Abstract
Both class switch recombination (CSR) and somatic hypermutation (SHM) of the Ig genes require the activity of activation-induced cytidine deaminase (AID). Expression of AID is restricted to B cells in the germinal centers of the lymphoid organs, where activated B cells undergo CSR and SHM. We previously showed that constitutive and systemic expression of AID leads to tumorigenesis in T cells and lung epithelium, but not in B cells. This finding led us to suspect that transgenic AID may be inactivated at least in part in B cells. To address this issue, we generated conditional AID-transgenic mice that constitutively express AID only in B cells. Studies on the cross between the AID-transgenic and AID-deficient mice showed that abundant AID protein accumulated by constitutive expression is inactivated in B cells, possibly providing an explanation for the absence of deregulation of CSR and SHM in AID-transgenic B cells.
Keywords: antibodies, protein modification, transgenic/knockout mice
Extensive proliferation of antigen-stimulated B lymphocytes is accompanied by two distinct genetic alterations, namely somatic hypermutation (SHM) and class switch recombination (CSR) (1). SHM accumulates point mutations in DNA encoding variable (V) regions of Ig and their immediate downstream flanks. B cells expressing Ig with high affinity to a given antigen are enriched when mutated B cells are subjected to selective expansion by limited amounts of antigen. On the other hand, CSR takes place between characteristic repetitive DNA sequences, switch (S) regions, located 5′ to Ig heavy chain constant-region genes. CSR diversifies effector functions of Ig by changing Ig isotypes from IgM to IgG, IgE, or IgA.
Activation-induced cytidine deaminase (AID) is required for both CSR and SHM because AID deficiency in mouse and human completely abolishes these two genetic alterations (2, 3). In addition to AID, CSR and SHM require transcription of target DNAs, S regions, and V regions, respectively (4–10). AID expression in non-B cells such as fibroblasts and T cells can induce CSR and SHM on transfected artificial constructs if the target DNA is actively transcribed (11–13). Therefore, AID and target transcription appear to be essential and sufficient for CSR and SHM in non-B as well as B cells. However, studies on transgenic (Tg) mouse lines ubiquitously expressing AID showed neither distortion of the B220+ B cell population nor increase in serum IgG levels (14). Although all mice died by 80 weeks because of T cell lymphomas, no B cell lymphomas were observed in the Tg mice (14). These observations have raised a possibility that Tg AID is negatively regulated in B cells. However, we could not exclude the possibility that dysregulation of T cells by a very early onset of T lymphomas may perturb B cell responses in AID-Tg mice.
It is therefore important to test the effects of constitutive AID expression in B cells using a more defined system. We thus generated Tg mice, which express AID constitutively only in B cells, and crossed them with AID-deficient mice. Using this system, we found that the Tg AID, despite its abundance, is much less efficient for CSR and SHM than the endogenous AID, suggesting that AID is negatively regulated in B cells.
Results
Generation of B Cell-Specific AID-Tg Mice.
To study the regulatory mechanism for induction of CSR and SHM in AID-expressing B cells, we generated AID conditional Tg mice that express AID specifically in B cells. Expression of the conditional AID transgene construct is designed in such a way that the AID protein synthesis is blocked by insertion of GFP flanked by two loxP sites, although its transcription is constitutive and ubiquitous by the regulation of the promoter pCAG (15) (Fig. 1A). AID can be expressed only after Cre-mediated deletion of the GFP cDNA from the transgene. These loxP GFP-AID-Tg (single-Tg, Tg/- - -) mice were crossed with the mice carrying the Cre gene knocked into the downstream of the CD19 promoter (CD19-cre mice) (16) so that GFP is deleted at the pro-B cell stage to express AID only in B lineage cells (17, 18).
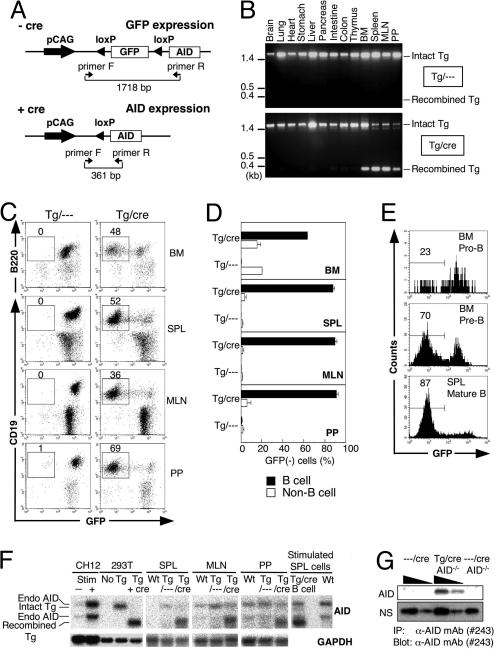
Fig. 1.
B cell-specific expression of AID in the double-Tg mice. (A) The conditional AID transgene construct that expresses AID after Cre-dependent recombination. The floxed GFP located upstream of AID cDNA is regulated under the transcriptional control of the constitutive and ubiquitous promoter pCAG. The position of primer pairs that detect both the intact and recombined forms of constructs are shown under the construct by arrowheads, and the sizes of amplified bands are shown. (B) PCR analysis of the recombination of the transgene DNA in various tissues of the Tg mice with or without CD19-cre. (C) Flow cytometric profiles of B lymphocytes from several lymphoid organs of the double-Tg mice after staining with antibodies for B cell markers (B220 or CD19). Numbers indicate percentages of B cell marker+ GFP− cells in lymphocytes. (D) The percentage of the GFP− cells in the B cell marker (B220 or CD19)-positive (B cell) and -negative cells (Non-B cell). Results obtained from three individual mice aged 8–9 weeks are summarized as column graphs with mean ± SD values. (E) GFP depletion in developing B cells of the double-Tg mice. Pro- and pre-B cells in BM are defined as c-kit+ B220+ and BP1+ B220+, respectively. Mature B cells in SPL are defined as IgM+ CD19+. (F) Northern blot analysis of endogenous and Tg AID expression in the lymphoid tissues of wild-type and Tg mice. Total RNA from unstimulated SPL, MLN, and PP were extracted and electrophoresed. Blots were probed for AID mRNA and rehybridized with a GAPDH probe after stripping the AID signal. RNA samples from stimulated and unstimulated CH12F3–2A (CH12) cells were prepared for endogenous AID control. RNAs from the HEK293T (293T) cells transfected with the transgene vector with or without Cre-expressing vector are used for controls of transgene AID before or after recombination, respectively. To compare the amounts of endogenous and Tg AID mRNA, RNA was prepared from purified GFP− B220+ SPL B cells of the double-Tg mice after stimulation by LPS for 3 days (stimulated SPL cells). (G) AID protein production from the double-Tg mice with AID−/− background. AID protein in splenocytes of the double-Tg × AID−/− mice and their controls after stimulation with LPS for 4 days was immunoprecipitated by an anti-AID monoclonal antibody (clone 243) and subjected to immunoblot by the same antibody after SDS/PAGE. Threefold dilutions of input amounts are shown. Nonspecific signal (NS) from the blot is shown as an internal control to indicate input protein amounts.
To examine CD19-cre-dependent deletion of GFP in loxP GFP-AID × CD19-cre (double-Tg, Tg/cre) mice, we performed PCR analysis of genomic DNA from several tissues of the double-Tg mice. PCR primers were designed to generate 1,718-bp and 361-bp products from the intact and recombined transgenes, respectively. CD19-cre-dependent recombination of the AID transgene was found in bone marrow (BM), spleen (SPL), mesenteric lymph nodes (MLN), and Peyer’s patches (PP) (Fig. 1B). We then quantitated loss of GFP signal in B cells of the lymphoid tissues by flow cytometry (Fig. 1C and D). We found constitutive GFP expression from the transgene in all of the B220+ or CD19+ cells in the single-Tg mice, whereas GFP expression is lost in 90% or more of the B220+ or CD19+ cells in the double-Tg mice. Percentages of GFP− cells in the B cell population gradually increased with the stage progression of B cell development (Fig. 1E).
Expression of the endogenous and Tg AID mRNA in the secondary lymphoid organs was measured by Northern blot analysis (Fig. 1F). Northern blot hybridization detected two bands for each of endogenous and Tg AID transcripts. Endogenous AID transcripts are known to have two variants because of differential polyadenylation site usage (19), whereas Tg AID transcripts are derived from the intact and recombined AID transgene. We confirmed that transcripts from the intact AID transgene did not produce AID protein by Western blot (data not shown). Consistent with loss of GFP signal in B cells, recombined Tg-derived AID mRNA was detected in SPL, MLN, and PP of the double-Tg mice together with endogenous AID mRNA. The expression level of endogenous AID mRNA in the LPS-stimulated B cells was augmented but still much less than that of Tg-derived AID (stimulated SPL cells in Fig. 1F). The protein amounts of endogenous and Tg AID in LPS-stimulated splenocytes were measured by immunoprecipitation by an anti-AID monoclonal antibody (clone 243) followed by immunoblot. Tg AID was readily detectable in the double-Tg mice of AID knockout background (Tg/cre AID−/−), whereas endogenous AID (- - -/cre) was undetectable in activated splenocytes by Western blot (Fig. 1G). The comparison of the samples indicates that the Tg AID protein is expressed far more abundantly (probably >9-fold) than the endogenous AID. We note that the entire sequence of the AID transgene was confirmed to be intact in this line of AID-Tg mice (data not shown).
B Cells of the Tg Mice Develop Normally.
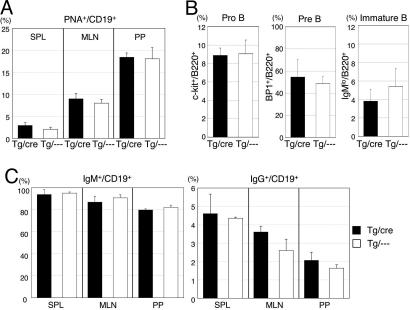
We examined whether the B cell development of the double-Tg mice is affected by constitutive expression of AID or not. The weights and/or sizes of the secondary lymphoid tissues such as SPL and MLN and the numbers and percentages of B cells in BM, SPL, MLN, and PP appeared to be normal (data not shown). We did not find any abnormality in the percentages of peanut agglutinin (PNA)-positive B cells in SPL, MLN, and PP of the double-Tg mice (Fig. 2A) and in the percentages of pro-B, pre-B, and immature B cells in BM (Fig. 2B). We did not observe any B cell lymphomas among 12 double-Tg mice in >20 months. These results suggest that constitutive expression of AID in B cells neither affected development of B cells nor induced B cell tumors.
Fig. 2.
Normal B cell development in BM and the secondary lymphoid organs in the double-Tg mice. (A) Flow cytometric analysis of B cells in SPL, MLN, and PP from the Tg mice. The percentages of PNA+ cells in the CD19+ population are indicated. Results obtained from three individual mice aged 8–9 weeks are summarized as column graphs with mean ± SD values. (B) Flow cytometric analysis of the BM cells from the Tg mice. The percentages of pro-B, pre-B, and immature B cells in the B220+ cell population in BM are indicated. Pro-B, c-kit+ B220+; pre-B, BP1+ B220+; immature B, B220lo IgM+. (C) The percentages of IgM+ and IgG+ cells in the CD19+ population are indicated. Results obtained from three individual mice aged 8–9 weeks are summarized as column graphs with mean ± SD values.
Tg AID Is Inefficient in CSR.
To examine whether CSR is augmented by constitutive expression of AID, we measured various isotype concentrations in sera and the IgA concentration in feces of the double-Tg mice, and we found that all of them were within the normal range (Fig. 3A and data not shown). We also analyzed the proportions of IgM+ and IgG+ B cells in the secondary lymphoid organs such as SPL, MLN, and PP, and again we found no differences between double- and single-Tg mice (Fig. 2C). These results indicate that constitutively expressed AID in B cells has shown little if any CSR activity in vivo. Then we measured in vitro CSR efficiency of splenocytes from the double-Tg mice (Fig. 3B). The double-Tg splenocytes showed a slightly higher CSR efficiency than the single-Tg and non-Tg mice splenocytes after stimulation with LPS and IL-4 (Fig. 3B), suggesting that Tg AID has some activity to induce CSR. It is noted that the CSR efficiency of splenocytes from the single-Tg mice was similar to those from non-Tg mice (Fig. 3B), excluding the possibility that the integration of the transgene itself affected CSR in B cells.
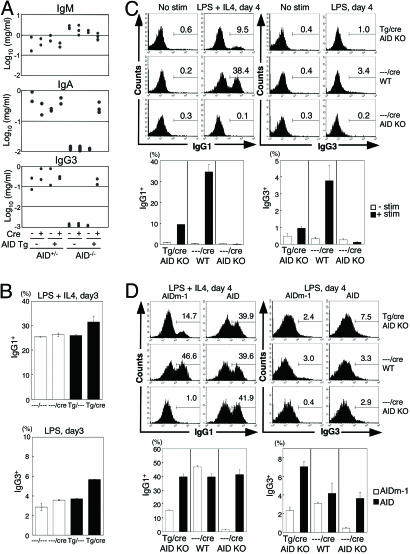
Fig. 3.
Analyses of CSR induced by Tg AID. (A) Serum Ig concentrations in the AID-Tg mice that lack endogenous AID and their control mice (aged 10–15 weeks) were determined by ELISA. (B) In vitro class switching of splenocytes from the AID-Tg mice in the presence of endogenous AID. Splenocytes after 3-day culture with LPS and IL-4 were stained with anti-B220, and anti-IgG1, or IgG3 antibodies and analyzed by flow cytometry. The percentages of IgG1+ or IgG3+ B cells in the B220+ fractions are indicated in each panel. Results obtained from two individual mice aged 13–14 weeks are summarized as column graphs with mean ± SD values. (C) In vitro class switching of splenocytes from the AID-Tg mice in the absence of endogenous AID. Splenocytes were stimulated with LPS and IL-4 for 4 days. The percentages of IgG1+ or IgG3+ B cells in the GFP− B220+ fractions are indicated in each panel. Results obtained from three individual mice aged 21–30 weeks are summarized as column graphs with mean ± SD values. (D) GFP− splenocytes from the AID-Tg mice were stimulated with LPS and IL-4 and infected with AID-expressing retrovirus 1 day after stimulation. Splenocytes with 4-day culture were analyzed as described in C. AIDm-1, a loss-of-function mutant of AID (36).
To examine whether Tg AID in the double-Tg mice is functional, we analyzed the activity of Tg AID in the absence of endogenous AID. First, we measured the serum levels of IgM, IgG3, and IgA in the double-Tg mice crossed with the AID-deficient mice (2) (Fig. 3A). AID-deficient mice that carry either loxP-GFP-AID-Tg or CD19-cre showed higher concentrations of serum IgM and undetectable levels of serum IgG3 and IgA, as previously shown in AID-deficient mice (2). On the other hand, AID-deficient mice carrying both loxP-GFP-AID-Tg and CD19-cre (double-Tg-AID−/−) showed a normal level of IgM, IgG3, and IgA in sera, indicating that Tg-derived AID is functional.
Next, we analyzed in vitro CSR efficiency of the double-Tg-AID−/− splenocytes. Naïve B cells of the double-Tg-AID−/− mice were negative for surface IgG. Only when subjected to stimulation by LPS and IL-4 to induce CSR in vitro, they switched to IgG1- or IgG3-expressing cells (Fig. 3C). However, the CSR efficiency of the double-Tg-AID−/− splenocytes was only <30% of wild-type control. This result could be either because the function of AID from the transgene is inactivated and thus less efficient than endogenous one or because other factors required for CSR are down-modulated in B cells that constitutively express AID. To examine these possibilities, GFP− splenocytes of the double-Tg-AID−/− mice were stimulated with LPS and IL-4 and infected with AID-expressing retrovirus 1 day after stimulation. The percentage of IgG1+ or IgG3+ cells in infected B220+ cells was measured by flow cytometry (Fig. 3D). AID expression by retrovirus infection enhanced the CSR efficiency of the B cells from the double-Tg-AID−/− mice to the level similar to or more than wild-type (Fig. 3D), indicating that other factors required for CSR were not limiting and that AID from the transgene, despite its abundance, is much less efficient than endogenous AID. These results indicate that constitutively expressed AID is somehow modified to down-modulate its activity of CSR.
Tg AID Is Inefficient in SHM.
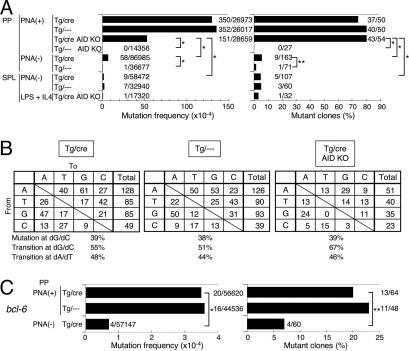
We also analyzed the frequency and pattern of SHM in the 3′ flanking region of the rearranged heavy-chain V exon of B cells in the double-Tg mice (20). First, we analyzed SHM in PNA+ B cells from PP of the double-Tg mice, in which B cells are activated to accumulate SHM by spontaneous antigen stimulation (21). The mutation frequency of B cells in the double-Tg mice was not at all higher than that in the control single-Tg mice (Fig. 4A). The mutation base profiles (transition vs. transversion proportion at dG/dC and dA/dT bases) of the double-Tg mice were also similar to those of the single-Tg mice (Fig. 4B). To see whether constitutive expression of AID alone can lead to SHM without activation by antigen, we analyzed SHM in the PNA− B cells. There was a low but significant level of SHM in PNA− PP B cells from the double-Tg B cells. However, we could not exclude the possible contamination of PNA+ PP B cells. We therefore analyzed SHM in PNA− SPL B cells, which accumulate little SHM under the normal condition (22). We detected no more than background levels of SHM in PNA− SPL B cells from the double-Tg mice (Fig. 4A). In vitro stimulation of SPL cells by LPS and IL-4, which activates transcription of the Ig locus and induces CSR to IgG1 and IgE, did not induce SHM (Fig. 4A). Because SHM was introduced to PNA+ B cells in the double-Tg-AID−/− mice (Fig. 4A), Tg AID is also functional for SHM. However, the efficiency of SHM in the double-Tg-AID−/− B cells was no more than 40% of the single-Tg B cells, which express only endogenous AID, again suggesting that constitutively expressed AID might have been drastically inactivated for the SHM activity.
Fig. 4.
Analyses of SHM in B cells of the AID-Tg mice. (A) Mutation frequency at the 3′-flanking region of the JH4 exon of the Ig heavy chain V region. Numbers of mutation/nucleotides analyzed (Left) and the numbers of mutated clones/Ig clones analyzed (Right) are indicated. Statistical significance was evaluated by Fisher’s exact tests for indicated sets of data. ∗, P < 0.01; ∗∗, P < 0.05. For each analysis, one to four individual mice aged 9–27 weeks were used. (B) Pattern of the base substitutions in the intron downstream of JH4 exon of PNA (+) B cells in PP of the AID-Tg mice. (C) Mutation frequency at the bcl-6 gene. Numbers of mutations/nucleotides analyzed (Left) and the numbers of mutated clones/clones analyzed (Right) are indicated. Statistical significance was evaluated as described in A.
It is worth noting that the base specificity of SHM in the double-Tg-AID−/− mice was similar to the wild-type or the single-Tg mice without strong bias to dG/dC bases (Fig. 4B). These results indicate that overexpression of AID in B cells does not perturb SHM base specificity in normal B cells. To test whether the target specificity of SHM by Tg AID is regulated similarly to the endogenous AID, we analyzed SHM on non-Ig genes: bcl-6, which is known to be subjected to SHM in normal germinal center B cells and GC-derived B lymphoma cells (23), and the genes for B220, CD19, GAPDH, and β-actin, which are known to be immune to SHM despite constitutive expression throughout B cell development. We also sequenced the AID transgene because overexpressed AID is known to mutate itself (13). SHM was accumulated on the bcl-6 gene in the PNA+ B cells of the double-Tg mice with a frequency similar to that of the single-Tg mice (Fig. 4C). No accumulation of SHM was found in the genes for the Tg AID (0 of 16,560 bp, <0.6 × 10−4 per bp), B220 (0 of 15,198 bp, <0.7 × 10−4 per bp), CD19 (0 of 16,299 bp, <0.6 × 10−4 per bp), GAPDH (1 of 13,791 bp, 0.7 × 10−4 per bp), and β-actin (1 of 18,386 bp, 0.5 × 10−4 per bp) even in the PNA+ B cells of the double-Tg mice. These results indicate that the Tg and endogenous AID proteins have a SHM target specificity similar to those previously reported in normal and malignant B cells and that the inefficiency of the Tg AID protein is not due to mutations in the AID transgene in activated B cells.
Limiting Factors for CSR Other than AID.
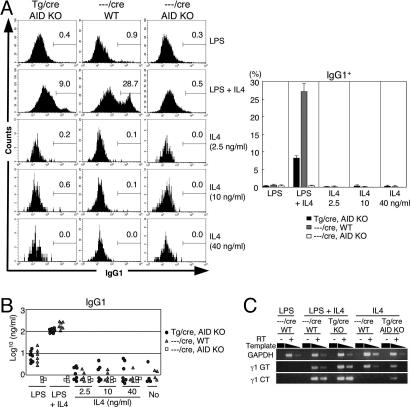
The above experiments demonstrated that constitutive expression of AID alone is not sufficient for CSR. To examine whether exogenous AID expression and γ1 transcription can drive CSR to IgG1, the percentage of IgG1+ cells was measured by flow cytometry after 4-day incubation of the double-Tg-AID−/− B cells with IL-4 alone, which is known to induce γ1 germ-line transcription and modest expression of endogenous AID, but not CSR to IgG1, in the absence of LPS (19, 24, 25) (Fig. 5A). No significant IgG1+ cells were detected in the GFP− B220+ fraction in the presence or absence of Tg AID. Concentrations of IgG1 in the culture supernatant from IL-4-stimulated and control samples were measured by ELISA (Fig. 5B). IgG1 secretion from IL-4-stimulated splenocytes from the double-Tg-AID−/− mice was not enhanced when compared with non-Tg or nonstimulated samples (Fig. 5B). To assess the ability of IL-4-stimulated splenocytes to carry out CSR to IgG1 in a more direct and sensitive manner, RT-PCR was performed to amplify the circle transcripts from the looped-out Iγ1-Cμ exon (γ1 circle transcripts) (26) (Fig. 5C). Although γ1 germ-line transcripts were detectable in all splenocytes stimulated with IL-4, γ1 circle transcripts were induced in splenocytes from the double-Tg-AID−/− mice but not from wild-type mice by stimulation with IL-4 alone (Fig. 5C). Because control LPS plus IL-4 stimulation induced both γ1 germ-line and γ1 circle transcripts in the splenocytes from either wild-type or the double-Tg-AID−/− mice, these results indicate that AID expression and γ1 germ-line transcription are sufficient to induce minimal CSR in the Sγ1 region and that additional stimulation signal, such as LPS stimulation, is necessary for more efficient switching.
Fig. 5.
Analyses of CSR induced by AID and γ1 germ-line transcription. (A) In vitro class switching of splenocytes from the AID-Tg mice. Splenocytes cultured for 4 days with LPS and/or IL-4 were analyzed as described in the legend for Fig. 3B. The percentages of IgG1+ B cells in the GFP− B220+ fractions are indicated in each panel. Results obtained from three individual mice aged 21–25 weeks are summarized as column graphs with mean ± SD values. (B) IgG1 titers of the culture supernatants of in vitro stimulated splenocytes described in A were determined by ELISA. (C) mRNA levels of GAPDH, γ1 germ-line transcripts, and γ1 circular transcripts of the splenocytes from AID-Tg mice. mRNAs were quantitated by semiquantitative RT-PCR with 10-fold dilutions after 2-day stimulation of the cells by LPS and/or IL-4.
Discussion
Negative Regulation of AID in B Cells.
The present study on B cell-specific AID-Tg mice showed that B cell development appears to be normal despite constitutive expression of AID throughout B cell development. The Cre-mediated recombination frequencies in the double-Tg mice and other loxP-flanked knock-in mice crossed with the same CD19-cre mice are indistinguishable (27), indicating that the number of AID-expressing B cells generated by CD19-cre recombination was not reduced. Furthermore, the fact that most of the B cells in the secondary lymphoid organs have completed recombination without reduction of cell numbers argues against AID-induced cell death, although we cannot exclude the possibility that a very limited number of B cells are affected and eliminated by constitutive expression of AID. We therefore concluded that constitutive AID expression does not perturb B cell development.
Although the amount of the Tg AID protein was expressed far more abundantly than endogenous AID, CSR and SHM in B cells expressing the Tg AID protein alone was only 30–40% compared with those expressing endogenous AID. The results indicate that the majority of constitutively expressed Tg AID is inactivated by unknown mechanisms. It is likely that accumulated AID in the double-Tg B cells is inactivated by protein modifications or interaction with other proteins and nucleic acids.
We have shown that CSR takes place in AID-expressing B cells even if B cells are not activated by antigen. This finding was demonstrated by the production of γ1 circle transcripts after IL-4 treatment of splenocytes of the double-Tg-AID−/− mice in vitro. The frequency of these CSR detected in naïve B cells is extremely low compared with those in fully activated B cells. There are several explanations for the requirement of B cell activation for efficient CSR. The simplest one is that transcription of the S and V regions in the resting B cells is weak and the access of a putative recombinase to the target S region is inefficient. Another explanation is modification of cis-acting components such as chromatin structure of Ig enhancers by B cell activation (28). It is also possible that B cell activation is required for an increase in transacting factors such as CSR- or SHM-specific cofactors (29, 30). There may be inhibitory factors for AID activity in nonactivated B cells, which are removed by B cell activation.
Safeguard Against Side Effects of AID in B Cells.
Constitutive expression of AID is potentially deleterious to B cells because AID may introduce double-strand breaks not only in the Ig V region genes, but also in other genes, including oncogenes, which further induces chromosomal translocations and mutations. To avoid such negative effects of AID, B cells may have adopted a strategy that limits the function of AID within a short time range after stimulation by antigen. The strategy appears to include two aspects, as revealed in the present study. One is inactivation of AID function. The other is requirement of activation for full function of AID. These strategies appear to be unique to B cells because AID expression in non-B cells including T cells, fibroblasts, and hybridomas can induce CSR and SHM in artificial assay constructs (11–13,31). However, the same SHM construct was very inefficient in the CH12 B cell line expressing AID (32). In addition, T lymphoma cells generated in AID-Tg mice continue to accumulate mutations (14, 33). This variation of SHM efficiency may reflect the absence and presence of SHM-specific cofactors in CH12 and T lymphoma cells, respectively.
SHM Induced by Tg and Endogenous AID Is Regulated Similarly in B Cells.
Overexpression of AID in non-B cells induces CSR and SHM in appropriate artificial assay constructs (11, 12) or SHM in the Ig locus (13). Multiple copies of artificial constructs inserted in various chromosomes are shown to be targets of SHM (34). In addition, the base specificity of SHM by overexpressed AID is >90% biased to dG/dC transition even in the Ig locus in hybridoma (12, 14, 31) whereas SHM in the Ig locus in normal mice is ≈60–40% dG/dC targeted. Thus, Tg AID has been considered to be regulated differently from the endogenous AID. In the present study, however, we have shown that Tg AID in B cells is regulated in a manner similar to the endogenous AID in terms of the base specificity and locus specificity. Because B cell activation is always accompanied by endogenous AID expression, it has been difficult to verify the existence of another layer of regulation for the function of AID. Studies on the double-Tg-AID−/− B cells may be useful for dissecting additional requirements for the AID function in B cells.
Materials and Methods
Mice.
A set of genes, EGFP cDNA flanked by loxP, polyA signal, murine AID cDNA (0.6 kb), and poly(A) signal, in this order, was cloned into the downstream of the chicken β-actin (CAG) promoter (15) of the modified pCXN2 vector. The vector was cut at the SalI site upstream of the CAG promoter and the HindIII site downstream of polyA signal after AID cDNA. The linearized fragment containing the AID transgene was purified and microinjected into C57BL/6 fertilized eggs to generate conditional AID-Tg mice. A single-copy Tg line was selected by Southern blot. The Tg mice were maintained in specific pathogen-free conditions in the animal facility of the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University. Recombination of the transgene in tissues was analyzed by PCR using primer F (5′-CTC TGC TAA CCA TGT TCA TGC CTT CTT C-3′) and primer R (5′-GGT CCC AGT CTG AGA TGT AGC GTA G-3′). Amplification was done by an initial denaturing step of 94°C for 5 min, followed by 40 cycles of PCR (94°C for 15 s, 60°C for 30 s, and 72°C for 2 min).
Flow Cytometry.
Single-cell suspensions from BM, SPL, MLN, and PP were stained with following antibodies: phycoerythrin-conjugated antibodies mouse CD19 (clone 1D3; Pharmingen), mouse c-kit (clone 2B8, Pharmingen), and mouse BP1 (clone BP1; Pharmingen); allophycocyanin-conjugated antibody mouse B220 (clone RA3-6B2; eBioscience); and biotin-conjugated antibodies mouse IgM (Cappel), mouse IgG (Pharmingen), mouse IgG1 (clone A85-1; Pharmingen), and mouse IgG3 (clone R40-82; Pharmingen). Biotin-conjugated PNA (Vector Laboratories) and phycoerythrin-conjugated (Vector Laboratories) and allophycocyanin-conjugated (Pharmingen) streptavidin were also used for staining. Flow cytometric analyses were performed on a FACSCalibur with cellquest software (Becton Dickinson). Alive lymphocytes were selected for the analyses by forward- and side-scatter intensity and propidium iodide gatings. Cell sorting was done by using FACSVantage SE (Becton Dickinson).
Northern Blot Analysis.
Total RNA was extracted from cultured cells by using TRIzol (Invitrogen) according to the manufacturer’s instructions. Total RNA (30 μg) was electrophoresed on a 1.2% denaturing agarose gel containing formaldehyde. Northern blots were hybridized with a 32P-labeled mouse AID cDNA and rehybridized with a GAPDH cDNA probe as an internal control, as described in ref. 19.
RT-PCR.
Total RNA was subjected to reverse transcription by using oligo-dT primer and SuperScript III reverse transcriptase after DNase I treatment, according to manufacturer’s instructions (Invitrogen). Semiquantitative PCR was done by using recombinant Taq polymerase (Takara) according to instructions. Amplification of endogenous AID, γ1 germ-line, γ1 circular, and GAPDH transcripts was done as described in refs. 2 and 26.
Preparation of Monoclonal Antibodies Specific for AID.
A monoclonal antibody (clone 243) for AID was prepared as described (35) except for the preparation of antigen. In brief, murine AID cDNA was cloned into expression vector pET16b (Novagen) to produce His-tagged recombinant AID protein in bacterial strain BL21(DE3). After induction by IPTG (1 mM) for recombinant AID protein at 37°C, the bacterial cells were collected, and the cell pellet was lysed in a lysis buffer containing 50 mM Tris·HCl (pH 7.4), 150 mM KCl, and 1% Nonidet P-40 by using Ultrasonic homogenizer US-300T (NIHON SEIKI). The lysate was centrifuged at 20,000 × g for 15 min, and then insoluble (pellet) fraction was collected and lysed in a denaturing buffer containing 10 mM Tris·HCl (pH 8.0), 50 mM sodium phosphate, 8 M urea, and 0.3% SDS. His-tagged AID protein was purified by using His Trap column (Amersham Pharmacia Biosciences) according to the manufacturer’s instructions. The purified AID protein was used as antigen for immunization. The culture supernatants of hybridomas after HAT selection were examined by ELISA and immunoblot for their reactivity to the recombinant AID protein. Selected clones were further examined by immunoblot, immunoprecipitation, and immunocytochemistry for reactivity to the AID protein produced by mammalian cells.
Immunoprecipitation and Western Blotting.
Cells were lysed in a buffer containing 10 mM Tris·HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and Complete Protein Inhibitor Mixture (Roche). Immunoprecipitation was performed with Protein G Sepharose (Amersham Pharmacia Biosciences) and the anti-AID monoclonal antibody (clone 243). Western blotting of the precipitate was done with monoclonal antibody 243 and horseradish peroxidase-conjugated goat anti-rat IgG antibody (Kirkegaard & Perry Laboratories) as secondary antibody.
ELISA.
Ig concentrations of sera and feces were measured as described in ref. 2 after serial dilution of the samples to find the linear range.
In Vitro Stimulation of Splenocytes.
Splenocytes were stimulated for 4 days as described in ref. 2, except that 25 μg/ml LPS (Sigma) and 2.5 ng/ml recombinant mouse IL-4 (e-Bioscience) were used, unless indicated otherwise. Retrovirus infection was performed as described in ref. 36.
SHM Analysis.
SHM frequency of the Ig (20) locus was determined as described except that Pyrobest DNA polymerase (Takara), which has the 3′ exonuclease activity and high fidelity, and a Topo cloning kit (Invitrogen) were used to clone the DNA fragment from the cells. Amplification of the bcl-6 locus was achieved by an initial denaturing step of 95°C for 5 min followed by 35 cycles of PCR (95°C for 10 s, 65°C for 30 s, 72°C for 1 min). Primers used were bcl-6 forward (5′-CTT TCT TGG TTG GAG TCG AGG C-3′) and reverse (5′-CGG GCT TGA GGT CAT TTC TC-3′).
Acknowledgments
We thank S. Fagarasan, A. Shimizu, and R. Shinkura for their useful suggestions; K. Rajewsky (Harvard Medical School, Boston) for the CD19-cre mice; J. Miyazaki (Osaka University Medical School, Suita, Japan) for the pCXN2 vector; T. Toyoshima, A. Kawamura, A. Takano, E. Inoue, and Y. Sasaki for their excellent technical assistance; and K. Fukui, Y. Shiraki, and T. Nishikawa for their excellent secretarial help. This work was supported by Center of Excellence Grant 1700201 (to T.H.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations
- SHM
somatic hypermutation
- CSR
class switch recombination
- AID
activation-induced cytidine deaminase
- Tg
transgenic
- CAG
chicken β-actin
- BM
bone marrow
- SPL
spleen
- MLN
mesenteric lymph node
- PP
Peyer’s patch
- V
variable
- PNA
peanut agglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Honjo T., Kinoshita K., Muramatsu M. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Bachl J., Carlson C., Gray-Schopfer V., Dessing M., Olsson C. J. Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 5.Fukita Y., Jacobs H., Rajewsky K. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 6.Peters A., Storb U. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 7.Tumas-Brundage K., Manser T. J. Exp. Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu H., Zou Y. R., Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 9.Jung S., Rajewsky K., Radbruch A. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 10.Xu L., Gorham B., Li S. C., Bottaro A., Alt F. W., Rothman P. Proc. Natl. Acad. Sci. USA. 1993;90:3705–3709. doi: 10.1073/pnas.90.8.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okazaki I. M., Kinoshita K., Muramatsu M., Yoshikawa K., Honjo T. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa K., Okazaki I. M., Eto T., Kinoshita K., Muramatsu M., Nagaoka H., Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 13.Martin A., Scharff M. D. Proc. Natl. Acad. Sci. USA. 2002;99:12304–12308. doi: 10.1073/pnas.192442899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki I. M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. J. Exp. Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niwa H., Yamamura K., Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 16.Rickert R. C., Rajewsky K., Roes J. Nature. 1995;376:352–355. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 17.Fearon D. T. Curr. Opin. Immunol. 1993;5:341–348. doi: 10.1016/0952-7915(93)90051-s. [DOI] [PubMed] [Google Scholar]
- 18.Tedder T. F., Zhou L. J., Engel P. Immunol. Today. 1994;15:437–442. doi: 10.1016/0167-5699(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 19.Muramatsu M., Sankaranand V. S., Anant S., Sugai M., Kinoshita K., Davidson N. O., Honjo T. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 20.Jolly C. J., Klix N., Neuberger M. S. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Fernandez A., Milstein C. Proc. Natl. Acad. Sci. USA. 1993;90:9862–9866. doi: 10.1073/pnas.90.21.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rada C., Gupta S. K., Gherardi E., Milstein C. Proc. Natl. Acad. Sci. USA. 1991;88:5508–5512. doi: 10.1073/pnas.88.13.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H. M., Peters A., Baron B., Zhu X., Storb U. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 24.Berton M. T., Uhr J. W., Vitetta E. S. Proc. Natl. Acad. Sci. USA. 1989;86:2829–2833. doi: 10.1073/pnas.86.8.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stavnezer J., Radcliffe G., Lin Y. C., Nietupski J., Berggren L., Sitia R., Severinson E. Proc. Natl. Acad. Sci. USA. 1988;85:7704–7708. doi: 10.1073/pnas.85.20.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita K., Harigai M., Fagarasan S., Muramatsu M., Honjo T. Proc. Natl. Acad. Sci. USA. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickert R. C., Roes J., Rajewsky K. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo C. J., Martin A., Scharff M. D. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 29.Ta V. T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., et al. Nat. Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 30.Shinkura R., Ito S., Begum N. A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. Nat. Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 31.Martin A., Bardwell P. D., Woo C. J., Fan M., Shulman M. J., Scharff M. D. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 32.Eto T., Kinoshita K., Yoshikawa K., Muramatsu M., Honjo T. Proc. Natl. Acad. Sci. USA. 2003;100:12895–12898. doi: 10.1073/pnas.2135587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotani A., Okazaki I. M., Muramatsu M., Kinoshita K., Begum N. A., Nakajima T., Saito H., Honjo T. Proc. Natl. Acad. Sci. USA. 2005;102:4506–4511. doi: 10.1073/pnas.0500830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C. L., Harper R. A., Wabl M. Proc. Natl. Acad. Sci. USA. 2004;101:7352–7356. doi: 10.1073/pnas.0402009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishida M., Iwai Y., Tanaka Y., Okazaki T., Freeman G. J., Minato N., Honjo T. Immunol. Lett. 2002;84:57–62. doi: 10.1016/s0165-2478(02)00142-6. [DOI] [PubMed] [Google Scholar]
- 36.Fagarasan S., Kinoshita K., Muramatsu M., Ikuta K., Honjo T. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]