Abstract
Spontaneous chromosomal breakages frequently occur at genomic hot spots in the absence of DNA damage and can result in translocation-related human disease. Chromosomal breakpoints are often mapped near purine–pyrimidine Z-DNA-forming sequences in human tumors. However, it is not known whether Z-DNA plays a role in the generation of these chromosomal breakages. Here, we show that Z-DNA-forming sequences induce high levels of genetic instability in both bacterial and mammalian cells. In mammalian cells, the Z-DNA-forming sequences induce double-strand breaks nearby, resulting in large-scale deletions in 95% of the mutants. These Z-DNA-induced double-strand breaks in mammalian cells are not confined to a specific sequence but rather are dispersed over a 400-bp region, consistent with chromosomal breakpoints in human diseases. This observation is in contrast to the mutations generated in Escherichia coli that are predominantly small deletions within the repeats. We found that the frequency of small deletions is increased by replication in mammalian cell extracts. Surprisingly, the large-scale deletions generated in mammalian cells are, at least in part, replication-independent and are likely initiated by repair processing cleavages surrounding the Z-DNA-forming sequence. These results reveal that mammalian cells process Z-DNA-forming sequences in a strikingly different fashion from that used by bacteria. Our data suggest that Z-DNA-forming sequences may be causative factors for gene translocations found in leukemias and lymphomas and that certain cellular conditions such as active transcription may increase the risk of Z-DNA-related genetic instability.
Keywords: DNA repair, genetic instability, non-B DNA, translocation
Chromosomal translocations leading to carcinogenesis may be associated with normal metabolic processes as well as DNA damage caused by endogenous or environmental genotoxic agents. The mechanism(s) responsible for these translocations are poorly understood, although the prevailing thought is that DNA double-strand breaks (DSBs) are required. In the majority of cases, translocations occur at common regions, or “hot spots,” that are sensitive to DNA breakage. We have previously found that an H-DNA-forming sequence from the human c-MYC gene induces genetic instability in mammalian cells (1), suggesting that non-B-DNA structures contribute to genetic instability and perhaps to tumor development. Furthermore, several reports have demonstrated a correlation between Z-DNA-forming sequences and chromosome breakage hot spots on tumor related-genes such as human BCL-2 (2, 3), c-MYC (4, 5), and the SCL gene (6), suggesting that Z-DNA may be involved in chromosomal breakage and translocation.
Z-DNA is a left-handed helix with a zigzag arrangement of the backbone of the DNA molecule that differs from the canonical Watson–Crick B-DNA. Because purine bases can adopt a syn conformation (7), specific sequences that have alternating purine–pyrimidine regions are typically the most readily converted into Z-DNA structures both in vitro and in vivo (8, 9), although many other sequences also have been shown to adopt this conformation (10). Formation of Z-DNA removes negative supercoiling in the surrounding region of the DNA, and the energy of supercoiling stabilizes the Z-DNA conformation. Sequences with the potential to adopt Z-DNA are abundant in eukaryotic genomes, occurring ≈1/3,000 bp in the human genome (11, 12).
Formation of Z-DNA has been found to stimulate homologous recombination by releasing the level of DNA supercoiling in human cells in culture (13–17). Z-DNA-forming CG repeats can induce small deletions in bacteria, likely generated by misalignment related to the formation of hairpin structures on individual DNA strands during either semiconservative DNA replication or refilling synthesis in the repair process (18–21). However, the mutagenic potential of Z-DNA in mammalian cells has not been reported, and the previously proposed mechanisms of slipped-strand misalignment and homologous recombination are not probable explanations for a potential role of Z-DNA in the generation of DNA DSBs leading to gene translocations.
In this work, we tested the ability of Z-DNA-forming sequences to induce genetic instability in both bacterial and mammalian cells. Based on the findings presented here, we propose that Z-DNA can be processed by repair mechanisms in mammalian cells, resulting in replication-independent DNA strand breakages and large-scale deletions or rearrangements. These types of genomic alterations are relevant to the chromosome breaks and gene translocations that map near Z-DNA-forming sequences in human leukemia and lymphoma.
Results
Inserts in Mutation Reporter Shuttle Plasmids.
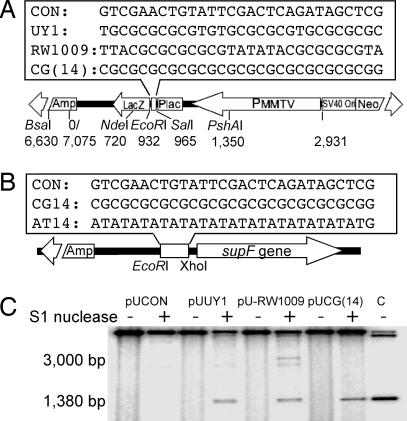
To test the mutagenic potential of Z-DNA structures in both bacterial and mammalian cells, we constructed a shuttle vector, pUCNIM, containing the lacZ′ gene expressing the amino-terminal fragment of β-galactosidase as a mutation reporter gene. Sequences to be tested were inserted into the pUCNIM plasmid after EcoRI/SalI digestion (between the promoter and the coding region of lacZ′) to maintain LacZ′ function (Fig. 1A), blue colonies containing a functional lacZ′ gene were screened by restriction analysis, and the inserts were verified by DNA sequencing (see Fig. 6, which is published as supporting information on the PNAS web site). The insertion of AT(14) inactivated the lacZ′ reporter gene; therefore, AT(14), CG(14), and control sequences were cloned into the pSP189 supF mutation-reporter plasmid after EcoRI/XhoI digestion (4 bp upstream of the supF reporter gene) (Fig. 2B). Thus, a series of reporter plasmids were constructed: pUCON, pUCG(14), pUUY1, and pU-RW1009 were prepared by using the pUCNIM backbone carrying lacZ′ as a reporter gene, and pSCON, pSCG(14), and pSAT(14) were constructed by using the pSP189 backbone carrying the supF gene as a mutation reporter.
Fig. 1.
Z-DNA structures formed at the inserts. (A and B) Schematic structure of the mutation reporter shuttle plasmid, pUCNIM (A) and pSP189 (B). (Upper) Positions and sequences of inserts are shown. (Lower) The diagnostic restriction sites used in this study are listed. (C) Identification of the Z-DNA conformation at the insertion site. The insert-containing plasmids were treated with single-stranded DNA-specific S1 nuclease followed by BsaI digestion. The ≈3,000-bp fragment and a shorter ≈1,380-bp fragment released by S1 nuclease cleavage from pUCON and Z-DNA forming plasmids, respectively, are indicated. Lane C is a 1,377-bp BsaI–EcoRI fragment as a size standard.
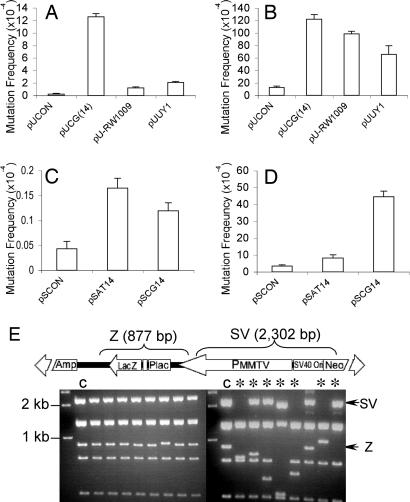
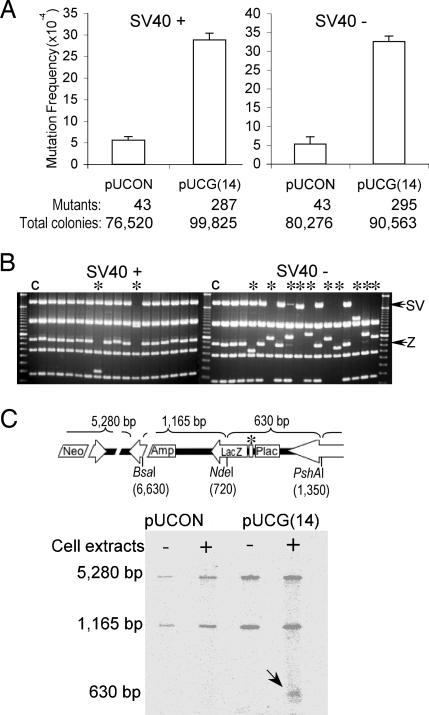
Fig. 2.
Z-DNA-induced mutations in bacterial cells and mammalian COS-7 cells. (A) Z-DNA-induced lacZ′ mutation frequencies in DH5α cells. (B) Z-DNA-induced lacZ′ mutation frequencies in mammalian COS-7 cells. (C) Z-DNA-induced supF mutation frequencies in DH5α cells. (D) Z-DNA-induced supF mutation frequencies in mammalian COS-7 cells. A total of >100,000 colonies were counted in each group. Error bars show the SEM of three or four independent experiments. (E) pUCG(14) mutants derived from DH5α cells (Left) and COS-7 cells (Right) were digested with EagI and BssSI, resulting in seven fragments. The 877-bp fragment containing the Z-DNA-forming sequence (Z) and the 2,302-bp fragment between the Z-DNA locus and SV40 Ori (SV) are shown schematically (Upper) and marked with arrows in the gel. ∗ indicates large-scale deletions (≥50 bp); C refers to control sample.
Probing the Z-DNA Structure in Mutation Reporter Plasmids.
To determine whether Z-DNA was formed in the plasmids, single-strand-specific S1 nuclease was used to cleave the unwound region exposed at the B–Z junction (22, 23) as described in ref. 24. The SV40 replication origin contains potential cruciform structures and is also accessible to S1 nuclease (25, 26). As shown in Fig. 1C, under the conditions of our assay, S1 nuclease cleaves the control plasmid, pUCON, at the SV40 Ori site ≈3 kb away from the BsaI site. However, in the presence of the Z-DNA-forming sequences, S1 nuclease cuts predominantly at the inserts, producing radiolabeled BsaI–S1 fragments of ≈1,380 bp, the same length as the BsaI–EcoRI fragment shown in lane C, indicating B–Z junctions formed near the EcoRI site.
Z-DNA-Forming Sequences Induce Small Deletions in Bacteria.
We determined the mutagenic potential of Z-DNA in bacterial cells to serve as a control for the background mutation frequency in the mutagenesis assay in mammalian cells. LacZ′ reporter plasmids, pUCG(14), pUUY1, pU-RW1009, and their control pUCON, and supF-based plasmids, pSCON, pSCG(14), and pSAT(14), were introduced into DH5α or MBM7070 cells, respectively, and the mutation frequencies were measured. As expected, an ≈50-fold increased mutation frequency was observed in the pUCG(14) plasmid in bacterial cells. Restriction and sequencing analyses showed that all characterized mutants induced by CG(14) in Escherichia coli were small deletions or expansions within the repeats (Fig. 2E Left; see also Table 2, which is published as supporting information on the PNAS web site), as previously reported in a different system (19). Interestingly, the sequences UY1 and RW1009, which have interruptions in the CG repeat to reduce (but not completely prevent) the slippage events, exhibited only 5- to 10-fold increased mutagenesis over that seen with pUCON (P < 0.001) in the lacZ′ gene, which is substantially less than that induced by CG(14) (Fig. 2A). In addition, the proportion of small deletions in UY1- and RW1009-induced mutants in bacteria were greatly reduced (50% and 60%, respectively; data not shown), consistent with their reduced length of direct repeat sequence. Because the supF gene encodes for a tRNA and not a protein, frameshift mutations cannot be detected; thus, it is not sensitive to small insertions or deletions near the insert. Both CG(14) and AT(14) induced only a slight increase (2- to 3-fold) in the frequency of supF mutations (Fig. 2C).
Z-DNA Does Not Block DNA Replication in HeLa Cell Extracts.
Because mutants produced during plasmid amplification in DH5α also were transfected into COS-7 cells and were detected as mutants in the assay, these events must be taken into account to calculate the Z-DNA-induced mutation frequency in mammalian cells. One prerequisite for discounting these E. coli-derived mutations from the total number found in COS-7 cells is that the mutants (containing ≤14 CG repeats) were replicated in mammalian cells at the same rate. In replication assays using HeLa cell extracts supplied with SV40 large T antigen, the incorporation of [α-32P]dCTP into pUCG(14) and pUCON was similar (≈1.2- to 1.3-fold above that of control plasmid, pSP189; P = 0.62) (Table 1). A similar result was seen with DpnI-digested plasmids (data not shown). The increase in [α-32P]dCTP incorporation above that in pSP189 likely reflects the differences in the length of pUCG(14)/pUCON and pSP189 (7 vs. 5 kb). This result indicates that Z-DNA does not block DNA replication and validates the subtraction of the number of small deletions within the repeats as background to correct for the mutation frequencies detected in mammalian cells.
Table 1.
Plasmid replication efficiency in HeLa cell extracts
| Plasmid | Experiment no. |
Mean | SE | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| pUCON/pSP189 | 1.27 | 1.16 | 1.33 | 1.25 | 0.086 |
| pUCG(14)/pSP189 | 1.19 | 1.15 | 1.31 | 1.21 | 0.078 |
The [α-32P]dCTP incorporation in nascent plasmids was quantified and normalized to that of the control plasmid pSP189, which is given a value of 1.0. The mean and SE from three independent repeats are shown.
Z-DNA Induces Large-Scale Deletions and Rearrangements in Mammalian Cells.
The Z-DNA-forming plasmids, pUCG(14), pUUY1, and pU-RW1009, or pSCG(14) and pSAT(14) and their controls, pUCON or pSCON, respectively, were introduced into mammalian COS-7 cells and recovered 48 h later. After DpnI treatment, to remove plasmids that had not replicated in mammalian cells, total mutants (including those occurring in both COS-7 cells and in bacterial cells) were measured by blue/white screening. Because the control plasmids replicated in mammalian cells at a similar rate as the Z-DNA-containing pUCG(14), we corrected the Z-DNA-induced lacZ′ mutation frequencies in COS-7 cells by subtracting the small deletion mutants that occurred in DH5α (Fig. 2A) from the total number of mutants generated. As shown in Fig. 2B, the corrected Z-DNA-forming sequence-induced lacZ′ mutation frequency was significantly greater than the control (P < 0.001, compared with pUCON). Sequencing and restriction analyses revealed the structures of the Z-DNA-induced lacZ′ mutants. Strikingly, ≈95% of the Z-DNA-forming sequence-induced mutants detected in COS-7 cells were large deletions or rearrangements (Fig. 2E Right). Most deleted regions spanned the Z-DNA-forming sequences and the SV40 replication origin, an area that has been shown to contain another S1 nuclease sensitive site (see Fig. 1C; see also Fig. 7, which is published as supporting information on the PNAS web site). Moreover, >85% of the junctions analyzed had 1–6 bp of homology (Fig. 7), a characteristic of nonhomologous end-joining repair (27), suggesting that these mutants are the products of DSB repair. Interestingly, although CG(14) induced an ≈13-fold higher mutation frequency in the supF system, consistent with the observation in the lacZ′ system, AT(14) resulted in only a 2-fold increase in mutation frequency above background (Fig. 2D), indicating that AT(14) is either quite stable or induces small deletions that are not detectable in the supF system.
Transcription Increases both the Spontaneous and the Z-DNA-Induced Genetic Instability.
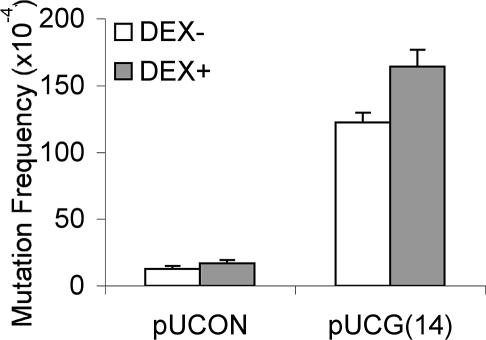
The topology of a given DNA molecule might be highly regulated in vivo, so that the supercoiling level of the DNA may vary temporally and with the rates of transcription and replication. Z-DNA forms transiently behind active RNA polymerases, stabilized by the negative supercoiling generated by an RNA polymerase moving through a gene (4). Sequences capable of adopting Z-DNA structures are more unstable (prone to small deletions) when actively transcribed in bacteria (28). Therefore, we examined whether transcription through CG repeats would increase the Z-DNA-induced mutagenesis in mammalian cells. By using the inducible mouse mammary tumor virus-LTR promoter on the plasmids, transcription through the CG repeat region was induced by addition of dexamethasone (confirmed by RT-PCR; data not shown). As shown in Fig. 3, we found that transcription increased the mutation frequency for the Z-DNA-containing plasmid from 122 × 10−4 to 164 × 10−4 (P < 0.001), as well as for the control plasmid from 13 × 10−4 to 17 × 10−4 (P < 0.05).
Fig. 3.
Transcription increases both spontaneous and Z-DNA-induced mutagenesis. After transfection with pUCON or pUCG(14) plasmids, COS-7 cells were incubated for 48 h in the absence or presence of 10 μM dexamethasone. Mutants in the recovered plasmids were screened. A total of >200,000 colonies were counted in each group. Error bars show the SEM of three or four independent experiments.
Z-DNA Induces DNA DSBs in Mammalian Cells.
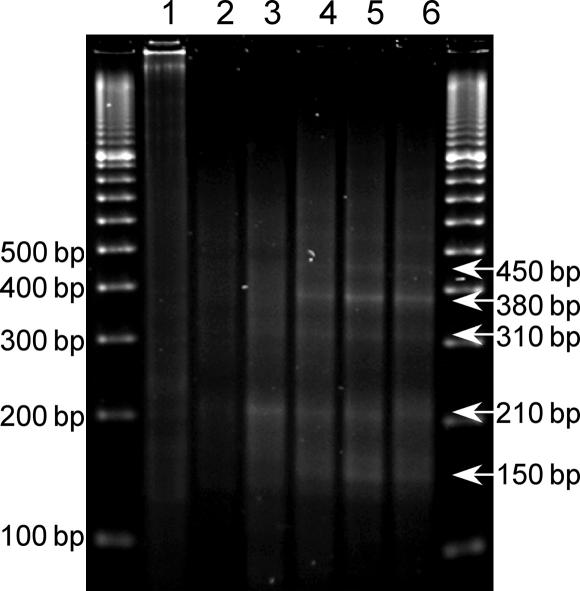
To test our hypothesis that the deletion mutants are the products of DSB repair, a linker-mediated PCR (LM-PCR) assay (1) was used to detect DSBs generated on plasmids recovered from COS-7 cells. By using a primer located 171 bp upstream of the CG(14) insert (EcoRI site), we mapped the DSBs induced on plasmids after replication in COS-7 cells for 4, 8, 24, and 48 h. On plasmids recovered 4 h after transfection, a major band of 210 bp was detected, indicating a breakage hot spot in the CG(14) sequence (Fig. 4, lane 3). This band decreased with increasing time of incubation. Products of 150, 310, 380, and 450 bp (Fig. 4, lanes 4–6), which were undetectable after 4 h of incubation, increased with increasing time of incubation (the corresponding breakpoints were located 60 bp upstream, and 100, 170, and 240 bp 3′ downstream of the EcoRI site, respectively). However, the CG(14) sequence did not induce DSBs in bacterial cells (Fig. 4, lane 1), consistent with the previous proposal that small deletions result from misalignment related to hairpin structure formation during replication in E. coli (19).
Fig. 4.
LM-PCR analysis of Z-DNA-induced DSBs. LM-PCR products were subjected to agarose gel electrophoresis and visualized by ethidium bromide staining. Lane 1, pUCG(14) prepared from DH5α cells; lane 2, pUCON replicated in COS-7 cells for 48 h; lanes 3–6, pUCG(14) replicated in COS-7 cells for 4, 8, 24, and 48 h, respectively. Sizes of detected bands are listed on the right.
DNA Replication-Independent Z-DNA-Induced DNA Breakages and Large-Scale Deletions in Mammalian Cells.
In bacterial cells, Z-DNA-forming CG repeats have been proposed to induce small deletions due to misalignment during DNA replication (19). To reveal the relationship between DNA replication and Z-DNA instability in mammalian cells, the mutagenic potential of pUCG(14) was determined in an in vitro mutagenesis assay by using SV40-negative HeLa cell-free extracts, in which replication of these reporter plasmids relies on the addition of SV40 large T antigen. As shown in Fig. 5A, the CG(14) sequence is mutagenic in HeLa cell-free extracts (P < 0.001), regardless of the presence or absence of SV40 T antigen. Approximately 88% of characterized mutants found in replicated plasmids are small deletions in the repeats, and ≈42% of mutants from unreplicated plasmids are small deletions (Fig. 5B). However, approximately half of all characterized mutants were generated in bacteria, as shown in Fig. 2A. After subtraction of the background mutations of small deletions (those that occurred in the bacteria), ≈70% of mutations occurring in replicated plasmids are small deletions, whereas >90% occurring in the absence of replication are large deletions (≥50 bp).
Fig. 5.
Replication-dependent and independent Z-DNA-induced genetic instability in mammalian cell extracts. (A) Replication-proficient (Left) and replication-deficient (Right) Z-DNA-induced mutagenesis in HeLa cell extracts. The number of mutants observed and the total number of colonies screened are listed below the corresponding bars. Error bars show the SEM of three independent experiments. (B) Restriction analysis of mutants generated in replicated (Left) and unreplicated (Right) systems. ∗ refers to large-scale deletions (≥50 bp); C refers to control sample. (C) Z-DNA-induced cleavages in HeLa cell extracts in the absence of replication. The positions of three restriction sites on the plasmid, pUCNIM, are shown schematically above. The Z-DNA locus is marked with ∗. A radiolabeled 630-bp fragment containing breakages generated in HeLa cell extracts is indicated by an arrow.
We also analyzed the Z-DNA-induced breakages in HeLa cell extracts. As shown in Fig. 5C, after incubation of the pUCG(14) plasmids with SV40-negative HeLa cell extracts, single-strand breaks in the 630-bp CG(14)-containing fragment were detected by postlabeling. This fragment was not detected on the control plasmids incubated with HeLa cell extracts or pUCG(14) prepared from DH5α cells (without incubation in HeLa cell extracts). These results demonstrate that DNA strand breaks were induced by the Z-DNA-forming sequence in HeLa cell extracts independently of replication.
Discussion
To date, at least 10 different types of non-B DNA conformations have been identified, and some conformations have been associated with various types of genetic instability in vivo, resulting in gene alterations, and disease development. For example, inverted repeats can result in deletions in E. coli (29, 30); the breakpoints of t(11;22) translocations in human cells localize at the center of AT-rich palindromic sequences on 11q23 and 22q11 (31–33); long CNG triplet repeats are unstable and can result in expansions and/or deletions in vivo (34); a 2.5-kbp polypurine–polypyrimidine sequence from the human PKD1 gene, known to form non-B DNA structures, can induce deletions and other instabilities (35). We have also demonstrated recently that H-DNA structures are intrinsically mutagenic in mammalian cells, leading to the generation of DSBs (1).
In this study, we found that Z-DNA-forming sequences are unstable in both bacterial and mammalian cells, but the induced mutation spectra are strikingly different. In mammalian cells, >95% of the mutations are large-scale deletions or rearrangements, whereas the majority of Z-DNA-induced mutations in bacteria are small deletions within the repeats. Z-DNA forms at alternating purine–pyrimidine repeats, particularly readily at CG repeats. However, the CG repeats (in theory) also can form hairpin or slippage structures, which are also potentially mutagenic. Comparing the stability of a series of sequences with different capabilities to adopt Z-DNA and/or slippage/hairpin structures can help distinguish which structure is involved in the mutagenesis. The Z-DNA-forming capability of the sequences used in this study are CG(14) > RW1009 (36) > AT(14) (37), and hairpin-structure-forming capability are AT(14) > RW1009 > CG(14). The fact that CG(14) induced more large-scale deletions in COS-7 cells than RW1009 and AT(14) suggests that it is Z-DNA, and not hairpin structures, that induces large-scale deletions in mammalian cells. Alternatively, small deletions within the repeats were the predominant mutations induced by the Z-DNA-forming sequences in bacteria. This finding is consistent with a previous study reporting that CG repeats induced small deletions within the repeats in the E. coli strain JM103 due to slippage events during DNA replication (19). Supporting this idea, UY1 and RW1009, which contain GT or AT inserts to interrupt the direct CG repeats, showed significantly lower frequencies of small deletions in bacteria, compared with pUCG(14). Furthermore, DSBs were found surrounding the CG(14) repeats in the plasmids recovered from COS-7 cells or in the plasmids incubated with HeLa cell extracts, but not in the plasmids prepared from DH5α cells. These data indicate that Z-DNA-forming sequences are processed in a strikingly different fashion in prokaryotic and eukaryotic cells, but the reason(s) for the difference is still not clear.
In a Z-DNA structure, the guanosine nucleotides are in syn position, where the bases are located over the sugar without protection and are more accessible to DNA-damaging factors. Conversely, the damages occurring to the DNA in a Z-DNA conformation are more resistant to processing by DNA repair enzymes (38, 39). Thus, it is possible that endogenous (e.g., oxidative) damage accumulates at Z-DNA regions, resulting in an increased mutation frequency. However, the majority of mutations found in COS-7 cells are large deletions or rearrangements, and not the typical spontaneous point mutations generated by oxidative damage. Sequencing analysis revealed that most (85%) large-scale deletion mutants generated in mammalian cells contain microhomologies at the junctions, indicating that they are the products of nonhomologous end-joining repair. Moreover, LM-PCR provided direct evidence that DSBs were generated surrounding the CG(14) repeats in the plasmids recovered from COS-7 cells. Thus, the accumulation of endogenous (oxidative) damage and/or resistance to repair are not likely reasons for the DSBs and large-scale deletions found in our study.
Alternatively, the breakages observed could be introduced by the stalling of DNA polymerase during replication due to the unusual non-B DNA structure. However, CG(14) repeats did not show significant effects on DNA replication in our replication assay. Furthermore, the Z-DNA-induced DNA breakages and large-scale deletions were detected in the HeLa cell extracts in the absence of DNA replication, making it unlikely that the breakages and mutations are the result of DNA polymerase stalling. Interestingly, we found that in the presence of SV40 antigen to support replication in HeLa cell extracts, more small deletions were generated, which is not the predominant mutation in COS-7 cells. This finding on one hand is consistent with the model that CG(14) induces small deletions/expansions during replication; on the other hand, it suggests that the large-scale deletions may be generated in vivo in the absence of replication. The observation that plasmids in HeLa cell extracts undergo more small deletions than in COS-7 cells, particularly when supplied with SV40 large T antigen, may reflect the difference of the extract versus cellular systems used. Thus, we propose that Z-DNA is processed by structure-directed nucleases or repair proteins in mammalian cells, resulting in DSBs in the CG repeat in an early stage (first 4 h after transfection), and is then further processed by repair factors to generate breakages surrounding the Z-DNA-forming sequence as the time after transfection increases from 4 to 48 h. However, the proteins responsible for the recognition and cleavage at the Z-DNA structure are not yet known. Several Z-DNA-specific binding proteins have been identified, such as double-stranded RNA adenosine deaminase (ADAR1), DLM-1, and E3L (reviewed in ref. 40), but these protein activities are not generally associated with nuclease activity. Further work is required to identify the proteins that cleave at these non-B DNA structure-forming sequences.
Z-DNA is a transient structure (40) that can be induced by both normal biological as well as pathological activities. The movement of RNA polymerase along the DNA generates negative supercoiling behind it, and Z-DNA can form near promoter regions in actively transcribed genes (4, 41, 42). Thus, the transcription-induced Z-DNA formation might be responsible for mutagenesis. Consistent with this idea, transcription initiated by a tetracycline-inducible promoter increases the frequency of small deletions at a Z-DNA-forming sequence in bacteria (28). Conversely, the positive supercoiling generated in front of RNA polymerase could unwind the Z-DNA structure and thus be expected to reduce its mutagenic potential. In the present study, we found that active transcription increases the mutation frequencies of both the control plasmid and the Z-DNA-containing plasmid proportionally in COS-7 cells [from 13 × 10−4 to 17 × 10−4 (P < 0.05), and 122 × 10−4 to 164 × 10−4 (P < 0.001), respectively]. This finding suggests that transcription has a general effect on genetic instability irrespective of the formation of Z-DNA.
Together, our results suggest that a previously undescribed replication-independent, structure-specific cleavage process at Z-DNA-forming sequences occurs in mammalian cells, resulting in DSBs followed by error-prone nonhomologous end-joining. This model provides an explanation for the colocalization of Z-DNA-forming sequences and breakpoints on translocated genes in many human diseases.
Materials and Methods
Construction of Z-DNA-Forming Plasmids.
An SV40 replication origin, a neomycin resistance gene, and a dexamethasone-inducible mouse mammary tumor virus-LTR promoter (restricted from pMAMneo) were cloned into pUC19 to generate a 7,075-bp mutation reporter shuttle plasmid, pUCNIM. Sequences used in this study are shown in Fig. 1. A CG repeat was selected as a model Z-DNA sequence in this study because it has been shown to adopt a Z-DNA structure more readily than other sequences of the same length (36, 37, 43). However, the CG(14) sequence is both an inverted and direct repeat and, in theory, also can form a hairpin or a slippage loop structure. To control for this possibility, a series of sequences with different capabilities to adopt Z-DNA or slippage/hairpin structures were included in this study. GT or AT interruptions were introduced in the pure CG repeating sequence to reduce the slippage events at the long direct repeat. RW1009 contains the sequence ATATAT within the CG repeats. AT repeats have a much higher propensity to adopt hairpin structures than Z-DNA; thus, in principle, RW1009 is more likely to form hairpins than is CG(14), although a Z-DNA conformation is still favored (36, 44). A sequence containing AT repeats [AT(14)] was selected as a sequence with a high propensity to adopt hairpin/slipped structures and a low propensity to adopt Z-DNA (37). The UY1 sequence contains GT interruptions to segregate CG repeats into three short blocks (≤4 repeats) to reduce the potential hairpin/slippage events occurring on the direct repeats. This sequence maintains the purine–pyrimidine alteration, so that a Z-DNA conformation is still favored. Oligonucleotides listed in Fig. 1 (Midland Certified Reagents, Midland, TX) were annealed with their complementary strands, phosphorylated with T4 polynucleotide kinase and ATP, and inserted at an EcoRI(932)–SalI(965) cassette between the promoter and the coding region of the lacZ′ gene on pUCNIM or into the pSP189 plasmid after EcoRI/XhoI digestion (4 bp in front of the supF mutation-reporter gene). Clones resulting from the transformation of DH5α cells were screened by restriction analysis and verified by sequencing. Recombinants were named according to the inserts (listed in Fig. 1A), for example, pUCNIM-derived pUCON, pUCG(14), pUUY1 and pU-RW1009, and pSP189-derived pSCON, pSCG(14), and pSAT(14). By using these mutation reporter shuttle plasmids, we were able to detect the mutations in the lacZ′ or supF reporter genes by facile blue/white screening in DH5α or MBM7070 cells, respectively.
Analysis of Z-DNA Conformations in the Modified Plasmids.
In the presence of Z-DNA, single-stranded areas are formed at the B-Z junctions (22, 23), thus the formation of Z-DNA structures can be examined by using S1 nuclease sensitivity assays, as described in ref. 24. After S1 nuclease digestion, the plasmids were digested with BsaI (at position 6,630), treated with calf intestinal phosphatase, radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP, separated on a 1.2% agarose gel, and subjected to autoradiography for visualization. The plasmid backbone labeled at the BsaI site served as a loading control. The lengths of the radiolabeled BsaI–S1 fragments indicated the positions of the S1 nuclease-sensitive sites relative to the BsaI restriction site.
Z-DNA-Induced Mutagenesis of the lacZ′ Gene in Mammalian COS-7 Cells.
COS-7 cells were transfected with the Z-DNA-forming plasmid, pUUY1, pU-RW1009, and pUCG(14), or the control plasmid pUCON, using the Nucleofector kit V (Amaxa, Cologne, Germany) according to manufacturer’s recommendations and incubated with medium alone or medium containing dexamethasone at a final concentration of 10 μM. After 48 h, the amplified plasmids were recovered by the method of Hirt (45). After treatment with DpnI to remove those plasmids that were not replicated in the COS-7 cells, the plasmids were transfected into DH5α cells to detect the total lacZ′ mutants, or into MBM7070 cells to detect the supF mutants, by using a blue/white screen. Z-DNA-induced mutation frequencies in COS-7 cells were corrected by subtracting the number of mutants that occurred in bacteria (measured by directly transforming the plasmids into E. coli) from the total mutants. Differences in mutation frequencies compared in this work were analyzed by using the χ2 test, a sample test of binomial proportion (two-tailed; P > 0.05).
LM-PCR Analysis of Z-DNA-Induced DSBs.
LM-PCR analysis was performed as described in ref. 1. Regions between the upstream primer [171–191 bp upstream of the Z-DNA (EcoRI) site] and the linkers were PCR amplified and separated on a 2% agarose gel. The length of each specific band (L) was measured on a Kodak 1D Imaging Station. The positions of the breakpoints relative to the Z-DNA loci (N) were calculated according to the length (L) of the PCR fragments: N = L − 20 (length of linker) − 20 (length of primer) − 171 (distance between the primer and the EcoRI site).
In Vitro Replication and Mutagenesis Assay.
In vitro replication of the SV40 Ori-containing plasmids in HeLa cell extracts was performed by using an SV40 DNA replication assay kit (CHIMERX, Madison, WI) according to manufacturer’s instructions. Briefly, a mixture of 25 ng of pUCG(14), or pUCON plasmid DNA, and 25 ng of pSP189 plasmid DNA was incubated with 180 μg of HeLa cell-free extract supplied with 1 μg of SV40 large T antigen in the reaction buffer {30 mM Hepes, pH 7.5/7 mM MgCl2/0.5 mM DTT/4 mM ATP/100 μM each of dNTPs/50 μM each of rNTPs/40 mM phosphocreatine/0.625 units of creatine phosphokinase/1 μCi [α-32P]dCTP (1 Ci = 37 GBq)} at 37°C for 8 h. Plasmids were purified from cell extracts and either digested with DpnI or left untreated. The plasmids were then linearized by XmnI digestion, separated on a 0.8% agarose gel, and then subjected to autoradiography. The [α-32P]dCTP incorporation into the pUCG(14) or pUCON plasmids was compared with that of pSP189, which has a similar SV40 Ori. For the in vitro mutagenesis assay, 50 ng of plasmid DNA was incubated in the HeLa cell extract with or without SV40 large T antigen in the replication buffer for 8 h without [α-32P]dCTP. Mutants derived during incubation were analyzed as described above, plus or minus DpnI digestion.
In Vitro Z-DNA-Induced Incision Assay.
Fifty nanograms of plasmid DNA were incubated with 180 μg of HeLa cell-free extract as described for the replication assay for 4 h in the absence of SV40 large T antigen. After purification from cell extracts, the plasmids were digested with BsaI(6, 630) to generate a breakpoint. The 5′ ends produced by both BsaI and HeLa extract then were radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP. The purified plasmid DNA was further digested with NdeI(720) and PshAI(1, 350) to release a 630-bp fragment containing either the Z-DNA-forming insert or control sequence. Products were separated on a 1.2% agarose gel and subjected to autoradiography. The 5,280- and 1,165-bp fragments that contain the BsaI-digested end provide a substrate for radiolabeling and can serve as a loading control. The radiolabel on the shorter, 630-bp fragment indicates that cleavage was induced by the Z-DNA-forming sequence.
Supplementary Material
Acknowledgments
We thank Drs. Rick Finch and Theodore Wensel for critical review of the manuscript and Sarah Henninger for manuscript preparation. This work was supported by National Cancer Institute Grant CA93729 (to K.M.V.), National Institute on Environmental Health Sciences Center Grant ES07784, and an Odyssey Special Fellowship (to G.W.) supported by the Odyssey Program and the Theodore N. Law Award for Scientific Achievement at The University of Texas M. D. Anderson Cancer Center.
Abbreviations
- DSB
double-strand break
- LM-PCR
linker-mediated PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wang G., Vasquez K. M. Proc. Natl. Acad. Sci. USA. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi M., Tsujimoto Y. Oncogene. 1990;5:1653–1657. [PubMed] [Google Scholar]
- 3.Seite P., Leroux D., Hillion J., Monteil M., Berger R., Mathieu-Mahul D., Larsen C. J. Genes Chromosomes Cancer. 1993;6:39–44. doi: 10.1002/gcc.2870060108. [DOI] [PubMed] [Google Scholar]
- 4.Wolfl S., Wittig B., Rich A. Biochim. Biophys. Acta. 1995;1264:294–302. doi: 10.1016/0167-4781(95)00155-7. [DOI] [PubMed] [Google Scholar]
- 5.Rimokh R., Rouault J. P., Wahbi K., Gadoux M., Lafage M., Archimbaud E., Charrin C., Gentilhomme O., Germain D., Samarut J., et al. Genes Chromosomes Cancer. 1991;3:24–36. doi: 10.1002/gcc.2870030106. [DOI] [PubMed] [Google Scholar]
- 6.Aplan P. D., Raimondi S. C., Kirsch I. R. J. Exp. Med. 1992;176:1303–1310. doi: 10.1084/jem.176.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rich A., Nordheim A., Wang A. H. Annu. Rev. Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 8.Malfoy B., Rousseau N., Vogt N., Viegas-Pequignot E., Dutrillaux B., Leng M. Nucleic Acids Res. 1986;14:3197–3214. doi: 10.1093/nar/14.8.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston B. H. Methods Enzymol. 1992;211:127–158. doi: 10.1016/0076-6879(92)11009-8. [DOI] [PubMed] [Google Scholar]
- 10.Feigon J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Science. 1985;230:82–84. doi: 10.1126/science.4035359. [DOI] [PubMed] [Google Scholar]
- 11.Hamada H., Kakunaga T. Nature. 1982;298:396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- 12.Schroth G. P., Chou P. J., Ho P. S. J. Biol. Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 13.Wahls W. P., Wallace L. J., Moore P. D. Mol. Cell. Biol. 1990;10:785–793. doi: 10.1128/mcb.10.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaho J. A., Wells R. D. Prog. Nucleic Acid Res. Mol. Biol. 1989;37:107–126. doi: 10.1016/s0079-6603(08)60696-0. [DOI] [PubMed] [Google Scholar]
- 15.Weinreb A., Collier D. A., Birshtein B. K., Wells R. D. J. Biol. Chem. 1990;265:1352–1359. [PubMed] [Google Scholar]
- 16.Wahls W. P., Moore P. D. Mol. Cell. Biol. 1990;10:794–800. doi: 10.1128/mcb.10.2.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juranic Z., Kidric M., Tomin R., Juranic I., Spuzic I., Petrovic J. Med. Hypotheses. 1991;35:353–357. doi: 10.1016/0306-9877(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 18.Casasnovas J. M., Ellison M. J., Rodriguez-Campos A., Azorin F. Eur. J. Biochem. 1987;167:489–492. doi: 10.1111/j.1432-1033.1987.tb13363.x. [DOI] [PubMed] [Google Scholar]
- 19.Freund A. M., Bichara M., Fuchs R. P. Proc. Natl. Acad. Sci. USA. 1989;86:7465–7469. doi: 10.1073/pnas.86.19.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells R. D. J. Biol. Chem. 1996;271:2875–2878. doi: 10.1074/jbc.271.6.2875. [DOI] [PubMed] [Google Scholar]
- 21.Sinden R. R. Nature. 2001;411:757–758. doi: 10.1038/35081234. [DOI] [PubMed] [Google Scholar]
- 22.Sinden R. R. DNA Structure and Function. San Diego: Academic; 1994. [Google Scholar]
- 23.Ha S. C., Lowenhaupt K., Rich A., Kim Y. G., Kim K. K. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 24.McLean M. J., Wells R. D. J. Biol. Chem. 1988;263:7370–7377. [PubMed] [Google Scholar]
- 25.Nordheim A., Rich A. Nature. 1983;303:674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- 26.Borowiec J. A., Hurwitz J. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sargent R. G., Brenneman M. A., Wilson J. H. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaworski A., Blaho J. A., Larson J. E., Shimizu M., Wells R. D. J. Mol. Biol. 1989;207:513–526. doi: 10.1016/0022-2836(89)90461-0. [DOI] [PubMed] [Google Scholar]
- 29.Collins J. Cold Spring Harbor Symp. Quant. Biol. 1981;45:409–416. doi: 10.1101/sqb.1981.045.01.055. [DOI] [PubMed] [Google Scholar]
- 30.Collins J., Volckaert G., Nevers P. Gene. 1982;19:139–146. doi: 10.1016/0378-1119(82)90198-6. [DOI] [PubMed] [Google Scholar]
- 31.Kurahashi H., Emanuel B. S. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 32.Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M. G., Shanske A., Goldberg R., Morrow B. E. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurahashi H., Shaikh T. H., Hu P., Roe B. A., Emanuel B. S., Budarf M. L. Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 34.Wells R. D., Dere R., Hebert M. L., Napierala M., Son L. S. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacolla A., Jaworski A., Larson J. E., Jakupciak J. P., Chuzhanova N., Abeysinghe S. S., O’Connell C. D., Cooper D. N., Wells R. D. Proc. Natl. Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean M. J., Blaho J. A., Kilpatrick M. W., Wells R. D. Proc. Natl. Acad. Sci. USA. 1986;83:5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. Cell. 1984;37:321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- 38.Lagravere C., Malfoy B., Leng M., Laval J. Nature. 1984;310:798–800. doi: 10.1038/310798a0. [DOI] [PubMed] [Google Scholar]
- 39.Boiteux S., Costa de Oliveira R., Laval J. J. Biol. Chem. 1985;260:8711–8715. [PubMed] [Google Scholar]
- 40.Rich A., Zhang S. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 41.Wittig B., Dorbic T., Rich A. Proc. Natl. Acad. Sci. USA. 1991;88:2259–2263. doi: 10.1073/pnas.88.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfl S., Martinez C., Rich A., Majzoub J. A. Proc. Natl. Acad. Sci. USA. 1996;93:3664–3668. doi: 10.1073/pnas.93.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaho J. A., Larson J. E., McLean M. J., Wells R. D. J. Biol. Chem. 1988;263:14446–14455. [PubMed] [Google Scholar]
- 44.McLean M. J., Lee J. W., Wells R. D. J. Biol. Chem. 1988;263:7378–7385. [PubMed] [Google Scholar]
- 45.Hirt B. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.