Abstract
Although it is known that the human genome contains hundreds of microRNA (miRNA) genes and that each miRNA can regulate a large number of mRNA targets, the overall effect of miRNAs on mRNA tissue profiles has not been systematically elucidated. Here, we show that predicted human mRNA targets of several highly tissue-specific miRNAs are typically expressed in the same tissue as the miRNA but at significantly lower levels than in tissues where the miRNA is not present. Conversely, highly expressed genes are often enriched in mRNAs that do not have the recognition motifs for the miRNAs expressed in these tissues. Together, our data support the hypothesis that miRNA expression broadly contributes to tissue specificity of mRNA expression in many human tissues. Based on these insights, we apply a computational tool to directly correlate 3′ UTR motifs with changes in mRNA levels upon miRNA overexpression or knockdown. We show that this tool can identify functionally important 3′ UTR motifs without cross-species comparison.
Keywords: gene expression, microarray, posttranscriptional control
Mature microRNAs (miRNAs) are short noncoding single-stranded RNAs that can posttranscriptionally regulate gene expression in plants and animals. Hundreds of distinct miRNA genes are now known to exist and to be differentially expressed during development and across tissue types. Overall, little is known about the biological function of animal miRNAs, but recent studies have suggested important regulatory roles for miRNAs in a broad range of biological processes including developmental timing, cellular differentiation, proliferation, apoptosis, oncogenesis, insulin secretion, and cholesterol biosynthesis (1–5). Even less is known about the specific role of miRNAs in gene regulation because only a few miRNA targets have been thoroughly analyzed experimentally. In animals, miRNAs usually inhibit gene expression through partially complementary elements in the 3′ UTRs of their target mRNAs. Although it has been suggested that animal miRNAs could play important roles in specifically modulating translational gene regulation (1, 2), there is also increasing evidence that miRNAs can directly induce mostly weak but significant negative effects on the steady-state mRNA levels of their targets. Evidence for these effects comes from a series of studies that used siRNA transfection, miRNA overexpression, or miRNA knockdown in cultured cell lines or entire organisms (5–8). It has been demonstrated that transfection of exogenous miRNA duplexes into HeLa cells can cause moderate down-regulation of hundreds of mRNAs, many of which contain the recognition motif of the overexpressed miRNA in their 3′ UTR (7). In Caenorhabditis elegans, the let-7 miRNA induces the degradation of lin-41 target mRNA. Furthermore, lin-14 and lin-28 mRNA levels decrease in response to lin-4 miRNA (8). Finally, in vivo knock-down of a liver-specific miRNA (miR-122) has shown that hundreds of mRNAs, many of them likely to be direct targets of this miRNA, were moderately up-regulated (5). In summary, these studies suggest that mRNAs containing partial miRNA complementary sites can be targeted for degradation in vivo, that miRNA-dependent regulation of mRNA stability may be more common than previously appreciated, and that this mode of gene regulation could be an important part of the biological function of miRNAs. However, all of these experiments involved only a few miRNAs, and the miRNA concentrations were changed dramatically, often in unnatural settings.
Here, we used computational methods to explore the effects of endogenous miRNA expression on endogenous steady-state mRNA levels. Using published microarray data (9, 10), we analyzed the expression of ≈7,000 human miRNA targets that were predicted by the algorithm PicTar (termed PicTar targets) (11) among ≈18,000 mRNAs whose expression was determined in ≈80 human tissues, cell types, and cancer lines. We then related the expression of miRNA targets to that of the miRNAs (12, 13) (P. Landgraf and T. Tuschl, personal communication).
Previously, miRNA targets have been mainly predicted computationally based on pure sequence analysis of 3′ UTRs. These algorithms suggested that conserved human miRNAs target at least 30% of all human genes (11, 14, 15) and that, on average, each miRNA targets ≈200 transcripts. Because these algorithms relied on cross-species comparisons, they almost certainly underestimated the number of miRNA targets. With the discovery of species-specific miRNAs (16–18) that may contribute to molding differences in the mRNA expression in closely related animal species, there is also a need to develop computational methods for identifying miRNA targets without relying on cross-species comparisons. In this article, we show that directly correlating mRNA expression levels with the 3′ UTR motif composition can improve the sensitivity problems of current target prediction algorithms and also indicates that the number of vertebrate miRNA targets seems to be larger than previously estimated.
Results
We extracted a set of altogether 12 miRNAs that seemed highly tissue-specifically expressed (12) (P. Landgraf and T. Tuschl, personal communication) or up-regulated in cancer (13) from miRNA expression profiles and analyzed, for each miRNA, the mRNA expression of its predicted targets across 79 human tissues (see Methods and Table 1). To assess the significance of tissue-specific up- or down-regulation of targets, we followed Lim and colleagues (7). In this analysis, the expression of each mRNA is compared across tissues against a background gene set. We termed this analysis “gene-centric.” The terms “down-regulation” and “up-regulation” in this analysis refer to the relative difference in expression levels of an mRNA across tissues. Fig. 1 shows the significance of tissue-specific up- or down-regulation of PicTar targets for three miRNAs: miR-122, miR-1, and miR-7. These miRNAs are highly specifically expressed in liver, heart/skeletal muscle, and pituitary, respectively. We found that for each of these miRNAs, the tissues in which their predicted targets were most highly significantly down-regulated (relative to all other tissues) matched precisely the tissues in which the miRNAs are specifically expressed. Significance scores for these results ranged between −8 and −17.5, corresponding to p values between 3 × 10−4 and 3 × 10−8. We next asked whether the expression of these miRNA targets was also significantly down-regulated compared with a background set of genes within each tissue (Fig. 4, which is published as supporting information on the PNAS web site). We termed this analysis “tissue-centric” (see Methods). We found that in all three cases, the expression levels of miRNA targets were not significantly lower compared with the background genes (comprising all PicTar targets) in the tissue where the cognate miRNA is expressed (p values of ≈0.5). Together, our results demonstrate that conserved miRNA targets for three highly specifically expressed miRNAs tend to have average expression levels in the tissue where the miRNA is expressed, but that compared with all other ≈78 tissues, their expression is significantly lower in the tissue where the cognate miRNA is expressed. We found similar results for other miRNAs whose expression seems specific to certain tissues, such as testis, heart, and brain, or are up-regulated in cancer (summarized in Table 1). We conclude that a number of highly tissue-specific human miRNAs seem to induce tissue-specific “signatures” of target mRNA expression. In these cases, the signatures are significant enough to computationally predict the tissue of cognate miRNA expression.
Table 1.
Targets of eight highly tissue-specific miRNAs and four miRNAs up-regulated in cancer are expressed at significantly lower levels in the tissue of miRNA expression compared with other tissues
| miRNA (ref.) | Tissue expression | Rank |
|---|---|---|
| miR-122a (12) | Liver | 1 |
| miR-1 (12) | Heart/skeletal muscle | 1, 2 |
| miR-133a (12) | Heart/skeletal muscle | 1, 2 |
| miR-9 (12) | Brain | 1, 4 |
| miR-7 | Pituitary | 1 |
| miR-216 (12) | Pancreas | 4 |
| miR-204 (12) | Testis | 1, 5 |
| miR-223 (12) | Bone marrow/lung | 6, 4 |
| miR-17-5p (13) | Cancer | 1, 2, 5 |
| miR-19a/b (13) | Cancer | 1, 2, 3 |
| miR-18a/b (13) | Cancer | 2, 5 |
| miR-25 (13) | Cancer | 1, 5 |
The second column shows the tissues where the miRNAs (first column) are expressed. The third column reports the rank of the tissues of miRNA expression when sorting all 79 tissues by the significance of how predicted miRNA targets were expressed at lower levels relative to other tissues compared with a background set of mRNAs (gene-centric analysis; see Methods). The last four miRNAs represent all unique families of miRNAs present in the mir-17–92 cluster (13) and in the PicTar data set. Expression data for miR-122a, miR-1, miR-133a, miR-9, miR-7, and miR-216 were provided by P. Landgraf and T. Tuschl (personal communication).
Fig. 1.
Cell-type-specific signatures of miRNAs on target mRNA expression. Shown is analysis of the mRNA expression of predicted targets for three highly tissue-specific miRNAs across 79 human tissues (miR-122, miR-1, and miR-7). Negative values (“scores”; y axis) indicate the significance of predicted miRNA targets to be expressed at lower levels in a tissue relative to all other tissues, compared with a background set of mRNAs (gene-centric analysis; see Methods). Analogously, positive values reflect the significance of miRNA targets to be expressed at high levels compared with other tissues. Tissues are sorted by these scores. Arrows indicate the tissue in which the miRNA is expressed.
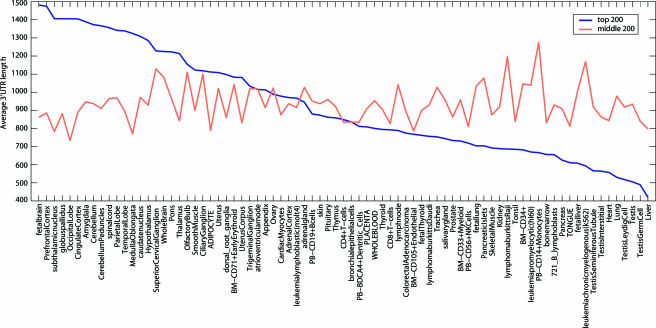
We then performed the gene-centric analysis separately for all human miRNAs in the PicTar data set and clustered miRNAs and tissues based on the significance scores (see Methods). The resulting heat map is shown in Fig. 5, which is published as supporting information on the PNAS web site. We observed that for many miRNAs, their predicted targets are up-regulated in neuronal tissues and blood cells compared with most other tissue types. The same overall effect was obtained by using the expression data by Johnson and colleagues (10) (data not shown). This effect can also be seen when plotting the number of PicTar targets amongst the top n specifically expressed mRNAs in a given tissue as a function of n (Fig. 2A). In neuronal tissues, 50–70% of the highly specifically expressed genes are PicTar targets, whereas in lung, heart, liver, and kidney, ≈25% are PicTar targets. The converse effect holds true for specifically down-regulated genes (Fig. 2B). This general pattern of miRNA target expression correlates with the pattern of 3′ UTR length distribution of specifically expressed genes in these tissues (Fig. 3). On average, a human 3′ UTR is ≈950 nt long. The average 3′ UTR length is ≈1,300 nt for highly expressed neuronal genes but only ≈700 nt for genes specific for a nonneuronal tissue. Again, the converse trend is seen for genes whose expression is specifically low (data not shown). Because fluctuations in 3′ UTR lengths are substantial, we performed a significance test of these trends in 3′ UTR lengths (Fig. 6, which is published as supporting information on the PNAS web site) and found that they are highly statistically significant. Using the data of Johnson and colleagues (10) produced similar results (data not shown), arguing that the observed effects are not artifacts of a particular expression data set.
Fig. 2.
The enrichment/depletion of predicted miRNA targets in highly/lowly expressed mRNAs is tissue-specific. (A) Number of transcripts predicted to be targeted by miRNAs (y axis) as a function of the top n specifically highly expressed genes (x axis) for three neuronal and four nonneuronal tissues. (B) Analogously, number of transcripts predicted to be targeted by miRNAs as a function of the top n specifically lowly expressed genes for three neuronal and four nonneuronal tissues.
Fig. 3.
The average 3′ UTR length of highly specifically expressed mRNAs is tissue-specific. Shown, for each tissue, is the average 3′ UTR length in nucleotides of the top 200 most highly specifically expressed mRNAs (blue curve). Tissues are sorted by these average 3′ UTR lengths. As a control, the average 3′ UTR lengths of 200 genes that were expressed at average levels in each tissue are also shown (red curve).
We then asked whether the mere presence of the central recognition motif, referred to as “nucleus” or “seed” sequence (11, 14, 19, 20), for each of these specific miRNAs in human 3′ UTRs, without any cross-species analysis, was sufficient for observing the tissue-specific “signature” of miRNAs. We note that for each miRNA, the number of 3′ UTRs with at least one nucleus for that miRNA (termed “nucleus 3′ UTR”) is typically a few thousand and thus larger by a factor of 5–20 compared with the number of predicted PicTar targets (i.e., 3′ UTRs with a conserved nucleus) for the miRNA. Similar to the results in Fig. 1, we found that mRNA levels of nucleus 3′ UTRs were significantly lower in the tissue of cognate miRNA expression compared with a background set simply comprising all genes. However, when removing PicTar targets from the set of nucleus 3′ UTRs, the significance of correlations between mRNA expression of nucleus 3′ UTRs and miRNA expression was weakened. For example, for miR-1, the p value dropped from 10−20 to 10−2. These data suggest that PicTar targets, compared with the entire pool of nucleus 3′ UTRs, make a strong contribution to the tissue-specific down-regulation of mRNA levels. However, many nonconserved targets, which are missed by PicTar and other algorithms (14, 21), can be functional [as also noticed in plants (22)] and appear to contribute to these miRNA “signatures.”
The result that nucleus 3′ UTRs correlate with tissue-specific down-regulation logically suggests that transcripts whose 3′ UTRs do not contain the nucleus of a miRNA (termed “nonnucleus 3′ UTRs”) tend to be more highly expressed in the tissues where the miRNA is expressed. We thus assessed the abundance of mRNAs with nonnucleus 3′ UTRs for genes that were highly and tissue-specifically expressed (see Methods). Indeed, for miR-1, miR-122, and miR-7, we found that the top 1,000 genes that were specifically expressed in the tissue where the miRNA is expressed were significantly enriched for genes with nonnucleus 3′ UTRs (p values in the range of 10−8 to 10−3). Even lower p values (10−20 to 10−9) were obtained when repeating the analysis simply on the top 1,000 genes expressed in each tissue.
Further insights into why miRNA targets have been missed by target prediction algorithms became apparent after more closely analyzing the 3′ UTR sequences of genes that had increased mRNA levels after in vivo knockdown of the liver-specific miR-122 in mice (5). In these murine 3′ UTRs, we had previously observed a significantly enhanced number of miR-122 nuclei (5). However, although an overall comparable number of miR-122 nuclei were present in the orthologous human 3′ UTRs, many of them were not aligned to the corresponding mouse nuclei in the available genome alignments, and thus these targets were not predicted by PicTar or other current miRNA target detection programs (14, 20, 21). Analysis of the miR-122 knockdown data (5) suggested that current algorithms (14, 20, 21) miss crudely 50% of likely miR-122 targets. Technical problems such as incomplete mRNA sequences and erroneous alignments, but possibly also compensatory mutations and species-specific gene regulation, are likely responsible for this false negative rate.
Whatever the problems are, the result that the mere presence (absence) of a nucleus sequence in a 3′ UTR seemed to be correlated with relative down-regulation (up-regulation) in the tissue where the miRNA is present led us to propose a simple quantitative model that exploits this correlation and attempts to explain changes in the mRNA levels in miRNA overexpression or knockdown experiments based on 3′ UTR motifs without any cross-species comparisons. In this model, changes in mRNA levels of a given gene (measured by the microarray experiment) are written as a sum over contributions from all sequence motifs in the 3′ UTR of that gene (for example, all possible 4,096 hexamer nucleotide motifs). The contribution of each motif to changes in expression of a gene is modeled as the raw count of the motif in the 3′ UTR multiplied by a fixed coefficient for that motif. Thus, both the presence and absence of all motifs in all 3′ UTRs are correlated with changes in mRNA levels. The motif coefficients are determined by fitting the model simultaneously to all mRNA data for all genes (see Methods). This algorithm is technically equivalent to REDUCE (23), an algorithm that has been previously used to detect functional transcriptional cis-regulatory motifs by correlating motifs in promoter sequences with microarray expression data. We tested this model by analyzing the mRNA microarray data from the miRNA overexpression experiments for miR-1 and miR-124 (7) and the in vivo miR-122 knockdown experiments (5) (Table 2, which is published as supporting information on the PNAS web site). In all three cases, the top-scoring hexamer of all 4,096 hexamers was the nucleus of the miRNA with a p value of 0. Interestingly, the second top scoring hexamer in both overexpression experiments was UAUUUA (p value of 0), which is part of a key AU rich mRNA degradation motif (24) that recently has been linked to miRNA function (25). Notably, three other highly significant hexamers precisely matched the recognition motifs (nucleus) of other known human miRNAs (miR-19a, let-7 family, and miR-27). However, our analysis of all three experiments also discovered several new hexamer motifs that were highly significant but did not match to the nucleus sequence of any currently known vertebrate miRNA. Finally, we estimated that the model can roughly explain changes in mRNA levels for 50% of all genes (see Methods). Because the expression of miRNAs in general can change dramatically during development, these data provide further evidence that miRNAs have a broad and functionally important impact on mRNA levels.
Discussion
By using a purely computational approach that analyzed correlations between existing mRNA expression data, 3′ UTR sequences, and miRNA expression data, we have shown that in natural conditions, several highly tissue-specific human miRNAs appear to specifically down-regulate a large number of mRNA targets. This down-regulation was correlated to previously predicted conserved mammalian miRNA targets, but also (although less significantly) more generally to the mere presence of the recognition motif of the miRNA, the nucleus (or “seed”) sequence in the human 3′ UTRs of target mRNAs. Absence of the nucleus, on the other hand, was positively correlated with expression of miRNAs. This phenomenon can be explained by simple evolutionary arguments: if miRNAs negatively regulate mRNA levels of targets, the mRNAs that should be highly expressed in the presence of the miRNA are under selection pressure to avoid binding sites for this miRNA. We found that although the down-regulation of any specific mRNA that is predicted to be a target of a miRNA is typically weak, the class of all predicted targets of a miRNA can be used to predict the tissue where the miRNA is expressed. We caution that we do not detect these tissue-specific signatures for all highly specific miRNAs. For example, we failed to detect signatures for miR-363 and miR-124 that seem to be highly specifically expressed in the adrenal gland and neuronal cells, respectively (P. Landgraf and T. Tuschl, personal communication) (12). Furthermore, the detection of such signatures becomes extremely complicated in cases where miRNAs are expressed in many tissues (i.e., for the majority of miRNAs) and in cases where different miRNAs could interact with each other. A topic of great interest will be to use refined computational analyses to analyze these cases. We also remark that our finding that many miRNA targets seem to be down-regulated in many nonneuronal tissues compared with neuronal tissues might hint at different modes of miRNA function in neuronal tissues, for example due to differences in Argonaute expression. In fact, it is even possible that this finding could be largely unrelated to miRNAs because we found that highly specifically expressed mRNAs tend to have much longer 3′ UTRs in neuronal tissues compared with most other tissues, and it is impossible for us to decide whether long 3′ UTRs are long because they contain many miRNA target sites or whether they are long for other reasons. We also caution that our analysis only considered mRNA levels of miRNA targets, and we cannot draw any conclusions about translational regulation of miRNA targets. Nevertheless, even just considering mRNA levels, our data suggest the existence of cell-type-specific “signatures” of miRNAs on gene expression. Moreover, although existing target predictions make a strong contribution to these signatures, the number of miRNA targets is larger than previously estimated, and we have shown that by exploiting correlations between mRNA levels of targets and changes in miRNA expression levels, a novel application of reduce (23) can successfully identify likely functional posttranscriptional 3′ UTR motifs. This tool does not rely on cross-species comparisons and thus has high sensitivity. Because experimental methods for stable and specific in vivo knockdown of miRNAs have recently become available (5), we envision that our results and methods will help to shed more light on the exciting universe of miRNA function.
Methods
Data Set of miRNA Targets.
We used human PicTar target transcripts as published by Krek and colleagues (11). These targets were predicted by using the RefSeq data set of mRNA sequences. We chose targets that were conserved between human, chimpanzee, mouse, rat, and dog. Multiple transcripts for the same gene were as best as possible removed, resulting in a set of ≈7,000 unique 3′ UTRs predicted to be targeted by at least one human miRNA.
Microarray Data.
To ensure that our results are not an artifact of how the microarray experiments were carried out, we used two independent datasets [retrieved from the Gene Expression Omnibus (www.ncbi.nih.gov/geo) and the SymAtlas web site (http://symatlas.gnf.org), Gene Expression Omnibus accession numbers GDS594, GDS596, and GSE740]. The first is a set of microarray experiments from Su and colleagues (9). The experiments are performed on custom chips (HG-U133A and GNF1H) that interrogate 44,775 probe sets. Of these, we could map ≈18,711 to the set of transcripts used for PicTar target predictions. The expression of the transcripts was measured across 79 human tissues. Each tissue was assayed twice. In our analysis, we averaged the logarithm of the expression values for probes of the same transcript and then averaged across tissue replicates. The second data set used was from microarray experiments performed by Johnson and colleagues (10). In this case, the probe set targeted exon–exon junctions for all human RefSeq mRNA sequences that have at least one exon–exon junction and have a genomic contig in the LocusLink database. Approximately 125,000 different 36-nt probes targeting exon–exon junctions were used. This data set described expression of ≈10,000 RefSeq transcripts across 52 tissues. The expression level of every transcript was measured by two replicates from each exon–exon junction. A gene’s expression level was given by the median intensity of its exon–exon junction probes. In our analysis, we removed three tissues, two in which 50% of the data were missing (heart interventricular septum and thymus normal) and one that was mislabeled. Furthermore, we removed 248 transcripts in which 25% of the values were missing, leaving 10,160 transcripts. To compute the expression level per transcript, we averaged the logarithm of the expression values, and then averaged across tissue replicates.
Statistical Methods for Analyzing the Expression of PicTar Targets Across Tissues.
We used two complementary ways of analyzing the expression of miRNA targets across mRNA microarray data. In the gene-centric method, the expression of miRNA targets was analyzed, gene by gene, across all tissues and compared with a background set of genes treated identically. In the tissue-centric method, the expression of miRNA targets in a particular tissue was compared only with expression values for a background set of genes in the same tissue. A nonparametric test, described in Nonparametric Statistical Test and Scores, was used to quantify the significance of the comparisons. To define background sets of genes, we experimented with three different large sets of genes. One contained all PicTar targets, another contained all genes, and the last comprised all genes after removing PicTar targets. We used the set of all PicTar targets as background because it gave the best results (Fig. 1 and Tables 1 and 2).
Nonparametric Statistical Test and Scores.
Tissue-centric application of the Wilcoxon rank sum test.
For each tissue, a vector of expression values was compared with a vector of expression values obtained from the background gene set. The nonparametric one-sided Wilcoxon rank sum test was used to assess the significance of the difference in the median distribution of values in the two vectors.
Gene-centric application of the Wilcoxon rank sum test.
Following Lim and colleagues (7), for each target gene, expression levels are ranked among tissues, resulting in a vector for each tissue that contains the tissue’s rank for every gene in the set. The significance of the difference in the median distribution in this tissue vector with that of a similarly generated vector obtained from the background set was assessed by using the Wilcoxon one-sided test.
The score was defined as the negative natural logarithm of the p value of the test. When the p value of the “less than” one-sided test was the smallest, the score was reported as a negative score. Thus, positive (negative) scores quantify the significance of a higher (lower) median mRNA expression of a given set of miRNAs targets compared with background. All statistical tests were carried out as implemented in R with default settings.
Clustering and Heat Map Generation.
Two-way clustering was performed based on the linear correlation between scores and displayed as a heat map by using default implementations in R. To make the heat map more readable, for each miRNA, scores within 1 SD from the median were removed after clustering.
3′ UTR Sequence Extraction and Length Analysis.
Human and mouse 3′ UTRs were extracted from the RefSeq database as described by Krek and colleagues (11) (human) and Krutzfeldt and colleagues (5) (mouse). The number of genes we could map to the probe tags of Johnson and colleagues (10) was 9,318, and 18,029 for the data of Su and colleagues (9). The Wilcoxon rank sum test was always performed against the background of all genes that could be mapped to a particular data set. The p values with the settings “less than” and “greater than” were calculated, and the smaller p value was chosen. Scores, based on p values, were determined as previously described.
Correlating mRNA Levels of 3′ UTRs with miRNA Recognition Motifs (Nucleus 3′ UTRs) to Expression Data.
For each gene, the median expression was calculated over all tissues, and each expression value associated with the gene was normalized by this median. For every tissue, the Wilcoxon test (see above) was used to assess the significance of the difference in the median of the expression levels of genes containing the nucleus for a given miRNA compared with that of all genes.
Correlating mRNA Levels of 3′ UTRs Without miRNA Recognition Motifs (Nonnucleus 3′ UTRs) to Expression Data.
Separately for miR-1, miR-122, and miR-7, we recorded the number of mRNAs that did not contain a single nucleus in 3′ UTRs (nonnucleus 3′ UTRs) among (i) the 1,000 genes most highly and specifically expressed in the tissues where these miRNAs are present and (ii) the 1,000 most highly expressed genes in the tissues where these miRNAs are present. The probability of observing these counts (or more) by chance was assessed by using the binomial distribution. The parameter p of this distribution (single-event probability) was the total number of nonnucleus 3′ UTRs divided by the number of all 3′ UTRs.
A Model for Correlating 3′ UTR Motifs with Changes in mRNA Levels in miRNA Knockdown or Overexpression Experiments.
The model is that motifs within 3′ UTRs make a linear contribution (either enhancing or inhibitory) to mRNA levels. The significant motifs are chosen one by one, by determining, in every iteration, which motif’s contribution brings about the greatest reduction in the difference between the model and the expression data. Motifs reported in Table 2 are ordered by these iteration rounds. Each of these motifs can be assigned a p value, and the procedure continues as long as the p value is lower than some chosen threshold. This iterative procedure for finding significant sequence motifs that correlate with changes in mRNA expression is entirely equivalent to the REDUCE algorithm as described by Bussemaker and colleagues (23) and was implemented in Perl. The occurrences of motifs of length 6 best modeled the data. The p-value cutoff used was 0.01. To estimate the extent to which changes in mRNA levels can be explained by the model, we calculated for each gene the difference between the logarithm of the ratio of the changes in mRNA expression values as measured in the experiment and the log ratio predicted by the model. We flagged the mRNA change of this gene as “explained” when this difference had the correct sign and was, by absolute value, smaller than 0.5 in logarithm base two units, corresponding to a fold-change error of 1.4. In all three experiments, ≈50% of all genes and 50–70% of up- or down-regulated genes were explained by these criteria.
Supplementary Material
Acknowledgments
We thank Tom Tuschl and Pablo Landgraf for providing unpublished miRNA expression data, Josep Ramon Goñi Macia for extracting overexpression microarray data that we analyzed, Sebastián Pueblas for preparing figures, Ken Kosik for discussions, and Tom Tuschl for helpful suggestions on the manuscript. This work was supported by a National Institutes of Health grant (to N.R.), Fondo Investimenti Ricerca di Base–Ministero Instruzione Universita Ricerca 2001 (RBNEO15MPB_001/RBNE01KXC9_006) (to G.M.), and Swiss National Fund Grant 205321-105945 (to M.Z.).
Abbreviation
- miRNA
microRNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Du T., Zamore P. D. Development (Cambridge, U.K.) 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 4.Chen C. Z. N. Engl. J. Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 5.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 6.Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 7.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson J. M., Castle J., Garrett-Engele P., Kan Z., Loerch P. M., Armour C. D., Santos R., Schadt E. E., Stoughton R., Shoemaker D. D. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 11.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., et al. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 12.Baskerville S., Bartel D. P. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., et al. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis B. P., Burge C. B., Bartel D. P. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V., Lindblad-Toh K., Lander E. S., Kellis M. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F. A., van Dyk L. F., Ho C. K., Shuman S., Chien M., et al. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 17.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., Cuppen E. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Lai E. C. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 20.Brennecke J., Stark A., Russell R. B., Cohen S. M. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Bussemaker H. J., Li H., Siggia E. D. Nat. Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 24.Zubiaga A. M., Belasco J. G., Greenberg M. E. Mol. Cell. Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S. C., Gram H., Han J. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.