Abstract
Preparedness for a possible influenza pandemic caused by highly pathogenic avian influenza A subtype H5N1 has become a global priority. The spread of the virus to Europe and continued human infection in Southeast Asia have heightened pandemic concern. It remains unknown from where the pandemic strain may emerge; current attention is directed at Vietnam, Thailand, and, more recently, Indonesia and China. Here, we report that genetically and antigenically distinct sublineages of H5N1 virus have become established in poultry in different geographical regions of Southeast Asia, indicating the long-term endemicity of the virus, and the isolation of H5N1 virus from apparently healthy migratory birds in southern China. Our data show that H5N1 influenza virus, has continued to spread from its established source in southern China to other regions through transport of poultry and bird migration. The identification of regionally distinct sublineages contributes to the understanding of the mechanism for the perpetuation and spread of H5N1, providing information that is directly relevant to control of the source of infection in poultry. It points to the necessity of surveillance that is geographically broader than previously supposed and that includes H5N1 viruses of greater genetic and antigenic diversity.
Keywords: genetics, human, influenza A, virus evolution, avian
Highly pathogenic avian influenza (HPAI) A subtype H5N1 virus was initially isolated from geese in Guangdong Province, China in 1996 (1). In the following years, this virus was repeatedly isolated from poultry and occasionally caused disease in humans in southern China (2, 3). Since late 2003, the H5N1 virus has expanded its geographical range to affect poultry in East and Southeast Asia, giving rise to >100 patients with severe human disease (4). The expansion of its geographical distribution in poultry and its repeated interspecies transmission to humans make it a significant pandemic threat, and preparedness for such a pandemic is a global priority (5, 6). To plan strategies for the control of H5N1 influenza in poultry and the consequent human health risk, it is critical that the mechanisms of virus transmission and perpetuation are well understood.
In May 2005, H5N1 virus caused an outbreak in migratory waterfowl in Qinghai Lake, western China (7, 8): the first observation of sustained transmission within migratory waterfowl. The likely source of this infection was thought to be from poultry in southern China, and it was predicted that HPAI H5N1 might spread to Europe through overlapping migratory flyways (7). Subsequently, H5N1 influenza outbreaks have been reported in Xinjiang and Tibet Autonomous regions of China, Kazakhstan, Mongolia, Russian Siberia, Turkey, Croatia, and Romania, apparently through the movement of migratory birds (9, 10). However, the source of infection for these new outbreaks, including the initial Qinghai outbreak, has not been fully determined. Whether or not migratory birds could survive infection and carry H5N1 over long distances also remains unanswered.
Our ongoing influenza virus surveillance in southern China shows that H5N1 influenza viruses have been persistently circulating in market poultry populations and also revealed that those viruses were present in apparently healthy migratory birds just before their migration. Genetic analyses reveal that the endemicity of the H5N1 viruses in domestic poultry has resulted in the establishment of distinct regional virus sublineages. The findings of this study demonstrate that H5N1 viruses can be transmitted over long distances by migratory birds. However, viruses in domestic poultry have evolved into distinct regional clades, suggesting that transmission within poultry is the major mechanism for sustaining H5N1 virus endemicity in this region.
Results
Surveillance.
To clarify the role of migratory birds in the spread of H5N1 virus, a total of 13,115 cloacal and fecal specimens of migratory birds (including 4,674 from captured migratory ducks) were collected during three winter seasons (2002–2005) in Mai Po Marshes, Hong Kong and Poyang Lake, Jiangxi, two sites in China at which large numbers of migratory birds over-winter (Fig. 1) (11). Forty-four influenza A viruses of six hemagglutinin (HA) subtypes were isolated (isolation rate, 0.34%), all of them from apparently healthy migratory ducks (Table 1). Six HPAI H5N1 viruses were isolated from apparently healthy migratory ducks at Poyang Lake on two sampling occasions (January and March 2005) just before their migration north. Two lowly pathogenic avian influenza (LPAI) H5N3 viruses were also isolated from fecal matter of migratory birds in Mai Po Marshes in November 2004. This finding shows that HPAI H5N1 virus can be found in apparently healthy migratory ducks.
Fig. 1.
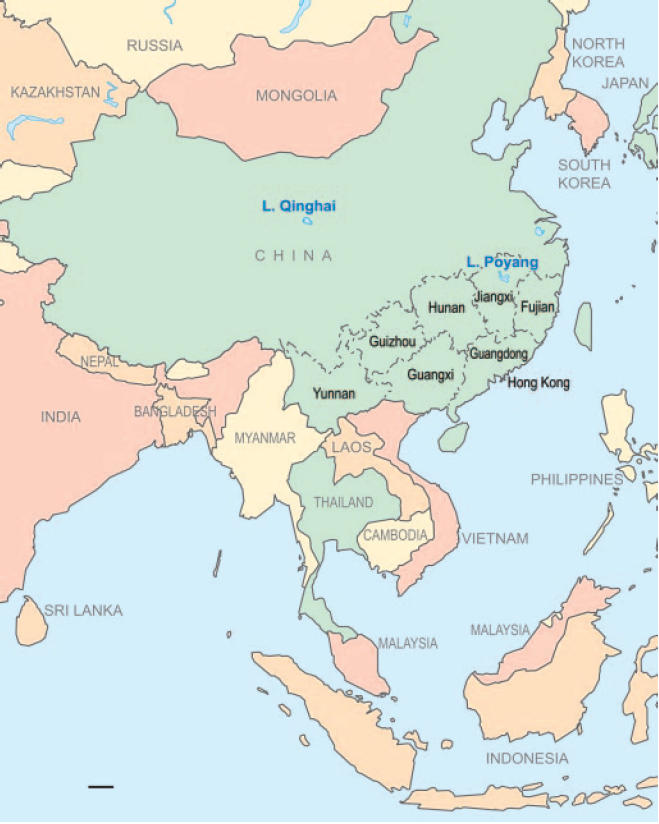
Map of eastern Asia showing Fujian, Guangdong, Guangxi, Guizhou, Hong Kong, Hunan, and Yunnan where influenza surveillance was conducted and the locations of Poyang and Qinghai Lakes. Following are the genotypes of H5N1 viruses tested in poultry from each Province in our surveillance program in southern China since 2004, plus data from migratory birds at Poyang and Qinghai Lakes: Guangxi, genotypes W (15), Z (5), and G (3); Hunan, genotypes Z (13) and G (3); Fujian, genotype Z (2); Yunnan, genotype Z (4); Guangdong, genotype Z (7); Poyang Lake, genotypes Z (3) and V (3); Qinghai Lake, genotype Z (20). (Scale bar: 500 km.)
Table 1.
Influenza viruses isolated from migratory birds in Mai Po Marshes and Poyang Lake, China
| Sampling period* | Bird† | No. of samples | HA subtype and no. of isolates‡ |
|||||
|---|---|---|---|---|---|---|---|---|
| H1 | H3 | H4 | H5 | H6 | H10 | |||
| Nov 2003–Jan 2004 (PL) | Migratory duck (C) | 205 | 3 | 0 | 2 | 0 | 2 | 0 |
| Migratory duck (F) | 587 | 0 | 1 | 2 | 0 | 2 | 2 | |
| Migratory goose (F) | 250 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Oct 2003–Mar 2004 (MP) | Migratory duck (F) | 6,005 | 0 | 0 | 5 | 2 | 2 | 5 |
| Oct 2004–Mar 2005 (PL) | Migratory duck (C) | 4,316 | 2 | 2 | 3 | 6 | 2 | 1 |
| Total | 13,115 | 5 | 3 | 12 | 8 | 8 | 8 | |
C, cloacal; F, fecal; MP, Mai Po Marshes; PL, Poyang Lake.
*Earlier sampling at Poyang Lake, December 2002 to February 2003 (153 tracheal swabs, 419 fecal swabs from migratory ducks; 602 fecal swabs from migratory geese; 578 fecal swabs from swans) yielded no influenza viruses.
†Migratory ducks included falcated teal (Anas falcata), mallard ducks (Anas platyrhynchos), and spot-billed ducks (Anas poecilorhyncha). Subtypes were identified by using World Health Organization reference anti-sera and H5 anti-sera available in our laboratory and were confirmed by PCR and sequencing reactions.
‡An additional 30 isolates from migratory ducks could not be identified with the reference anti-sera.
As part of pandemic preparedness, systematic influenza surveillance of poultry markets has been ongoing since July 2000 but has expanded to include a further three provinces in southern China since July 2004. A total of 512 H5N1 influenza viruses (1.0%), along with 3,051 (6.0%) other influenza A virus of diverse subtypes (H3, H6, H9, H11, and others), were isolated from 51,121 samples collected from apparently healthy birds in live-poultry markets of southern China since January 2004 (Table 4, which is published as supporting information on the PNAS web site). Domestic poultry tested positive for H5N1 virus in 16 of the past 18 months between January 1, 2004 and June 30, 2005. H5N1 virus was most frequently isolated from ducks and geese (isolation rates, 1.8% and 1.9%, respectively), followed by minor poultry (0.46%) and chicken (0.26%) (Table 4). Therefore, H5N1 virus has persisted during this period in various types of poultry in the markets of southern China.
Serological Analysis.
To understand whether any H5 subtype-infected migratory ducks have recovered from infection, neutralization assays of 1,092 sera from captured migratory ducks, with a representative HPAI H5N1 isolate from migratory duck (MDk/JX/1653/05) and a low pathogenic H5N2 virus from a nearby domestic duck farm (Dk/JX/3345/05), revealed 34 (3.1%) and 47 (4.3%) samples, respectively, to be positive. Most positive sera showed neutralizing titers to both viruses, usually with 2- to 4-fold higher titers to Dk/JX/3345/05 (H5N2) in comparison with MDk/JX/1653/05 (H5N1) (data not shown). These findings suggest that migratory ducks at Poyang Lake may have first been exposed to low pathogenic H5-subtype viruses that may have provided partial protection against HPAI H5N1 infection.
Antigenic Analysis.
Antigenic analysis with mAbs and polyclonal antisera against representative H5N1 viruses demonstrated a diversity of reaction patterns that differed according to the geographical origins of the viruses and generally corresponded to their phylogenetic sublineages (Table 2). Notably, a ferret antiserum against the current H5N1 virus human vaccine candidate VNM/1203/04 (12) was reactive with most isolates from Vietnam but reacted weakly with those from Guangdong (GD), Guangxi (GX), Hunan (HN), Yunnan (YN), and Indonesia. Similarly, ferret antiserum to the Indonesian H5N1 isolate, Dk/IDN/MS/04, reacted weakly with viruses from Vietnam and with the migratory bird viruses from Jiangxi (JX) and Qinghai (QH). It was also noted that MDk/JX/2136/05 from migratory duck at Poyang Lake had a similar reaction pattern as bar-headed goose, BH goose/QH/65/05. The mAb reaction profiles confirmed the diversity of antigenic crossreactions (Table 2).
Table 2.
Antigenic analysis of H5N1 influenza viruses from different regions in Asia by HA inhibition titer
| Virus | Sublineage* | mAb to Ck/PA/1370/83 |
mAb to Ck/HK/YU22/02 |
mAb to VNM/1203/04 |
Antisera |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP25 | CP58 | CP176/26 | 3C8 | 7C6 | 8H11 | 10H4D2 | 15A3 | 1203/04 | MS/04 | 437.4/99 | ||
| Ck/PA/1370/83 | Reference | 12,800 | 12,800 | 12,800 | 12,800 | 12,800 | 12,800 | 12,800 | 12,800 | 160 | 80 | 80 |
| Gs/HK/437.10/99 | 3,200 | 12,800 | 12,800 | 800 | 12,800 | 12,800 | 12,800 | 12,800 | 1,280 | < | 1280 | |
| Dk/HK/2986.1/00 | 1,600 | 6,400 | 1,600 | 6,400 | 12,800 | 12,800 | 12,800 | 12,800 | 1,280 | < | 640 | |
| Ck/HK/YU22/02 | 200 | < | 200 | 6,400 | 1,600 | 1,600 | 12,800 | 12,800 | < | 80 | 640 | |
| Dk/HN/5806/03 | HN | < | 1,600 | 12,800 | 12,800 | 800 | 12,800 | 12,800 | < | < | 80 | 320 |
| Ck/HN/999/05 | < | 200 | 12,800 | 6,400 | 1,600 | 6,400 | 12,800 | < | < | 80 | 80 | |
| Dk/YN/6445/03 | YN | < | < | 200 | 3,200 | 400 | 800 | < | 200 | < | 160 | 320 |
| Ck/YN/115/04 | < | 200 | 400 | 6,400 | 800 | 1,600 | 400 | 400 | < | 80 | 320 | |
| Ck/YN/447/05 | < | 1,600 | 1,600 | 1,600 | < | 400 | < | < | < | < | < | |
| Ck/Indonesia/BL/03 | IDN | < | < | 800 | 12,800 | 1,600 | 12,800 | < | 800 | < | 320 | 640 |
| Dk/Indonesia/MS/04 | < | < | 400 | 12,800 | 1,600 | 3,200 | < | 400 | < | 160 | 640 | |
| Ck/Wajo/BBVM/05 | < | < | 400 | < | 400 | 1600 | < | < | < | 160 | 320 | |
| Vietnam/1194/04 | VTM | 400 | 200 | < | 12,800 | 6,400 | < | 12,800 | 12,800 | 2,560 | < | 640 |
| Vietnam/1203/04 | 800 | 400 | < | 6,400 | 6,400 | < | 12,800 | 12,800 | 1,280 | < | 1,280 | |
| Thailand/1(Kan-1)/04 | < | < | < | 12,800 | 12,800 | < | 12,800 | 12,800 | 5,120 | < | 1,280 | |
| Dk/Vietnam/11/04 | 400 | 12,800 | < | 6,400 | 3,200 | 800 | 12,800 | 12,800 | 1,280 | < | 640 | |
| Dk/Vietnam/S654/05 | < | < | < | 800 | 400 | < | 1,600 | 200 | 160 | < | 160 | |
| Dk/Vietnam/568/05 | VNM2 | < | < | < | < | < | < | < | < | < | < | < |
| HK/212/03 | GD | 800 | 200 | 200 | 12,800 | 12,800 | 800 | 12,800 | 12,800 | 160 | 320 | 2,560 |
| Gs/GX/345/05 | Mixed | < | < | < | 6,400 | < | 200 | < | 200 | < | < | < |
| Ck/ST/810/05 | < | < | < | 800 | < | < | < | < | < | < | < | |
| MDk/JX/1653/05 | MB | 400 | 800 | 200 | < | 3,200 | 3,200 | 6,400 | 6,400 | < | < | 640 |
| MDk/JX/2136/05 | < | 800 | 12,800 | 12,800 | 400 | 12,800 | 200 | 200 | < | < | 160 | |
| BH goose/QH/65/05 | < | 800 | 12,800 | 12,800 | 400 | 12,800 | < | < | < | < | < | |
<, lowest dilution tested: 1:100 for monoclonal antibody titres, 1:40 for ferret and chicken anti-sera. 437.4/99, Gs/HK/437.4/99; 1203/04, VNM/1203/04; GD; Guangdong; HN, Hunan; IDN, Indonesia; MB, migratory bird; MS/04, Dk/IDN/MS/04; VNM2, recent Vietnam introduction; VTM, Vietnam/Thailand/Malaysia; YN, Yunnan. Underlined numbers indicate titers to prototype viruses.
*Viruses were selected from each of the sub-sublineages as defined by phylogenetic analysis of the HA gene (Fig. 2A).
Pathogenicity Tests.
Most of the ducks infected with MDk/JX/1653/05 and MDk/JX/1657/05 survived (seven of nine and five of nine, respectively) and continued to shed virus for up to 3 days postinfection (Table 3). All nine ducks infected with BH goose/QH/65/05 survived, and three ducks shed this virus for up to 7 days postinfection. All geese infected with BH goose/QH/65/05 died before day 7 (Table 3). Experimentally infected ducks shed virus throughout the experiment. Because these viruses, isolated from migratory duck, are not invariably fatal for ducks, there is a possibility that migratory ducks could harbor the virus and have the potential to transmit interregionally during migration.
Table 3.
Pathogenicity of H5N1 viruses from migratory birds
| Strain | Day 3 p.i. |
Day 7 p.i. |
Day 11 p.i. |
|||
|---|---|---|---|---|---|---|
| Ducks | Geese | Ducks | Geese | Ducks | Geese | |
| MDk/1653/JX/05 | 2/9 (7)* | ND | 2/9 (3) | ND | 2/9 (2) | ND |
| MDk/1657/JX/05 | 2/9 (7) | 1/6 (5) | 4/9 (0) | 6/6 (0) | 4/9 (0) | † |
| BH goose/QH/65/05 | 0/9 (9) | 0/6 (6) | 0/9 (3) | 6/6 (0) | 0/9 (0) | † |
ND, not determined; p.i., postinfection.
*Number of birds died/number tested (number shedding).
†All geese died.
Phylogenetic Analysis.
To elucidate the evolution and ecology of H5N1 viruses in Asia, we sequenced 69 viruses isolated in China since January 2004 and 52 viruses isolated in Indonesia, Malaysia, and Vietnam between August 2003 and May 2005. Phylogenetic analysis of the HA gene revealed that the HA of all viruses tested had originated from the Gs/GD/1/96-like (Gs/GD-like) lineage and generally formed distinct sublineages that corresponded to their geographic origins (Fig. 2A). The viruses isolated in Vietnam, Thailand, and Malaysia (VTM) in 2003–2005 belonged to a sublineage closely related to the GD sublineage of viruses isolated from domestic and migratory birds in Hong Kong in late 2002 and early 2003. Similarly, viruses isolated in HN, YN, and IDN from 2003 onward formed independent sublineages (Fig. 2A). Phylogenetic analysis of the nucleoprotein (NP) gene, along with the neuraminidase (NA) and other internal gene segments, confirmed that these sublineages had been established from H5N1 viruses isolated from 2002 onward (Fig. 2B; and also see Figs. 4–9, which are published as supporting information on the PNAS web site). These regionally distinct sublineages of H5N1 influenza virus demonstrate the long-term endemicity of the viruses in those regions of Asia.
Fig. 2.
Phylogenetic relationships of the HA (A) and NP (B) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in the paup* program (35) (maximum-likelihood and Bayesian analysis revealed the same relationships). Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities. Not all supports are shown due to space constraints. Analysis was based on nucleotides 1–1696 of the HA gene and 1–990 of the NP gene. The HA tree was rooted to Gs/GD/1/96 and the NP tree to Dk/HK/Y280/97. (Scale bar: 0.01 substitutions per site.) Purple, viruses from Indonesia; red, viruses from Vietnam, Thailand, and Malaysia; green (genotype V) and blue (genotype Z), viruses isolated from migratory birds; brown, viruses isolated from poultry in Hunan and Yunnan in late 2004 and early 2005. GD, Guangdong; HN, Hunan; IDN, Indonesia; MB, migratory bird; YN, Yunnan; VNM2, recent Vietnam introduction; VTM, Vietnam/Thailand/Malaysia. ∗, Because 90% of the poultry consumed in Hong Kong was imported from Guangdong, the viruses isolated from domestic and migratory birds in Hong Kong were designated as belonging to the GD sublineage.
Remarkably, one of the viruses isolated in May 2005 in Vietnam (Dk/Vietnam/568/05) was not closely related to other viruses isolated after 2003 in Vietnam and Thailand; instead, all of their gene segments clustered with those of from a virus isolated in January 2005 in Guangxi, China (Gs/GX/345/05). This finding suggests that the virus had likely been recently introduced to Vietnam from Guangxi through poultry movement. These viruses had seven genotype Z-like genes and a genotype W-like (Gs/GD-like) PB2 gene. This reassortant genotype had not been recognized previously, and so it was designated as genotype G (Fig. 3) (2, 13).
Fig. 3.

Genotypes of H5N1 influenza reassortants in Asia. The eight gene segments are (horizontal bars starting at the top downward): PB2, PB1, PA, HA, NP, NA, M, and NS. Each color represents a virus lineage, with red indicating origin from Gs/GD/1/96. The definition of genotypes and the generation of genotypes Z, Z+, V, and W have been described (13). Novel genotype G seems to have resulted from reassortment between genotypes Z and W circulating in southern China.
Phylogenetic analysis also revealed that the viruses isolated from migratory ducks at Poyang Lake belonged to two different H5N1 genotypes: Z (MDk/JX/1653/05, MDk/JX/1657/05, MDk/JX/1701/05) and V (MDk/JX/2136/05, MDk/JX/2295/05, MDk/JX/2300/05). The matrix (M), nonstructural (NS), polymerase acidic (PA), polymerase basic (PB) 1, and PB2 genes of isolates from an outbreak at Qinghai Lake in central China (represented by BH goose/QH/65/05; genotype Z) clustered with those of MDk/JX/1653/05 (genotype Z), whereas the HA, NA, and NP genes of the Qinghai Lake isolates grouped with those of MDk/JX/2136/05 (genotype V), both of which were isolated from Poyang Lake in January and March 2005, respectively. Thus, all eight gene segments of viruses from the Qinghai Lake outbreak in central China can be traced to the H5N1 viruses isolated from migratory ducks at Poyang Lake in southeast China, ≈1,700 km distant, indicating that migratory birds can disseminate the virus over long distances.
Molecular Characterization.
All viruses characterized in this study maintained the motif of multiple basic amino acids at the HA cleavage site characteristic of HPAI. The receptor binding pocket of HA1 retains amino acid residues Gln 222 and Gly 224 (H5 numbering used throughout) that preferentially bind to 2,3-NeuAcGal linkages of avian cell-surface receptors (14). Other amino acid residues relevant to receptor binding were identical to those of HK/156/97 and Gs/GD-like viruses (15), with the exception of two viruses from Yunnan (Ck/YN/447/05 and Ck/YN/493/05) that had an Ala-149-Ser mutation. Comparison of the deduced amino acid sequences of the H5N1 viruses in this study with sequence data available from GenBank identified two HA residues (Ile-99 and Asn-268) and one NA residue (Arg-110) that seemed to be unique to viruses isolated from migratory birds at Qinghai and Poyang Lakes. Furthermore, the presence of an R→G mutation at position −8 of the HA1 connecting peptide was observed only in MDk/JX/2136/05, MDk/JX/2295/05, MDk/JX/2300/05, and all isolates from Qinghai Lake. This result was also reported in H5N1 viruses isolated from the recent Siberian outbreak (9, 10). It remains to be determined whether these amino acid substitutions are host adaptation signatures.
Of the viruses characterized in this study, only three from China (Dk/GX/351/04, Dk/GX/380/04, and Dk/ST/4610/03) and three from Southeast Asia (Ck/MYS/5858/04, Ck/Salatiga/BBVet-I/05, and Dk/VNM/S654/05) had the Ser-31-Asn mutation in the M2 protein that confers resistance to amantadine (16). All other viruses have residues indicating sensitivity to amantadine. No viruses characterized in this study had Lys-627 in the PB2 gene or Glu-92 in the NS1 protein, both mutations that are reportedly associated with increased virulence of H5N1 and H7N7 viruses (17–19).
Discussion
The likelihood of an H5N1 influenza pandemic seems high, and the consequences could be catastrophic. Recent findings suggest that the 1918 “Spanish flu” pandemic may have resulted from a similar interspecies transmission event in which a purely avian virus adapted directly to human-to-human transmission without prior reassortment (20). Our results demonstrate that long-term endemicity of H5N1 viruses in different regions of Asia has resulted in the establishment of regional sublineages.
This study confirms the continued dominance of H5N1 genotype Z virus in most regions in Asia (in the sublineages VTM, GD, HN, YN, and IDN). However, viruses isolated from some provinces of southern China (Guangdong, Guangxi, and Hunan; in the sublineages Mixed) are more diverse, containing genotypes Z, V, W, and G (Figs. 1 and 3) (13). This diversity is likely a reflection of the movement of poultry and the continued circulation of these viruses among these adjacent provinces (Fig. 1). Because the precursor of these H5N1 viruses, Gs/GD/1/96, was originally detected in southern China in 1996 (1) and because H5N1 viruses in this region have the greatest genetic diversity, southern China is the most likely source of the emergent and reemergent HPAI H5N1 influenza viruses. This finding supports the “influenza epicenter” hypothesis (21), which argues that southern China is the epicenter from which influenza pandemics emerge.
Phylogenetic analysis of all gene segments of the H5N1 viruses in this study allowed us to track the origins of the viruses and the mechanisms of their transmission (poultry transport or bird migration). Genetic relatedness of gene segments of H5N1 viruses isolated at Poyang and Qinghai Lakes (Figs. 1, 2, and 4–9) strongly argues that migrating birds can transfer the virus over long distances. As revealed by virus neutralization and pathogenicity tests (Table 3), these HPAI H5N1 viruses isolated from apparently healthy migratory ducks are not invariably fatal, and surviving ducks may shed virus for as long as 7 days after infection in those experimental ducks. This result was also observed in previous experiments with different H5N1 strains (22). Therefore, migratory ducks at Poyang Lake could have survived infection with the HPAI H5N1 virus and transmitted the virus over long distances during migration. This possibility may provide insight into reported H5N1 outbreaks in Mongolia, Siberian Russia, and Europe that have been linked to migratory birds (10). Previously, it has been shown that LPAI could be transmitted from migratory birds to domestic poultry and subsequently acquire a multibasic amino acid cleavage site and gain virulence (23). However, because precursors of both the genotype V and Z viruses isolated from migratory ducks at Poyang Lake were identified in domestic poultry in southern China (Figs. 1, 2, and 4–9), it is more likely that the migratory ducks were infected by local poultry while over-wintering in southern China.
The establishment of regional virus sublineages suggests that H5N1 virus is perpetuated in poultry largely through the movement of poultry and poultry products rather than by continued reintroduction of viruses by migrating birds. The direct precursor of the dominant genotype Z viruses of the VTM sublineage has not been clearly identified. Phylogenetic analysis suggests that the VTM sublineage is derived from southern China, but systematic surveillance data from this period are lacking. Another introduction of H5N1 viruses from Guangxi to adjacent Vietnam seems to have occurred in early 2005, as demonstrated by the genotype G precursor virus Gs/GX/345/05 and the derivative Dk/Vietnam/345/05 and Dk/Vietnam/568/05 isolates. A similar introduction may have occurred in 2001, because the HA gene of the viruses Gs/Vietnam/113/01 and Gs/Vietnam/324/01, isolated from live-poultry markets in Hanoi in October 2001, was derived from the HA of Dk/GX/22/01 (Fig. 2A) (24, 25).
We have shown that H5N1 virus has persisted in its birthplace, southern China (1), for almost 10 yr and has been repeatedly introduced into neighboring (e.g., Vietnam) and distant (e.g., Indonesia) regions, establishing “colonies” of H5N1 virus throughout Asia that directly exacerbate the pandemic threat. The best approach to avert the threat is to control H5N1 virus infection at its source, domestic poultry. Experience in Hong Kong (26), Japan (27), and South Korea (28) has shown that early detection and large-scale culling of infected poultry, combined with other measures, is effective in controlling this HPAI H5N1 influenza (10). Control measures in China, Indonesia, Thailand, and Vietnam have been less effective, allowing the establishment of virus endemicity and repeated interspecies transmission to humans (4, 29). Our results indicate that H5N1 virus has been introduced into Vietnam from southern China on multiple occasions; in 2001, 2003, and 2005. Therefore, control of this regional epizootic and its attendant pandemic threat requires that the source of virus in southern China be contained.
The antigenic diversity of viruses currently circulating in Southeast Asia and southern China challenges the wisdom of reliance on a single human vaccine candidate virus for pandemic preparedness; the choice of candidate viruses for development of human vaccines must reflect the antigenic diversity observed across this wider region. Furthermore, antigenic drift observed over time within those H5N1 sublineages highlights the necessity of continually updating the candidate virus chosen for future H5N1 vaccines. These concepts are critical for the control of this pandemic threat.
Methods
Virological Surveillance, Isolation, and Characterization.
Cloacal, tracheal, and fecal samples were collected once every 7–10 days from apparently healthy poultry in live poultry markets in Fujian, Guangdong, Guangxi, Guizhou, Hong Kong, Hunan and Yunnan. Surveillance of migratory birds was conducted at Poyang Lake and Mai Po Marshes. Fecal swabs were collected at islets where birds aggregated; birds were briefly captured for cloacal sampling and released. Poyang Lake, located in north Jiangxi Province, is the largest freshwater lake in China. Mai Po Marshes are in northwestern Hong Kong, at the border with mainland China. Both are major over-wintering sites for migratory birds in eastern Asia (11). Migratory birds usually arrive in September and leave in April.
Viruses were grown in embryonated eggs, and isolates were identified and subtyped by using reference antisera as described (30). Sera of migratory ducks (n = 1,092) from Poyang Lake, collected from November 2004 to January 2005, were tested by neutralization assay for antibodies against the representative isolates MDk/JX/1653/05 (H5N1) and Dk/JX/3345/05 (H5N2), both viruses isolated from samples from Poyang Lake during the same season and with HA homology of 90%. Sera were screened at a 1:4 dilution against 100 TCID50 (50% tissue culture infective dose) of the viruses MDk/JX/1653/05 (H5N1) and MDk/JX/3345/05 (H5N2) to exclude negative samples. Titers ≥16 were regarded as positive.
Antigenic Analysis.
The antigenic characteristics of the H5N1 influenza viruses from different regions were compared by HA inhibition (HI) assay with mAbs and polyclonal antisera to H5 subtype viruses as described (2). The mAbs 3C8, 7C6, 8H11, and 10H4D2 to the HA of Ck/HK/YU22/02 were produced in our laboratories, and mAb 15A3 to the HA of VNM/1203/04 was produced by the Department of Infectious Diseases at St. Jude Children’s Research Hospital.
Pathogenicity Tests.
Nine 4-week-old mallard ducks (Anas platyrhynchos) and four 4-week-old Chinese geese (Anser cygnoides) were intranasally inoculated as described (31) with 8.75 log10 EID50 (50% egg infective dose) of the viruses MDk/JX/1653/05, MDk/JX/1657/05, or BH goose/QH/65/05 in a 1-ml volume. Birds were monitored daily for signs of illness; cloacal and tracheal samples were collected on days 3, 7, and 11 after inoculation. All pathogenicity tests were conducted in biosafety level 3 facilities.
Phylogenetic Analysis and Molecular Characterization.
To understand the interrelationship of H5N1 viruses isolated from migratory birds and poultry, we analyzed 20 isolates from a 2005 Qinghai Lake outbreak in migratory waterfowl, all six isolates from migratory ducks at Poyang Lake, 46 of 512 (9%) isolates from live poultry markets in seven provinces in southern China in 2004–2005, 52 viruses isolated from poultry in Indonesia and Vietnam in 2004–2005, and two viruses from Malaysia. Genetic characterization and analysis of all eight gene segments of these H5N1 viruses, together with virus sequence data available in GenBank, were conducted.
RNA extraction, cDNA synthesis, and PCR were carried out as described (32). Sequencing was performed by using BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI PRISM 3700 DNA Analyzer (Applied Biosystems) following the manufacturer’s instructions. All eight gene segments of these viruses were characterized and phylogenetically analyzed together with virus sequence data available in GenBank. All sequences were assembled and edited with lasergene 6.0 (DNASTAR, Madison, WI); bioedit 7 was used for alignment and residue analysis (33). The program mrmodeltest 2.2 (34) was used to determine the appropriate DNA substitution model and γ rate heterogeneity. The generated model was used in all subsequent analyses. Neighbor-joining (NJ) and maximum-likelihood trees were constructed by using paup* 4.0 (35). Bayesian analysis was conducted with mrbayes 3.0 (36) by using two replicates of 1 million generations. Estimates of the phylogenies were calculated by performing 1,000 NJ bootstrap replicates, and Bayesian posterior probabilities were calculated from the consensus of 14,000 trees after excluding the first 3,000 trees as burnin. All eight genes were sequenced for each virus isolate.
Supplementary Material
Acknowledgments
We acknowledge Emeritus Professor Ken Shortridge for his valuable comments and discussion of the manuscript and Sharon Naron for editorial assistance. This work was supported by the Li Ka Shing Foundation, the Research Grants Council, and the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government, and the Ellison Foundation.
Abbreviations
- BH goose
bar-headed goose
- GD
Guangdong
- GX
Guangxi
- HA
hemagglutinin
- HN
Hunan
- HPAI
highly pathogenic avian influenza
- IDN
Indonesia
- JX
Jiangxi
- NA
neuraminidase
- NP
nucleoprotein
- PB
polymerase basic
- QH
Qinghai
- YN
Yunnan
- VTM
Vietnam/Thailand/Malaysia.
Footnotes
References
- 1.Xu X., Subbarao K., Cox N., Guo Y. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 2.Guan Y., Poon L. L. M., Cheung C. Y., Ellis T. M., Lim W., Lipatov A. S., Chan K. H., Sturm-Ramirez K. M., Cheung C. L., Leung Y. H. C., et al. Proc. Natl. Acad. Sci. USA. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subbarao K., Klimov A., Katz J., Regnery H., Lim W., Hall H., Perdue M., Swayne D., Bender C., Huang J., et al. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . Influenza A/H5N1 in humans in Asia. Geneva: World Health Organization; 2005. May 6–7, available at www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_GIP_2005_7_04.pdf. [Google Scholar]
- 5.Webster R. G., Hulse D. Nature. 2005;435:415–416. doi: 10.1038/435415a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Global Influenza Program Surveillance Network Emerg. Inf. Dis. 2005;11:1515–1521. doi: 10.3201/eid1110.050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H., Smith G. J. D., Zhang S. Y., Qin K., Wang J., Li K. S., Webster R. G., Peiris J. S. M., Guan Y. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 8.Liu J., Xiao H., Lei F., Zhu Q., Qin K., Zhang X.-w., Zhang X.-l., Zhao D., Wang G., Feng Y., et al. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 9.World Animal Health Organization . Disease Information. No. 32. Vol. 18. Paris: OIE; 2005. available at http://www.oie.int/eng/info/hebdo/aIS_58.htm#Sec6. [Google Scholar]
- 10.Food and Agricultural Organization of the United Nations Avian Influenza Disease Emergency Bulletin. 2005;(Issue 35) available at www.fao.org/ag/againfo/subjects/en/health/diseases-cards/avian_update.html. [Google Scholar]
- 11.Li Z. W. D., Mundkur T. Numbers and Distribution of Waterbirds and Wetlands in the Asia-Pacific Region: Results of the Asian Waterbird Census: 1997–2001. Kuala Lumpur, Malaysia: Wetlands International; 2004. [Google Scholar]
- 12.World Health Organization . Available Evidence Suggests No Need to Change the WHO Recommended Influenza A/H5N1 Vaccine Prototype Strains. Geneva: World Health Organization; 2005. available at http://www.who.int/csr/disease/avian_influenza/statement_2005_07_20/en/index.html. [Google Scholar]
- 13.Li K. S., Guan Y., Wang J., Smith G. J. D., Xu K. M., Duan L., Rahardjo A. P., Puthavathana P., Buranathai C., Nguyen T. D., et al. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 14.Ha Y., Stevens D. J., Skehel J. J., Wiley D. C. Proc. Natl. Acad. Sci. USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claas E. C., Osterhaus A. D., van Beek R., De Jong J. C., Rimmelzwaan G. F., Senne D. A., Krauss S., Shortridge K. F., Webster R. G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 16.Scholtissek C., Quack G., Klenk H. D., Webster R. G. Antiviral Res. 1998;37:83–95. doi: 10.1016/s0166-3542(97)00061-2. [DOI] [PubMed] [Google Scholar]
- 17.Hatta M., Gao P., Halfmann P., Kawaoka Y. Science. 2001;293:1773–1775. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 18.Seo S. H., Hoffmann E., Webster R. G. Nat. Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 19.Fouchier R. A., Schneeberger P. M., Rozendaal F. W., Broekman J. M., Kemink S. A., Munster V., Kuiken T., Rimmelzwaan G. F., Schutten M., Van Doornum G. J., et al. Proc. Natl. Acad. Sci. USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taubenberger J. K., Reid A. H., Lourens R. M., Wang R., Jin G., Fanning T. G. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 21.Shortridge K. F., Stuart-Harris C. H. Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 22.Hulse-Post D. J., Sturm-Ramirez K. M., Humberd J., Seiler P., Govorkova E. A., Krauss S., Scholtissek C., Puthavathana P., Buranathai C., Nguyen T. D., et al. Proc. Natl. Acad. Sci. USA. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banks J., Speidel E. S., Moore E., Plowright L., Piccirillo A., Capua I., Cordioli P., Fioretti A., Alexander D. J. Archiv. Virol. 2001;146:963–973. doi: 10.1007/s007050170128. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D. C., Uyeki T. M., Jadhao S., Maines T., Shaw M., Matsuoka Y., Smith C., Rowe T., Lu X., Hall H., et al. J. Virol. 2005;79:4201–4212. doi: 10.1128/JVI.79.7.4201-4212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Deng G., Li Z., Tian G., Li Y., Jiao P., Zhang L., Liu Z., Webster R. G., Yu K. Proc. Natl. Acad. Sci. USA. 2004;101:10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis T. M., Bousfield R. B., Bissett L. A., Dyrting K. C., Luk G. S., Tsim S. T., Sturm-Ramirez K., Webster R. G., Guan Y., Malik Peiris J. S. Avian Pathol. 2004;33:492–505. doi: 10.1080/03079450400003601. [DOI] [PubMed] [Google Scholar]
- 27.Mase M., Tsukamoto K., Imada T., Imai K., Tanimura N., Nakamura K., Yamamoto Y., Hitomi T., Kira T., Nakai T., et al. Virology. 2005;332:167–176. doi: 10.1016/j.virol.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Lee C. W., Suarez D. L., Tumpey T. M., Sung H. W., Kwon Y. K., Lee Y. J., Choi J. G., Joh S. J., Kim M. C., Lee E. K., et al. J. Virol. 2005;79:3692–3702. doi: 10.1128/JVI.79.6.3692-3702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization . Avian Influenza: Situation in Indonesia. Geneva: World Health Organization; 2005. Update 26; available at www.who.int/csr/don/2005_07_29c/en/index.htm. [Google Scholar]
- 30.Guan Y., Shortridge K. F., Krauss S., Chin P. S., Dyrting K. C., Ellis T. M., Webster R. G., Peiris M. J. Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturm-Ramirez K. M., Hulse-Post D. J., Govoroka E. A., Humberd J., Seiler P., Puthavathana P., Buranathai C., Nguyen T. D., Chaisongh A., Long H. T., et al. J. Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan Y., Peiris J. S., Lipatov A. S., Ellis T. M., Dyrting K. C., Krauss S., Zhang L. J., Webster R. G., Shortridge K. F. Proc. Natl. Acad. Sci. USA. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall T. A. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 34.Nylander J. A. A. mrmodeltest 2. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 35.Swofford D. L. paup*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 36.Huelsenbeck P. J., Ronquist F. R. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



