Abstract
Helicobacter pylori persistently colonizes about half the human population and contributes to the development of peptic ulcer disease and gastric cancer. This organism has evolved means to structurally alter its surface characteristics to evade innate and adaptive immune responses. H. pylori produces LPS O-antigen units that can be posttranslationally fucosylated to generate Lewis antigens, structures also found on human epithelial cells. We demonstrate an extensive diversity of Lewis x and Lewis y expression in LPS O-antigen units, occurring over time and in different regions of the human stomach. Lewis expression patterns were correlated with the on/off status of the three fucosyltransferases (FucT), FutA, FutB, and FutC, which are regulated via slipped-strand mispairing in intragenic polyC tract regions of the corresponding genes. The α1,3-FucT, FutA and FutB, each contain a C-terminal heptad repeat region, consisting of a variable number of DD/NLRV/INY tandem repeats. Variations in the number of heptad repeats correlated to the sizes of O-antigen polymers to become decorated by fucose residues. Our data support a molecular ruler mechanism for how H. pylori varies its LPS fucosylation pattern, where one heptad repeat in the enzyme corresponds to one N-acetyl-β-lactosamine unit in the O-antigen polysaccharide.
Keywords: chronic, human stomach, phase variation
Helicobacter pylori colonization of the gastric mucosa is established during childhood and persists for decades without clearance by the immune system (1, 2). Infection is associated with inflammation that, in most cases, is asymptomatic but develops into severe gastric pathology in a subset of infected individuals (3, 4). Although coinfection with multiple strains has been described, most individuals are monoinfected and carry a single strain (5, 6). Microdiversity is, however, common between single H. pylori cells within a host, although these clones constitute a population with similar characteristics that originates from a common ancestral strain (7–10).
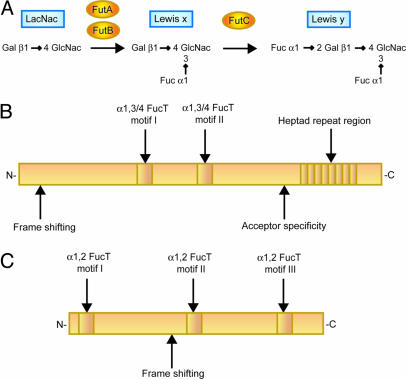
The most variable features of bacterial pathogens are those structures present on the surface of the microbe. Changing these surface molecules is a way to evade the immune system or to alter characteristics important for interaction with host cells (11). A major surface structure of Gram-negative bacteria is LPS, composed of a polysaccharide O-antigen chain; a core oligosaccharide; and a lipid moiety, lipid A, that anchors the LPS molecule to the outer membrane (12). The O-antigen of H. pylori has been suggested to be synthesized in a stepwise glycosylation manner (13–15). This is in contrast to the well described biosynthesis of O-antigens in other Gram-negative bacteria that polymerize preformed oligosaccharide units (12). A typical H. pylori O-antigen consists of polymeric N-acetyl-β-lactosamine (LacNAc) units that are partly fucosylated, showing mimicry to Lewis antigens, structures also found on human erythrocytes and epithelial cells (15–17). Such Lewis antigen glycosylation occurs on a highly diverse substrate consisting of O-antigen polymers with varying number of disaccharide units, each with a nonreducing end. With two α1,3-fucosyltransferases (α1,3-FucT), FutA and FutB, H. pylori is capable of expressing monofucosylated Lewis x antigens on O-antigen polymers of different sizes, both internal in the polymers as well as at the nonreducing ends, providing a substrate for the FutC α1,2-FucT to yield difucosylated Lewis y (Fig. 1) (18). Once a terminal O-antigen unit is Lewis y-glycosylated by FutC, further polymerization is blocked (13, 15). Less frequently, H. pylori O-antigens may display Lewis a, Lewis b, and H-1 antigen, but such mimicry occurs in association with expression of Lewis x and Lewis y (14, 19).
Fig. 1.
H. pylori FucT. (A) Structures of the Lewis antigens discussed in this paper and the pathway and enzymes involved in their synthesis in H. pylori. (B) Schematic representation of the α1,3-/α1,4-FucT in H. pylori (FutA and FutB). Expression of the genes is regulated via frame shifts in a 5′ polyC tract. Discrimination between types 1 and 2 antigen chains as acceptors is determined in a divergent C-terminal region. A repetitive region with a variable number of a 7-aa sequence, which is present in the C terminus, forms a coil-coiled motif. (C) Schematic representation of the α1,2-FucT in H. pylori (FutC). Frame shifts in a polyC tract, situated in the middle of the gene, regulate the expression status.
Expression of the three FucT in H. pylori is regulated via slipped-strand mispairing in intragenic polyC tract regions, resulting in alternate reading frames and an on/off switch that gives rise to subclones with different Lewis glycosylation patterns (20, 21). In addition, FutA and FutB contain a C-terminal tandem repeat region, consisting of a variable number of a 7-aa sequence, DD/NLRV/INY. The heptad-repeat pattern forms an α-helical coiled-coil motif (22). This motif is not present in mammalian FucT and its function has not been elucidated.
The objective of the present study was to describe H. pylori LPS variability in a chronic human infection and to clarify the underlying molecular mechanisms. By assessing Lewis antigen expression patterns and FucT genotypes, we demonstrate a key role for the C-terminal heptad repeat region of FutA and FutB in the generation of such diversity.
Results
Temporal and Spatial Phenotypic Diversity Within a Bacterial Population.
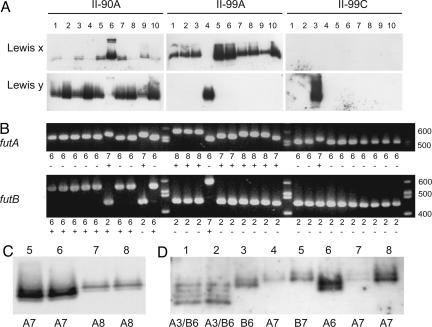
To study the diversity of LPS fucosylation in vivo, we isolated H. pylori single colonies from two patients, each infected by a single strain. Gastric biopsies were collected at two occasions with a 9-year interval, with samples obtained from antrum (90A) at the initial endoscopy and from both antrum (99A) and corpus (99C) at the followup endoscopy (10). All isolates expressed smooth LPS, as seen by the appearance of a typical ladder pattern when extracted LPS was separated on SDS/PAGE gels and stained with silver (data not shown). Isolated LPS preparations were found to carry varying amounts of fucosylated Lewis x and Lewis y antigens (Figs. 2A and 3A), although Lewis a, Lewis b, and H-1 antigens were absent (data not shown). On both occasions, all patient I isolates from antrum expressed Lewis x and only traces of or no Lewis y (Fig. 2A). In contrast, H. pylori populations with different overall Lewis phenotypes existed at the two occasions in patient II. At the initial endoscopy (90A), most isolated bacteria expressed predominantly Lewis y, whereas the population that was isolated at the followup endoscopy (99A) was mainly of the Lewis x phenotype (Fig. 3A).
Fig. 2.
Lewis phenotype and fut genotype in isolates from patient I. (A) Western blotting was performed on extracted LPS with antibodies detecting Lewis x and Lewis y, respectively. (B) PCR amplification of the 3′-repetitive region in futA and futB, respectively. The number of heptad repeats and their on/off (±) status are shown below the lanes. (C and D) Enlargement of Lewis x expression in isolates I-99A:2–3 (C) and Lewis y expression in isolate I-99C:5 (D) with a lower exposure time compared with A. The number of heptad repeats in in-frame FutA and FutB enzymes is shown below lanes. (E and F) Lewis expression in mutants from parental strains I-99C:4 (E) and I-99C:5 (F), where knockout mutants were constructed by the insertion of resistance markers in futA and/or futB, respectively. Wt, wild-type parental strain; A−, futA knockout; B−, futB knockout; AB−, futA, futB knockout.
Fig. 3.
Lewis phenotype and fut genotype in isolates from patient II. (A) Western blotting was performed on extracted LPS with antibodies detecting Lewis x and Lewis y, respectively. (B) PCR amplification of the 3′-repetitive region in futA and futB, respectively. The number of heptad repeats and their on/off (±) status are shown below lanes. (C) Lewis x-expression in isolates II-99A:5–8 with a lower exposure time compared with A. (D) Lewis y glycosylation in various isolates with different FutA/FutB phenotype. Lane 1: I-99C:5 (FutA3/FutB6), lane 2: I-99C:10 (FutA3/FutB6), lane 3: II-90A:5 (FutB6), lane 4 II-90A:9 (FutA7), lane 5: II-99A:4 (FutB7), lane 6: II-99A:4 futB− (FutA6), lane 7: II-99A:4 futB− (FutA7), lane 8: II-99C:3 (FutA7).
We further investigated the diversity among H. pylori cells isolated from two separate compartments of the stomach, i.e., the antrum and the corpus, at the same time of biopsy sampling (99A and 99C). Patient I was colonized by a bacterial population that predominantly expressed Lewis x, both in antrum and corpus (Fig. 2A). In patient II, 9 of 10 antrum isolates (99A) expressed Lewis x on the LPS (Fig. 3A). Remarkably, the same phenotype was not found in any of the 10 patient II clones from the corpus (99C), where most isolates were Lewis x/y negative (Fig. 3A) and instead expressed i-antigen (N-acetyl-β-lactosamine, LacNAc) units (data not shown).
Sequence Variation in PolyC Tracts of FucT Genes.
To understand the genetic background of the various Lewis phenotypes, we analyzed the FucT genes in the isolates. The on/off status of futA and futB was assessed by sequencing and translating the 5′ ends of the genes that contain a polyC tract and are subject to slipped-strand mispairing events (Figs. 2B and 3B and Tables 2 and 3, which are published as supporting information on the PNAS web site). Both genes were in-frame in all isolates from patient I, implying two active α1,3-FucT. In patient II, all but one isolate (II-90A:6) contained either one or no active α1,3-FucT. Notably, isolates from the three different biopsies from patient II showed different patterns of on/off status in the studied FucT. In the 90A and 99A samples from patient II, most isolates contained active FutB or FutA, respectively, whereas both genes were out of frame in all but one isolate (II-99C:3) from the 99C samples.
Sequencing of futC in isolates from 99A and 99C from patient I showed that only the two Lewis y positive isolates, I-99C:5 and I-99C:10 (Fig. 2A), harbored futC in-frame (Table 2). All other isolates contained an insertion of one nucleotide in the polyC tract that rendered futC truncated. Sequencing of futC in 90A isolates from this patient as well as in isolates from patient II showed that the polyC tract consisted of 13–16 C residues, which could not be accurately sequenced by conventional methods.
Variations in the C-Terminal Repetitive Region of FutA and FutB Correlate to Polymer Sizes of O-Antigen Units Recognized for Lewis x Glycosylation.
We used PCR to monitor potential variations in the in-frame 3′-repetitive region of futA and futB, encoding a heptad repeat (DD/NLRV/INY) of unknown function. Both genes exhibited substantial genetic variation in this region (Figs. 2B and 3B). Sequence analysis of the PCR fragments confirmed that the variation in size of the fragments corresponded to an alteration in the number of heptad repeats. This finding suggests that recombination between homologous repeat sequences in vivo creates subpopulations with different numbers of heptads.
All 90A isolates from patient I were in-frame for both futA and futB, and all possessed FutA with one heptad repeat and FutB with eight heptad repeats (FutA1/FutB8) (Fig. 2B). Nine years later, all isolates were still able to express both enzymes. At this time point, one 99A isolate was FutA3/FutB6, four FutA1/FutB7, and two FutA1/FutB8. Of the 99C isolates, six were FutA3/FutB6, three FutA1/FutB7, and only one had the same FutA1/FutB8 constellation as all of the isolates from the earlier time point. When comparing Lewis x-glycosylation patterns for FutA3/FutB6 isolates (e.g., I-99A:3) with FutA1/FutB7 isolates (e.g., I-99A:2) from patient I, the former fucosylated four polymers, each differing from one another by one O-antigen unit, whereas as many as seven polymer sizes were fucosylated by the latter (Fig. 2 A and C). The difference in size between these respective polymers was confirmed as one O-antigen unit by comparing the electrophoretic mobility of the polymers with LPS standards possessing O-antigen units of known structure (data not shown). FutA3/FutB6 isolates lacked Lewis x glycosylation of the two shortest O-antigen polymers that were fucosylated in isolates expressing FutA1/FutB7. Moreover, FutA1/FutB7 isolates fucosylated an O-antigen polymer one unit larger than the largest polymer fucosylated by the FutA3/FutB6 isolates.
Possession of only one active α1,3-FucT in patient II was associated with a restricted number of O-antigen polymer sizes being Lewis x-glycosylated. Thus, Lewis x-decorated isolates expressing FutA7 (e.g., II-99A:5 and II-99A:6) preferentially fucosylated an O-antigen polymer, which was one unit shorter than FutA8 isolates (e.g., II-99A:7 and II-99A:8) (Fig. 3 B and C).
Taken together, these data suggest that there appears to be a correlation between the numbers of heptad repeats in both FutA and FutB and the sizes of O-antigen polymers being fucosylated. One heptad repeat seemingly corresponds to one O-antigen unit.
FutC Substrates Are Generated by Both FutA and FutB, and Their Heptad Repeat Number Correlates to Lewis y O-Antigen Polymer Sizes.
In patient I, Lewis y was markedly expressed in only two isolates (I-99C:5 and I-99C:10), both expressing FutA3/FutB6. Four O-antigen polymer sizes were terminated with Lewis y. However, in contrast to the Lewis x pattern of FutA3/FutB6 isolates that had futC out of frame, the shortest O-antigen polymer was preferred for fucosylation by FutC in Lewis y-decorated isolates (Fig. 2 A and D). This may be due to a relatively high activity of FutC in these isolates, resulting in α1,2-fucosylation of most O-antigen polymers once a Lewis x substrate is presented, through the activity of an α1,3-FucT with three heptad repeats.
From patient II isolates, we conclude that fucosylation by either FutA or FutB may provide a substrate for FutC fucosylation, leading to synthesis of Lewis y. Thus, possession of an in-frame FutA7 and FutB6, as well as FutB7, was associated with Lewis y expression among these isolates (Fig. 3 A and B). When comparing the glycosylation pattern of the various Lewis y-expressing isolates, it was apparent that the number of heptad repeats in FutA and/or FutB also affected FutC-mediated fucosylation. In patient II isolates, fucosylated by FutA/FutB enzymes with six or seven heptad repeats, FutC fucosylated larger O-antigen polymers, as compared with FutA3/FutB6 isolates in patient I (Fig. 3D). Isolates possessing FutA and FutB with the same amount of heptad repeats preferentially Lewis y-glycosylated O-antigen polymers of identical size, whereas an alteration in numbers of heptad repeats resulted in a corresponding change in the sizes of the Lewis y-glycosylated O-antigen polymers (Fig. 3D).
Mutations in futA and futB Highlight a Role for the C-Terminal Repeat Region in Enzyme Target Specificity.
Using two parental isolates from patient I, mutants that were deficient in futA and/or futB were constructed. The first isolate (I-99C:4; Fig. 2E) that synthesized Lewis x had futC out of frame and expressed FutA1/FutB7. A futA knockout mutant in I-99C:4 retained Lewis x glycosylation, although fucosylation, now solely catalyzed from FutB7, was restricted to high-molecular-weight O-antigen polymers. Somewhat surprisingly, a futB knockout was devoid of Lewis x expression, even though the futA gene remained in-frame and instead expressed i-antigen (N-acetyl-β-lactosamine, LacNAc) units (data not shown). Thus, FutA1 was not mediating in vivo fucosylation in the absence of FutB7. The second isolate (I-99C:5; Fig. 2F) possessed FutA3/FutB6, had futC in-frame, and produced Lewis y. Disruption of futB in this isolate had no effect on Lewis x and y glycosylation, showing that FutA3, in the absence of FutB6, was capable of synthesizing Lewis x that could act as substrate for FutC to yield Lewis y. In this isolate, a futA knockout was deficient in Lewis y production but instead displayed Lewis x. Fucosylation was, however, restricted to two polymers, both larger than the polymer preferentially fucosylated by the FutC enzyme in the parental strain. Sequencing of the futC polyC tract revealed that the gene was out of frame in the mutant (data not shown).
FucT mutants were also generated in isolates from patient II, having only one of the futA or futB genes in frame. Two futB mutants were created in isolate II-99A:4 that produced Lewis y through the expression of FutB7. The futA gene was turned on in both mutants but, whereas the first retained the six 3′ repeats from the silent parental futA (FutA6/FutB−), the second had acquired one additional repeat (FutA7/FutB−). Both mutants still expressed Lewis y, and the numbers of heptad repeats in the active enzyme was correlated with sizes of Lewis y-glycosylated O-antigen polymers. Although the predominant Lewis y-glycosylated O-antigen polymer was of the same size in the FutA7/FutB− mutant and the parental FutB7 isolate, the FutA6/FutB− mutant preferentially Lewis y-glycosylated a polymer that was one unit shorter (Fig. 3D). These data suggest that inactivation of the single expressed futA/B gene in an isolate expressing FutC may select for an on switch in the intact futA/B gene, thereby linking FutC expression to substrate availability.
Discussion
Our data suggest a direct role of the C-terminal tandem heptad-repeat region in FutA and FutB when creating diversity in LPS fucosylation. Variation in the C-terminal heptad-repeat number appears to constitute a mechanism for recognition of O-antigen polymers of different sizes as substrate. Our data are compatible with one heptad corresponding to one O-antigen disaccharide unit. The heptad-repeat region in FutA and FutB is predicted to adopt an extended coiled-coil structure (22). If this region can mediate homo- as well as heterodimers, one would expect a number of different dimers to be formed, depending on the number of heptads and which heptads are engaged in dimerization (Fig. 4). Our finding that FutA1 in the absence of FutB is not able to catalyze α1,3-fucosylation, even though it is required for fucosylation of shorter polymers in the presence of FutB7, supports the idea of heterodimer formation.
Fig. 4.
Model of Lewis x glycosylation in H. pylori. FutA and FutB may form homodimers or heterodimers that differ between isolates, reflecting translational frame and number of C-terminal heptad repeats in the enzymes. In this model, fucosylation occurs at the active site, whose distance from a fixed point depends on the number of C-terminal heptads, and which affects the sizes of the O-antigen polymer that is fucosylated. Addition of one heptad repeat in the FucT shifts the fucosylation to an O-antigen polymer that is one unit larger. (A) An isolate expressing FutA3/FutB6 may form heterodimers in four different constellations, depending on which heptad repeats mediate the dimerization. Fucosylation by FutA3 and FutB6 occurs at O-antigen units 3–6, respectively. (B) In FutA1/FutB7 isolates, the single heptad of FutA1 may dimerize with any of the heptads found in FutB7. These variants create seven different appearances of heterodimers (only the two extreme positions are shown) that lead to fucosylation of O-antigen units 1–7. In the model in A and B, the O-antigen units (brown squares) are Lewis x-glycosylated in a polymeric manner. Some strains may display fewer Lewis x epitopes, e.g., only at the nonreducing terminus. The simultaneous presence of O-antigen polymers of different lengths, each with a terminal Lewis x, will nevertheless exhibit the same ladder pattern in Western blot experiments as polymeric Lewis x-bearing O-antigen polymers. (C) In the case of isolates with either FutA or FutB present, only homodimers can be formed. The strongest dimerization involves all heptad repeats, which thus is the dominating form. However, dimerization excluding some heptads may form at lower frequencies. In such isolates, one O-antigen unit is preferably fucosylated, but O-antigen polymers differing in size by a few units may also become fucosylated at lower frequencies.
FutC does not possess a variable C-terminal region and, except for insertion or deletion of a minor number of nucleotides in the central polyC tract, the genes in different isolates are of constant length. This suggests that, in contrast to FutA and FutB, fucosylation by FutC is not restricted by the size of the enzyme. Nevertheless, the number of heptad repeats in FutA and FutB appears to affect subsequent α1,2-fucosylation by FutC, probably reflecting availability of terminal Lewis x substrates on O-antigens. We did not detect Lewis x expression on polymers that were decorated with Lewis y. This may be due to capping of O-antigens with terminal Lewis y that could obscure internal Lewis x units for detection by anti-Lewis x antibodies (23, 24).
Varying the overall frequency of Lewis antigen expressors within a H. pylori population would be one mechanism whereby H. pylori modulates the inflammatory response within the stomach (25, 26). We envision that, in a persistent infection of a human stomach, a number of variants of a strain exist, and each isolate is more or less unique with respect to surface molecules, such as LPS. Such diversity may be of importance when surrounding conditions change, for example, during colonization of a new host or a different compartment of the stomach. Furthermore, the surroundings may continuously change within an infected individual, such as during the development of atrophic gastritis with reduced acid production in older individuals. This is consistent with observations that Lewis antigen expression in H. pylori LPS can vary in response to environmental pH (27). The diversity of a bacterial population is essential during such environmental changes and promotes adaptation of the best-fit isolates. It may therefore be the phenotype of the whole bacterial population and the capacity of the collected pool of individual cells that are important for the ability of H. pylori to colonize, persist, and induce pathological changes in the gastric mucosa.
In conclusion, H. pylori extensively varies its Lewis antigen expression pattern in vivo, and phenotypically distinct populations may be present at different sites in the stomach simultaneously. The diversity is accounted for at the molecular level by modifications in genes coding for FucT. The genes may be switched on and off by slipped-strand mispairing, as previously described in vitro (20, 21), and variation in the number of tandem heptads in FutA and FutB will direct the O-antigen polymer sizes that can become fucosylated. This sophisticated mode of generating Lewis antigen diversity likely constitutes an important means by which H. pylori adapts to the host environment during persistent infection and evades the host immune system.
Materials and Methods
Bacterial Strains.
H. pylori were isolated from two Swedish patients from a study by Gustavsson et al. (28). When entering the study, patients I and II were 43 and 29 years old and suffered from epigastralgia and gastric reflux, respectively. Endoscopy was performed twice, in 1990 and 1999, and gastric biopsies were obtained from the antrum on the initial occasion and from both the antrum and corpus at the followup endoscopy. The different biopsies are referred to as 90A, 99A, and 99C, respectively (10). H. pylori were grown on chocolate agar plates, as described (29). Ten single colonies were isolated from the primary culture of each biopsy, with the exception of 99A in patient I, where only seven isolates were obtained. The isolates included in this study have been analyzed genetically elsewhere (10). All isolates within a patient share genetic characteristics and constitute subclones with a common ancestor.
LPS Purification.
H. pylori was grown in Brucella broth (Becton Dickinson) supplemented with 5% FBS (Sigma) and 1% IsoVitaleX Enrichment (Becton Dickinson) to exponential phase. An equal amount of bacteria (as measured by OD600) from each strain was harvested by centrifugation and washed once in PBS and once in PBS supplemented with 0.15 mM CaCl2/0.5 mM MgCl2. LPS was extracted twice with equal volumes of water and phenol at 70°C with repeated vortexing for 15 min. After centrifugation at 16,000 × g for 15 min at 4°C, the aqueous phases were pooled and precipitated at −20°C overnight in 10 volumes of 99.5% ethanol and sodium acetate (final concentration 0.5 M). The LPS was precipitated by centrifugation (16,000 × g for 20 min at 4°C), washed in 70% ethanol, dried in air, and resuspended in water.
Polyacrylamide Gel Electrophoresis and Immunoblotting.
LPS preparations were fractionated by SDS/PAGE by using a 4% polyacrylamide stacking gel and a 15% polyacrylamide separating gel, and blotted onto polyvinylidene difluoride membrane (Bio-Rad). Membranes used for immunoblots were blocked in PBS supplemented with 1% BSA (Sigma) and 0.1% Tween-20 (blocking buffer) overnight. Mouse anti-Lewis x (MCA1762), anti-Lewis y (MCA1091) (both from Serotec), anti-Lewis a (T174), anti-Lewis b (T128), or anti-H-1 antigen (17–206) (all from Signet Laboratories, Dedham, MA) were used as primary antibodies and horseradish peroxidase-conjugated rabbit anti-mouse Ig (DAKO) as secondary antibody. An antibody against i-antigen was generated in mice by using synthetic N-acetyl-β-lactosamine (LacNAc) glycoconjugate and also used as a primary antibody. Membranes were incubated with antibodies at a dilution of 1/500 (anti-Lewis x), 1/5,000 (anti-Lewis y and secondary antibody), or 1/1,000 (anti-Lewis a, anti-Lewis b, anti-H-1 antigen, and anti-i-antigen), in blocking buffer for 1 h, washed in PBS supplemented with 0.1% Tween-20, developed with enhanced chemiluminescence (ECL) (Amersham Pharmacia Biosciences), and exposed to Hyperfilm ECL (Amersham Pharmacia Biosciences) for chemiluminescence detection. The specificities of the antibodies were validated by their ability to bind the respective antigen from a panel of synthetic Lewis and blood group antigens (Isosep, Tullinge, Sweden, and Dextra Laboratories, Reading, U.K.) and the LPS of H. pylori strains of known structure (27).
PCR and Sequencing.
H. pylori DNA was purified with DNeasy Tissue Kit (Qiagen, Hilden, Germany). PCR was performed under standard conditions with DyNAzyme Taq polymerase and buffer (Finnzymes, Espoo, Finland). The genes encoding FutA, FutB, and FutC [HP0379, HP0651, and HP0093–94, according to The Institute for Genomic Research nomenclature of H. pylori 26695 (30)] were amplified by PCR and sequenced with the primers shown in Table 1. Cycle-sequencing reactions were performed by using a BigDye Terminator, Ver. 3.1, Cycle Sequencing Kit (Applied Biosystems). The sequencing analysis was performed on ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Table 1.
Primers used in this study
| Primer | Sequence, starting from 5′ | Specificity* | Application† |
|---|---|---|---|
| HP0379 F | CTC TCG TGA TCT TGG CTT ATT | futA | PCR |
| HP0379 R | AAG TAG CGT CTG CGA TGA | futA | PCR |
| C-tract up 379 | CTT GGC TTA TTT CAA ACG C | futA | Seq |
| C-tract up 651 | GCC CTA ATC AAG CCT TTG | futB | PCR, Seq |
| C-tract down 3 | CCG GTG TAA AAC ACT CGT TTA G | futA, futB | Seq |
| C-tract down 4 | TCA TCA AAG CCT ATG GCG TA | futA, futB | Seq |
| Repeat up-2 | CGC ACC CAA ACG CTT ATT TA | futA, futB | PCR, Seq |
| Repeat down 379 | GAT GAT AGC GCA AGG GGT TT | futA | PCR, Seq |
| Repeat down 651 | AAA ACC CCA CGC TCA AAA A | futB | PCR, Seq |
| HP93/94 F1 | CTC ACA CGC GTC TTT TTC AA | futC | PCR, Seq |
| HP93/94 R1 | GAC GCT CGC TAT AAA GAA ATC C | futC | PCR |
| HP93/94 F2 | CAA CAC CTC CCC AAG CTA GT | futC | Seq |
| HP93/94 R2 | CAA GAT TGC ATG AGC AGC AT | futC | Seq |
| HP93/94 R3 | GCG GCT AAA ATC AAA GAA AGC | futC | Seq |
| FutMBamF | GAT CGT TAT TTG AGG ATC CCT TTG TAT TAT GCC C | futA, futB | Mut |
| FutMBamR | GGG CAT AAT ACA AAG GGA TCC TCA AAT AAC GAT C | futA, futB | Mut |
*Gene specificity for the primer.
†Methods in which the primers were used in this study. Seq, sequencing; Mut, mutagenesis primer used for generating a BamHI restriction site (shown in bold).
Construction of Knockout Mutants.
PCR fragments of futA and futB were amplified from H. pylori 26695, cloned into pGEM-T Easy Vector (Promega), and transferred into pGEM-5Zf(+) Vector (Promega) by using NotI sites. A BamHI restriction site was created in futA and futB at nucleotides 365–370 by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with primers FutMBamF and FutMBamR (Table 1). A BamHI fragment containing aphA3 from pJMK30 (31), conferring kanamycin resistance, was cloned into the constructed BamHI sites. For chloramphenicol resistant mutants, the cat gene was cleaved out from pDT2548 (32) by using HincII sites and ligated into the constructed BamHI site that was treated with Pfu DNA polymerase to create blunt-end restriction sites. The plasmids, containing futA and futB disrupted by kanamycin or chloramphenicol resistance cassettes, were transformed into H. pylori strains by natural transformation, thereby allowing integration into the chromosome. Transformants were selected on chocolate agar plates, supplemented with appropriate antibiotics (20 μg/ml kanamycin and/or 20 μg/ml choramphenicol). Because of sequence similarity between futA and futB, homologous recombination occurred randomly in either gene with the different constructs. Knockout mutants were analyzed by PCR and sequencing to confirm the insertion of the resistance marker into the targeted genes, as well as to verify that no genetic alterations were acquired in the nondisrupted FucT.
Supplementary Material
Acknowledgments
We thank Magnus Unemo and Dan Danielsson (Örebro University Hospital, Örebro, Sweden) for providing the H. pylori strains used in this study. This work was supported by the Swedish Foundation for Strategic Research (the Infection and Vaccinology Program), the Swedish Cancer Foundation, and the Irish Health Research Board.
Abbreviations
- FucT
fucosyltransferase
- α1,3-FucT
α1,3-fucosyltransferase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Mitchell H. M., Li Y. Y., Hu P. J., Liu Q., Chen M., Du G. G., Wang Z. J., Lee A., Hazell S. L. J. Infect. Dis. 1992;166:149–153. doi: 10.1093/infdis/166.1.149. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell H. M., Hu P., Chi Y., Chen M. H., Li Y. Y., Hazell S. L. Gastroenterology. 1998;114:256–261. doi: 10.1016/s0016-5085(98)70475-5. [DOI] [PubMed] [Google Scholar]
- 3.Ernst P. B., Gold B. D. Annu. Rev. Microbiol. 2000;54:615–640. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M. J., Atherton J. C. J. Clin. Invest. 2004;113:321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enroth H., Nyrén O., Engstrand L. Dig. Dis. Sci. 1999;44:102–107. doi: 10.1023/a:1026658301825. [DOI] [PubMed] [Google Scholar]
- 6.Wong B. C., Wang W. H., Berg D. E., Fung F. M., Wong K. W., Wong W. M., Lai K. C., Cho C. H., Hui W. M., Lam S. K. Aliment. Pharmacol. Ther. 2001;15:493–503. doi: 10.1046/j.1365-2036.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 7.Kuipers E. J., Israel D. A., Kusters J. G., Gerrits M. M., Weel J., van Der Ende A., van Der Hulst R. W., Wirth H.-P., Höök-Nikanne J., Thompson S. A., et al. J. Infect. Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falush D., Kraft C., Taylor N. S., Correa P., Fox J. G., Achtman M., Suerbaum S. Proc. Natl. Acad. Sci. USA. 2001;98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Israel D. A., Salama N., Krishna U., Rieger U. M., Atherton J. C., Falkow S., Peek R. M., Jr Proc. Natl. Acad. Sci. USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundin A., Björkholm B., Kupershmidt I., Unemo M., Nilsson P., Andersson D. I., Engstrand L. Infect. Immun. 2005;73:4818–4822. doi: 10.1128/IAI.73.8.4818-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallet B. Curr. Opin. Microbiol. 2001;4:570–581. doi: 10.1016/s1369-5274(00)00253-8. [DOI] [PubMed] [Google Scholar]
- 12.Raetz C. R., Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aspinall G. O., Monteiro M. A. Biochemistry. 1996;35:2498–2504. doi: 10.1021/bi951853k. [DOI] [PubMed] [Google Scholar]
- 14.Monteiro M. A., Chan K. H., Rasko D. A., Taylor D. E., Zheng P. Y., Appelmelk B. J., Wirth H.-P., Yang M., Blaser M. J., Hynes S. O., et al. J. Biol. Chem. 1998;273:11533–11543. doi: 10.1074/jbc.273.19.11533. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro M. A., Appelmelk B. J., Rasko D. A., Moran A. P., Hynes S. O., MacLean L. L., Chan K. H., Michael F. S., Logan S. M., O’Rourke J., et al. Eur. J. Biochem. 2000;267:305–320. doi: 10.1046/j.1432-1327.2000.01007.x. [DOI] [PubMed] [Google Scholar]
- 16.Aspinall G. O., Monteiro M. A., Pang H., Walsh E. J., Moran A. P. Biochemistry. 1996;35:2489–2497. doi: 10.1021/bi951852s. [DOI] [PubMed] [Google Scholar]
- 17.Appelmelk B. J., Simoons-Smit I., Negrini R., Moran A. P., Aspinall G. O., Forte J. G., De Vries T., Quan H., Verboom T., Maaskant J. J., et al. Infect. Immun. 1996;64:2031–2040. doi: 10.1128/iai.64.6.2031-2040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G., Ge Z., Rasko D. A., Taylor D. E. Mol. Microbiol. 2000;36:1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- 19.Heneghan M. A., McCarthy C. F., Moran A. P. Infect. Immun. 2000;68:937–941. doi: 10.1128/iai.68.2.937-941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appelmelk B. J., Martin S. L., Monteiro M. A., Clayton C. A., McColm A. A., Zheng P., Verboom T., Maaskant J. J., van den Eijnden D. H., Hokke C. H., et al. Infect. Immun. 1999;67:5361–5366. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G., Rasko D. A., Sherburne R., Taylor D. E. Mol. Microbiol. 1999;31:1265–1274. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- 22.Mason J. M., Arndt K. M. Chem. Bio. Chem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 23.Hynes S. O., Moran A. P. FEMS Microbiol. Lett. 2000;190:67–72. doi: 10.1111/j.1574-6968.2000.tb09264.x. [DOI] [PubMed] [Google Scholar]
- 24.Hynes S. O., Keenan J. I., Ferris J. A., Annuk H., Moran A. P. Helicobacter. 2005;10:146–156. doi: 10.1111/j.1523-5378.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 25.Bergman M. P., Engering A., Smits H. H., van Vliet S. J., van Bodegraven A. A., Wirth H.-P., Kapsenberg M. L., Vandenbroucke-Grauls C. M. J. E., van Kooyk Y., Appelmelk B. J. J. Exp. Med. 2004;200:979–990. doi: 10.1084/jem.20041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamri W., Moran A. P., Worku M. L., Karim Q. N., Walker M. M., Annuk H., Ferris J. A., Appelmelk B. J., Eggleton P., Reid K. B. M., et al. Infect. Immun. 2005;73:7677–7686. doi: 10.1128/IAI.73.11.7677-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran A. P., Knirel Y. A., Senchenkova S. N., Widmalm G., Hynes S. O., Jansson P. E. J. Biol. Chem. 2002;277:5785–5795. doi: 10.1074/jbc.M108574200. [DOI] [PubMed] [Google Scholar]
- 28.Gustavsson A., Unemo M., Blomberg B., Danielsson D. Dig. Dis. Sci. 2005;50:375–380. doi: 10.1007/s10620-005-1613-1. [DOI] [PubMed] [Google Scholar]
- 29.Björkholm B., Lundin A., Sillén A., Guillemin K., Salama N., Rubio C., Gordon J. I., Falk P., Engstrand L. Infect. Immun. 2001;69:7832–7838. doi: 10.1128/IAI.69.12.7832-7838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomb J. F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S., Dougherty B. A., et al. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 31.van Vliet A. H., Wooldridge K. G., Ketley J. M. J. Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Taylor D. E. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.