Development (Cambridge, U.K.) 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D., Penttila T. L., Morris P. L., Teichmann M., Roeder R. G. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 6.Dikstein R., Zhou S., Tjian R. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 7.Freiman R. N., Albright S. R., Zheng S., Sha W. C., Hammer R. E., Tjian R. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 8.Vanderhyden B. Front. Biosci. 2002;7:d2006–d2022. doi: 10.2741/A895. [DOI] [PubMed] [Google Scholar]
- 9.Falender A. E., Freiman R. N., Geles K. G., Lo K. C., Hwang K., Lamb D. J., Morris P. L., Tjian R., Richards J. S. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein L. S., Stoica G., Tilley R., Burghardt R. C. Cancer Res. 1991;51:696–706. [PubMed] [Google Scholar]
- 11.Mengus G., May M., Carre L., Chambon P., Davidson I. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 12.Shao H., Revach M., Moshonov S., Tzuman Y., Gazit K., Albeck S., Unger T., Dikstein R. Mol. Cell. Biol. 2005;25:206–219. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattori K., Angel P., Le Beau M. M., Karin M. Proc. Natl. Acad. Sci. USA. 1988;85:9148–9152. doi: 10.1073/pnas.85.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks A. R., Shiffman D., Chan C. S., Brooks E. E., Milner P. G. J. Biol. Chem. 1996;271:9090–9099. doi: 10.1074/jbc.271.15.9090. [DOI] [PubMed] [Google Scholar]
- 15.Dean D. C., Bowlus C. L., Bourgeois S. Proc. Natl. Acad. Sci. USA. 1987;84:1876–1880. doi: 10.1073/pnas.84.7.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyanaga K., Shimasaki S. Mol. Cell. Endocrinol. 1993;92:99–109. doi: 10.1016/0303-7207(93)90080-4. [DOI] [PubMed] [Google Scholar]
- 17.Tanimoto K., Yoshida E., Mita S., Nibu Y., Murakami K., Fukamizu A. J. Biol. Chem. 1996;271:32760–32769. doi: 10.1074/jbc.271.51.32760. [DOI] [PubMed] [Google Scholar]
- 18.Matrisian L. M., Leroy P., Ruhlmann C., Gesnel M. C., Breathnach R. Mol. Cell. Biol. 1986;6:1679–1686. doi: 10.1128/mcb.6.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rittling S. R., Baserga R. Mol. Cell. Biol. 1987;7:3908–3915. doi: 10.1128/mcb.7.11.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. EMBO J. 2005;24:2753–2767. doi: 10.1038/sj.emboj.7600748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y., Lu Y., Hu Y., Li R. J. Cell. Biochem. 2005;96:751–759. doi: 10.1002/jcb.20577. [DOI] [PubMed] [Google Scholar]
- 22.Matzuk M. M., Burns K. H., Viveiros M. M., Eppig J. J. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 23.Pugh B. F., Tjian R. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi Y., Rayman J. B., Dynlacht B. D. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.