Abstract
Membrane fusion in the secretory pathway is mediated by soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins. Different fusion steps are thought to be effected by independent sets of SNAREs, but it is unclear whether specificity is determined by an intrinsic specificity of SNARE pairing or by upstream factors. Using a newly developed microscopy-based assay, we have investigated the SNARE specificity of homotypic early endosomal fusion. We show that early endosomes contain multiple sets of SNAREs, including, in addition to the putative early endosomal SNAREs, those involved in exocytosis and in fusion of late endosomes. We demonstrate that fusion is largely mediated by a complex formed by syntaxin 13, syntaxin 6, vti1a, and VAMP4, whereas the exocytic and late endosomal SNAREs play little or no role in the reaction. In contrast, proteoliposomes reconstituted with early endosomal SNAREs promiscuously fuse with liposomes containing exocytotic or late endosomal SNAREs. We conclude that the specificity of SNARE pairing does not suffice to determine the specificity of organelle fusion.
Keywords: endocytosis, exocytosis, PC12 cells
Soluble N-ethylmaleimide (NEM)-sensitive factor attachment receptor (SNARE) proteins are represented by a superfamily of small membrane-associated proteins that mediate membrane fusion in the secretory pathway. All SNAREs possess a SNARE motif that is usually flanked by a C-terminal transmembrane domain (for recent reviews, see refs. 1–3). SNAREs assemble into stable bundles of four α-helices, forming coiled coils with the transmembrane domains located at one end of the bundle (4–6). Each helix is contributed by a different SNARE motif belonging to separate subfamilies, termed Qa-, Qb-, Qc-, and R-SNARE motifs, respectively (7, 8). All functional SNARE complexes are thought to contain one member of each subfamily (QabcR-rule), and, despite limited sequence homologies, the structure of SNARE complexes is highly conserved (6).
Assembly among SNAREs bridging the two fusing membranes is considered to be the key step in membrane fusion. It is probably initiated at the N terminus of the SNARE motifs and proceeds toward the C terminus (referred to as “zippering”), resulting in a strained “trans”-complex. During membrane merger, the intermediate relaxes into a “cis”-complex in which the transmembrane domains are aligned so that they are parallel (1–3).
There are 25 SNAREs in yeast and >35 in mammals, and it was proposed that the SNAREs directly encode the specificity of organelle fusion (9–12). According to this view, each SNARE complex functions in only one intracellular fusion step and pairs only with its “cognate” binding partners. For instance, the SNAREs involved in regulated exocytosis of neurons and neuroendocrine cells, including syntaxin 1 (Qa), soluble NSF attachment protein (SNAP)-25 (Qbc), and synaptobrevin/VAMP2 (R), are thought to function exclusively in regulated exocytosis. Similarly, unique SNARE complexes have been assigned for most trafficking steps in yeast.
However, certain SNAREs function in multiple trafficking steps, and some SNAREs are known to substitute for each other (13–17). Which are the factors determining, for a given SNARE, the fusion steps in which it operates and from which it is excluded? In in vitro experiments involving soluble SNAREs, little discrimination was observed between SNAREs of the same subfamily in their capacity to form core complexes (18, 19). However, experiments involving fusion of SNARE-containing liposomes suggested a high degree of pairing specificity, raising the possibility that SNARE pairing is much more specific when the SNAREs are integrated in membranes (10–12).
In the present study, we have investigated the SNARE specificity of homotypic fusion of early endosomes from neurosecretory cells. Early (or sorting) endosomes are central relay stations for membrane recycling. Early endosomes are connected to late endosomes and lysosomes, to the Golgi apparatus, and to the plasma membrane, and they are involved in recycling of secretory and synaptic vesicles (20, 21). Consequently, the early endosomes would contain not only the SNAREs involved in homotypic fusion, but also SNAREs involved in fusion of the organelles with which they interact. Thus, homotypic fusion of early endosomes is suited as a model to investigate to which extent the SNAREs involved in this fusion step are differentiated from other SNAREs.
Homotypic fusion of early endosomes is one of the best-characterized fusion reactions (22). Recognition, tethering, and docking, the first steps in the reaction sequence, are orchestrated by the small GTPase Rab5, which recruits a variety of effector molecules (23–25). However, the identity of the SNARE proteins responsible for early endosome fusion has not yet been unambiguously determined. Proteins involved in Rab5-effector complexes interact with syntaxin 13 (Qa), syntaxin 16 (Qa), and syntaxin 6 (Qc). Furthermore, antibodies directed against syntaxin 13 inhibit fusion (26). Reports about a functional involvement of syntaxin 6 are contradictory, however (27). Coimmunoprecipitation studies revealed an association between syntaxin 6, vti1a (Qb), and VAMP4 (R) and syntaxin 16 instead of syntaxin 13 (28). In another set of immunoprecipitation experiments, syntaxin 13 was found to be associated with synaptobrevin 2 (R) and SNAP-25 (Qbc) (29). Moreover, botulinum neurotoxin E, which selectively cleaves SNAP-25, has recently been reported to block early endosome fusion (29), whereas, in an earlier study, cleavage of cellubrevin and synaptobrevin by tetanus toxin had no effect (30).
Results
Characterization of an in Vitro Assay for Homotypic Fusion of Early Endosomes.
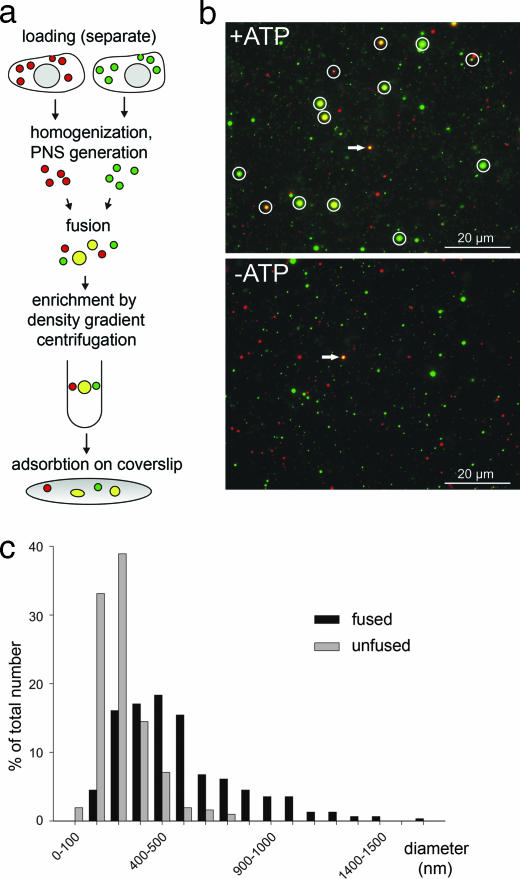
To measure homotypic fusion of early endosomes, we labeled two separate sets of PC12 cells for 5 min with dextrans (10-kDa molecular mass) conjugated to the dyes Alexa 488 and Alexa 594, respectively, as fluid phase markers. Postnuclear supernatants (PNSs) were combined and incubated for the times indicated, and the reaction was stopped by chilling on ice. Fused endosomes were enriched by Nycodenz density gradient centrifugation and then adsorbed on coverslips by low-speed centrifugation (Fig. 6, which is published as supporting information on the PNAS web site), followed by image analysis (Fig. 1a).
Fig. 1.
Microscopic assay for measuring fusion of early endosomes from PC12 cells. (a) Schematic overview of the assay. (b) Representative micrographs of fusion reactions (45 min of incubation) after adsorption of endosomes on coverslips (green channel, dextran-Alexa 488; red channel, dextran-Alexa 594). Fusion assays were carried out in the presence of either an ATP-regenerating system (+ATP) or an ATP-depleting system (−ATP). Images acquired in the red and green channels were aligned by using fluorescent beads (arrows) as reference. Fused endosomes are identified by colocalization (yellow, circles). (c) Size distribution of labeled organelles after incubation for 45 min. Sizes of fused (black) and unfused (gray) organelles were measured by linescan analysis (corrected for the point spread function, see Materials and Methods for details), binned in 100-nm classes, and plotted as percentage of total number (n = 311 for each population).
In vitro fusion of early endosomes is known to be ATP-dependent, and numerous bright spots labeled with both dyes were visible in the presence of ATP (Fig. 1b). The degree of colocalization ranged from 7% to 27% in 80 independent experiments, with much less variability in parallel experiments performed on the same day (particles were scored as fused if their intensity centers in the two channels were not >100 nm apart, and all values were corrected for accidental colocalization). No colocalization was observable in the absence of ATP. Similar results were obtained with labeled transferrin, which is selectively internalized via clathrin-coated vesicles (in three independent experiments, 6–16% colocalization was observed in the presence of ATP, and no colocalization was observed in the absence of ATP). Fusion results in size increase of the organelles (Fig. 1c). The size increase exceeded what would be expected if each endosome would fuse only once, indicating multiple rounds of fusion.
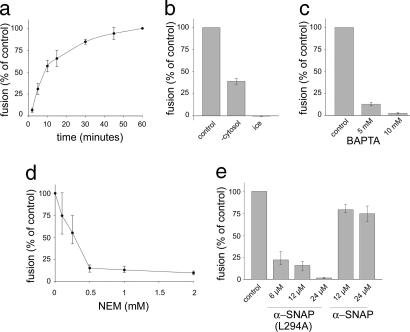
Fusion showed an almost linear increase within the first 10 min and plateaued after ≈1 h (Fig. 2a). Omission of rat brain cytosol reduced fusion, and no fusion was observed even after prolonged incubation on ice (Fig. 2b). Fusion was inhibited by the fast Ca2+-chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (Fig. 2c). These findings are in close agreement with data obtained with a conventional content mixing assay (31–33). Moreover, fusion was progressively inhibited by increasing concentrations of NEM (Fig. 2d), which is attributed to the inhibition of the NEM-sensitive factor (34). To confirm this finding, we used a dominant negative variant of the cofactor α-SNAP (α-SNAP L294A) (35). A dose-dependent inhibition of fusion was observed, whereas wild-type α-SNAP was ineffective (Fig. 2e).
Fig. 2.
Characterization of early endosome fusion by using the microscopic assay. (a) Time course of endosome fusion. Values are means ± SEM of eight independent experiments. Fusion is given as percentage of the 60-min value. (b) Endosome fusion is accelerated when supplemented with rat brain cytosol and is blocked at 0°C. Values are means ± SEM of eight (cytosol) or seven (ice) independent experiments. (c) Endosome fusion is Ca2+-dependent. Fusion assays were performed in the presence of 5 mM or 10 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA) as Ca2+-chelator. Values are means ± SEM of 14 independent experiments. (d) NEM inhibits fusion activity. Both PNSs and rat brain cytosol were preincubated separately with indicated concentrations of NEM for 15 min on ice. Values are means of four independent experiments. Error bars indicate the range of values. (e) Inhibition of endosome fusion by the dominant negative α-SNAP mutant L294A. PNS fractions were preincubated separately with the indicated concentrations of α-SNAP (L294A) or of wild-type α-SNAP for 15 min on ice. Values are means of two to four independent experiments. Error bars indicate the range of values.
Localization of SNAREs on Early Endosomes.
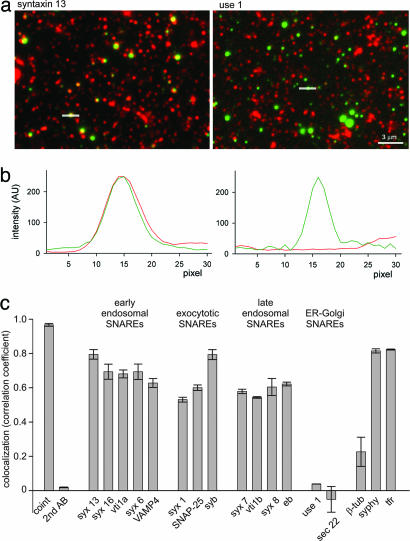
To localize SNAREs, early endosomes were pulse-labeled and isolated as above, fixed, and immunolabeled for a variety of SNAREs (Fig. 3). Not surprisingly, there were many more immunoreactive organelles than labeled endosomes. To determine the degree of colocalization, linescans were performed after image alignment, followed by correlation analysis (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for details). Correlation coefficients of 1 represent complete colocalization, whereas correlation coefficients of 0 represent independent distribution. As a positive control, two differently colored dextrans were cointernalized by the same cells, resulting in a correlation coefficient of 0.97. Immunostaining for the early endosomal SNARE candidates, including syntaxin 13, syntaxin 16, vti1a, syntaxin 6, and VAMP4, resulted in a high degree of colocalization, similar to the transferrin receptor, a bona-fide resident of early endosomes (36). Similarly high correlation coefficients were obtained for both exocytotic and late endosomal SNAREs and for the synaptic vesicle protein synaptophysin (Fig. 3c). In contrast, no correlation was observed for sec 22 and use 1, two SNAREs operating in trafficking between the endoplasmic reticulum and the Golgi apparatus, although immunopositive organelles were of comparable abundance (Fig. 3 a and c).
Fig. 3.
Localization of SNAREs on early endosomes by using immunocytochemistry. (a) Fluorescence micrographs showing endosomes labeled with dextran-Alexa 488 (green channel) immunostained for syntaxin 13 (Left) and use 1 (Right) (red channel). To determine colocalization, linescans were performed through the intensity centers of green endosomes (examples indicated by white lines). (b) Representative linescan analysis, obtained from the images shown in a, showing intensity profiles of green (endosomes) and red (antibody-staining) signals. (c) Colocalization between early endosomes and SNARE proteins as determined by linescan analysis and correlation (see Materials and Methods). A correlation coefficient of 1 represents complete colocalization, whereas a correlation coefficient of ≈0 represent independent distribution. Coint, simultaneous labeling with dextran-Alexa 488 and dextran-Alexa 594 (positive control); 2nd AB, omission of the primary antibody (negative control); syphy, synaptophysin; syx, syntaxin; eb, endobrevin/VAMP8; β-tub, β-tubulin; tfr, transferrin receptor. Values are means of two independent experiments with 60 analyzed endosomes each. Error bars indicate the range of values.
Inhibition of Early Endosome Fusion by Competition with Soluble SNAREs.
To investigate which SNAREs are involved in early endosome fusion, we added recombinant soluble SNAREs to compete with assembly of the endogenous SNAREs and thus inhibit fusion (37). Inactive cis-complexes formed between endogenous membrane-resident SNAREs and exogenously added SNAREs are probably disassembled by NEM-sensitive fusion, which counteracts inhibition; thus, sufficiently high concentrations of the competing SNARE are required.
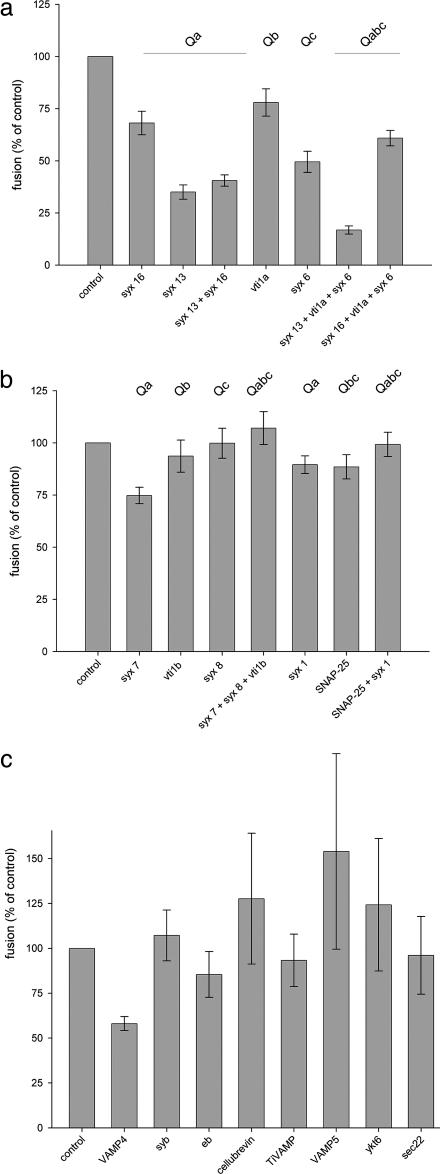
Fig. 4a shows the effects of the candidate early endosomal Q-SNAREs, including the two Qa-SNAREs syntaxin 13 and 16, the Qb-SNARE vti1a, and the Qc-SNARE syntaxin 6, either alone or in various combinations. Inhibition was observed in all cases. However, syntaxin 13 was consistently more potent than syntaxin 16, and no additivity was observed. vti1a and syntaxin 6 caused partial inhibition, and a combination of vti1a, syntaxin 6, and syntaxin 13 almost abolished fusion. No such synergism was observed with syntaxin 16, suggesting that syntaxin 16 does not interact functionally with vti1a and syntaxin 6. In contrast, the neuronal and late endosomal Q-SNAREs were largely ineffective in inhibiting fusion (Fig. 4b). As an independent means to exclude the SNAP-25 functions in early endosome fusion (29), we treated the PNSs with the light chain of botulinum neurotoxin E, a SNAP-25-specific protease. No inhibition was observed (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 4.
Effects of adding recombinant SNAREs on fusion of early endosomes. (a) Effects of Q-SNAREs considered to be involved in the fusion of early endosomes, including syntaxin 16 (syx 16), syntaxin 13 (syx 13), vti1a, and syntaxin 6 (syx 6). Values are means ± SEM of 7–10 independent experiments. (b) Effects of late endosomal and neuronal Q-SNAREs, including syntaxin 7 (syx 7), vti1b, syntaxin 8 (syx 8), syntaxin 1 (syx 1), and SNAP-25. Values are means ± SEM of 7–13 independent experiments. (c) Effects of R-SNAREs, including VAMP4, synaptobrevin/VAMP2 (syb), endobrevin/VAMP8 (eb), cellubrevin/VAMP3, Ti-VAMP/VAMP7, VAMP5, ykt6, and sec22. Values are means ± SEM of three to seven independent experiments. Individual Q-SNAREs were used at 12 μM, with R-SNAREs tested at 25 μM.
We also tested eight different R-SNAREs (Fig. 4c). VAMP4 inhibited fusion to a higher level than all of the other candidates (>40% inhibition, P < 0.001). Endobrevin resulted in only a minor effect (≈14% inhibition, P < 0.05), whereas synaptobrevin, cellubrevin, Ti-VAMP, VAMP5, Ykt6, and Sec22 failed to elicit an inhibitory effect, despite some scatter in the results. Combining endobrevin and VAMP4 did not result in a higher block of fusion than that obtained with VAMP4 alone (data not shown).
Fusion of Proteoliposomes Containing SNAREs.
The results described so far have established (i) that early endosomes contain at least three complete sets of SNAREs and (ii) that only one of these sets is mainly used for homotypic fusion. To test whether this specificity is encoded by the SNAREs themselves, we performed fusion experiments using liposomes coreconstituted with recombinant SNARE proteins. Previous work has established that liposomes reconstituted with appropriate sets of SNAREs spontaneously fuse with each other and that fusion is associated with the formation of SNARE core complexes (38).
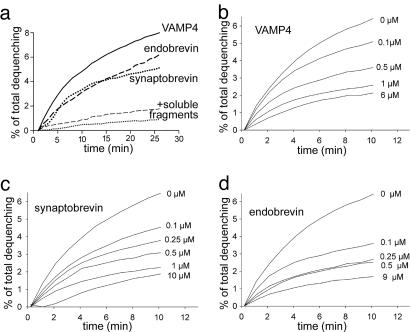
Fusion was monitored by lipid dequenching (39). First, we investigated whether liposomes containing all three early endosomal Q-SNAREs (syntaxin 13, syntaxin 6, and vti1a) fuse exclusively with liposomes containing VAMP4 or whether they are also capable of fusing with liposomes containing synaptobrevin 2 and endobrevin. Robust fusion signals were observed in each case (Fig. 5a); fusion was sensitive to competition by the corresponding soluble R-SNARE. Fusion activity was observed regardless of whether the Q- or R-SNARE liposomes were labeled (Fig. 5a and data not shown). Similarly, liposomes carrying neuronal Q-SNAREs fused equally well with liposomes containing either endobrevin or synaptobrevin (data not shown). Second, we investigated to what extent fusion of Q-SNARE liposomes with VAMP4-containing liposomes is competed for by soluble VAMP4, synaptobrevin, and endobrevin. Again, all three R-SNAREs were capable of competing with fusion in a similar concentration range (Fig. 5 b–d).
Fig. 5.
Proteoliposomes containing the Q-SNAREs syntaxin 13, syntaxin 6, and vti1a show no specificity for R-SNAREs in fusion. (a) Fusion with liposomes containing VAMP4, endobrevin, or synaptobrevin. Fusion was monitored by fluorescence dequenching due to dilution of labeled phospholipids with unlabeled phospholipids during fusion (see Materials and Methods) and normalized to maximal fluorescence measured after adding detergent at the end of the reaction. As control, Q-SNARE liposomes were preincubated for 1 h at room temperature with purified endobrevin or synaptobrevin lacking the transmembrane domain (soluble fragment, final concentration of 30 μM) before starting the fusion reaction. (b–d) Dose-dependent inhibition by soluble R-SNAREs of fusion between liposomes containing the Q-SNAREs syntaxin13, vti1a, and syntaxin 6 and liposomes containing the R-SNARE VAMP4. Measurements were performed at 37°C with an overall protein concentration of 3 μM in the liposomes. The soluble R-SNAREs were added simultaneously with the acceptor liposomes at the start of the reaction (concentrations represent final assay concentrations).
Discussion
In the present study we have identified the SNAREs syntaxin 13, vti1a, syntaxin 6, and VAMP4 as the main SNAREs responsible for homotypic fusion of early endosomes. Neither the SNAREs involved in exocytosis nor the SNAREs involved in the fusion of late endosomes participate to significant levels in the fusion reaction, although both sets of SNAREs are abundantly present on the organelles. In contrast, proteoliposomes reconstituted with the same set of SNAREs promiscuously fuse with each other, showing that the specificity of organelle fusion is not determined by the specificity of SNARE pairing.
Our assay for early endosome fusion is more sensitive than conventional content mixing assays, and it allows for performing immunocytochemistry on isolated organelles. However, the percentage of fusion determined by particle counting is lower than that determined by classical contents-mixing assays. This divergence is to be expected because our assay does not discriminate between single and multiple fusion events, with the latter occurring abundantly, as demonstrated by the size increase during fusion. We were unable to identify a dilution-resistant state of the reaction, because dilution invariably caused an immediate inhibition of fusion (D.B. and R.J., unpublished observations). Moreover, attempts to define a docked but not fused state by measuring adjacent but nonoverlapping dots were not successful even when the putatively postdocking inhibitor 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate was used.
Previous studies have shown that cell-free fusion assays are preferentially inhibited by cognate SNAREs although some crosstalk between cognate and noncognate SNAREs was observed. For instance, in cracked PC12 cells, exocytosis is more sensitive to inhibition by synaptobrevin/VAMP2 than by VAMP7 and endobrevin/VAMP8 (40). However, specificity was not absolute because VAMP4 was almost as effective as synaptobrevin. Similarly, syntaxin 4 was almost as potent in inhibiting exocytosis as syntaxin 1, whereas several other syntaxins, including syntaxin 6, syntaxin 8, and syntaxin 13, were ineffective. In our hands, a higher degree of specificity was observed. This observation closely agrees with the fact that the early endosomes form a main sorting platform through which many different SNAREs travel; accurate sorting probably relies on effective mechanisms to select one set of SNAREs for the homotypic fusion while silencing all others.
Our results show that the SNARE specificity of intracellular fusion reactions is not controlled by their ability to form complexes or by their ability to fuse liposomes, suggesting that pairing specificity is not a main determinant for the specificity of organelle fusion (at least in early endosome fusion). Rather, it is likely that specificity is achieved by additional layers of control that may involve regions outside the SNARE motif or the outer surface of the helical SNARE motifs. Recently, it has been shown that the SM protein Sly1p increases the specificity of SNARE complex formation in the yeast endoplasmic reticulum-to-Golgi SNARE complex (41), raising the possibility that SM proteins are an integral part of the SNARE proofreading mechanism. Another possibility is the direct interaction of specific SNAREs with organelle-specific tethering factors, as exemplified by the binding of syntaxin 13 and syntaxin 6 to EEA1 (27, 42). The contrast between the ability of recombinant SNARE motifs to inhibit endosome and liposome fusion asks for mechanisms that differentiate between individual SNARE motifs of the same subclass, which remain to be identified.
Materials and Methods
Antibodies.
The following antibodies were described previously: rabbit sera specific for syntaxin 7 and syntaxin 8 (37), endobrevin (18), and vti1b (43), mouse monoclonal antibodies specific for synaptophysin Cl 7.2 (44), synaptobrevin Cl 69.1 (45), and SNAP-25 Cl 71.1 (46) (all available from Synaptic Systems, Göttingen, Germany), and syntaxin 1 HPC-1 (47). Antibodies for the transferrin receptor were obtained from Zymed, and antibodies for β-tubulin (TUJ1) were obtained from Babco (Richmond, CA). Secondary antibodies (goat anti-rabbit and goat anti-mouse, Cy3-labeled) were purchased from Jackson ImmunoResearch.
Rabbit antisera were generated by using purified cytosolic parts of VAMP4, syntaxin 6, syntaxin 13, and vti1a as antigens. All antisera recognized single bands of the expected molecular mass in immunoblots of PC12 cell homogenates (data not shown). Where indicated, antibodies were affinity-purified by using the respective purified proteins bound to CNBr-Sepharose (Amersham Pharmacia). Affinity-purified rabbit antibodies for the following proteins were kind gifts: Sec 22, use 1 (S. Verrier, Max Planck Institute for Biophysical Chemistry), and vti1b (G. Fischer von Mollard, University of Bielefeld, Bielefeld, Germany).
Molecular Cloning and Recombinant Proteins.
Syntaxin 6 (residues 1–232) cDNA in pGEX vector was provided by R. H. Scheller (Genentech, San Francisco) (48). The transmembrane region was attached by PCR using standard procedures. Syntaxin 13 pGEX 1–232 (37) was subcloned in pET28a. DNA encoding syntaxin 16 and Vti1a was provided by G. Fischer von Mollard (28). cDNA encoding full-length VAMP4 (residues 1–141) was amplified by RT-PCR using AccessQuick RT-PCR System kit from Promega from a rat brain library (provided by S. Takamori, Tokyo Medical and Dental University, Tokyo). All constructs were subcloned into the pET28a vector. Each clone was verified by DNA sequencing. For details, see Supporting Materials and Methods.
Recombinant proteins were expressed as His6-tagged or GST-tagged fusion proteins and purified by Ni2+–agarose or glutathione–Sepharose, respectively. The tags of all proteins were removed by using thrombin cleavage. All proteins were further purified by ion-exchange chromatography.
Syntaxin 7 (residues 1–236), vti1b (residues 1–206), syntaxin 8 (residues 1–213), and endobrevin (residues 1–74) were cloned, expressed, and purified as described in ref. 37. Syntaxin 1a (residues 1–262), SNAP-25a (residues 1–206), and synaptobrevin/VAMP2 (residues 1–96) were described in refs. 18 and 49.
Wild-type α-SNAP and α-SNAP L294A (35) were provided by D. Fasshauer (Max Planck Institute for Biophysical Chemistry). Sec22 was provided by S. Verrier. VAMP5, ykt6, and sec22 were used as GST-tagged constructs, with GST alone used as a negative control. All proteins were 95% pure as judged by SDS/PAGE and Coomassie blue staining.
Cell Culture and Internalization of Marker.
PC12 cells [clone 251 (50)] were grown to confluence on 15-cm-diameter culture dishes in DMEM (with 5% FCS, 10% horse serum, 4 mM glutamine, and 100 units/ml each of penicillin and streptomycin) at 37°C in 10% CO2.
Cells were harvested by washing once with ice-cold PBS (150 mM NaCl/200 mM Na2HPO4, pH 7.4), adding trypsin/EDTA (2 ml per plate; Cambrex, East Rutherford, NJ). Fifteen confluent plates resulted in a cell pellet of ≈2 ml volume. Cells were washed with internalization medium (OptiMEM; Invitrogen, supplemented with 10 mM glucose), prewarmed, and incubated for 5 min with marker (10-kDa dextran labeled with Alexa 488 or Alexa 594, respectively; Molecular Probes) dissolved in internalization medium. After the internalization had been stopped by diluting the cells in 10 ml of ice-cold PBS with 5 mg/ml BSA, the cells were washed three times.
Preparation of Subcellular Fractions and Rat Brain Cytosol.
PC12 cells were homogenized as described in ref. 33 with slight modifications. Briefly, the cell pellet was resuspended 1:4 in homogenization buffer (250 mM sucrose/3 mM imidazole·HCl, pH 4.7) with protease inhibitors (0.2 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 0.7 μg/ml pepstatin) and homogenized by 10 passages through a stainless-steel ball homogenizer with a clearance of 0.0008 inch (20 μm). The homogenates were centrifuged for 15 min at 1,200 × g, and the resulting PNSs were divided into aliquots and snap frozen in liquid N2. PNS fractions labeled with Alexa 488 and Alexa 594 were used for the cell-free fusion assay. Rat brain cytosol was prepared from fraction S2 (51) by centrifugation at 300,000 × g for 30 min.
Cell-Free Fusion Assay.
Reaction mixtures (50 μl final volume) contained, as final concentrations, 4 mg/ml PNSs, 2 mg/ml cytosol, 11.25 mM Hepes at pH 7.0, 1.35 mM magnesium acetate, 0.18 mM DTT, 45 mM potassium acetate, as an ATP-regenerating system, 3.2 mM ATP, 26 mM creatine phosphate, and 0.132 mg creatine kinase (800 units/mg; Roche, Basel, Switzerland) or, as an ATP-depleting system, 5 μl of hexokinase (1,500 units/ml dissolved in 250 mM glucose; Roche). The reaction mixtures were incubated in polycarbonate centrifuge tubes for 45 min at 37°C. After the reaction had been stopped by cooling on ice, reaction mixtures were overlaid with 100 μl of ice-cold 30% Nycodenz solution [30% Nycodenz (Nycomed Pharma, Unterschleissheim, Germany) in 0.5 mM EDTA/3 mM imidazole, pH 7.4] and 100 μl of ice-cold 12% Nycodenz solution (12% Nycodenz in 0.5 mM EDTA/3 mM imidazole, pH 7.4) and then centrifuged for 90 min at 170,000 × g at 4°C. This purification step was necessary to reduce background fluorescence in the Alexa 488 channel. After removal of the top 130 μl, 10 μl was harvested from the top of the remaining solution and transferred into 1 ml of PBS on coverslips (18-mm diameter) in 12-well plates. After centrifugation for 2 h at 1,500 × g at 4°C, coverslips were analyzed with a fluorescence microscope (see below). Before use of the coverslips, TetraSpeck beads (200-nm diameter, dilution 1:100,000 in 1 ml PBS; Molecular Probes) were bound to the surface by centrifugation for 1 h at 1,500 × g.
For treatment with NEM, PNSs and cytosol were incubated separately with indicated final concentrations of NEM for 15 min on ice. For the experiments presented in Fig. 4, PNS fractions were preincubated with recombinant proteins for 15–20 min. Coverslips were analyzed by using a Zeiss Axiovert 200M fluorescence microscope with a 1.4 numerical aperture ×100 objective and appropriate filter sets. Several controls were performed to ensure that the centrifugation/adsorption steps that followed the fusion reaction did not distort the results (see Supporting Materials and Methods for details).
Size Determination of Early Endosomes.
Fusion assays with ATP-regenerating and -depleting systems were performed as described above. Images were analyzed by using the linescan function in metamorph (Universal Imaging, West Chester, PA). Linescans with a width of 1 pixel and a length of 31 pixels were performed through the intensity centers of 311 fused and unfused endosomes, respectively. Values were corrected by the point spread function that was experimentally defined by measuring TetraSpeck beads of 200-nm size in the red and green channel and divided into subclasses of 100-nm size.
Immunocytochemistry.
PNSs obtained from labeled cells were incubated by using an ATP-depleting system as described above. Gradient centrifugation and adsorption to a glass coverslip were carried out as above except that the coverslips were preincubated with 10 mg/ml BSA overnight at 37°C. Adsorbed organelles were fixed with 4% paraformaldehyde in PBS for 15 min, quenched for 15 min in PBS containing 50 mM NH4Cl, and washed two times with PBS. Primary antibodies were used in a 1:100 dilution in PBS containing 5% FCS for 1 h. Subsequently, endosomes were washed three times with PBS for 10 min each, followed by incubation with secondary antibodies (Cy 3-labeled, dilution 1:100 in PBS/5% FCS) for 45 min. After washing three times with PBS, coverslips were imaged as described for the fusion assays.
Other Methods.
Liposome preparation and liposome fusion assays were performed as described in ref. 52, except that the Hepes buffer contained 1 M KCl and further purification of liposomes on Nycodenz-gradients was omitted. Liposomes were adjusted to yield a final protein concentration of 3 μM, and all fusion experiments were carried out at 37°C. SDS/PAGE and immunoblotting were performed as described in refs. 53 and 54, and protein determination was performed according to ref. 55.
Supplementary Material
Acknowledgments
We thank Drs. P. Küster (Max Planck Institute for Biophysical Chemistry) and C. Schütte (ProSciencia, Lübeck, Germany) for their help in image and data analysis; Drs. G. Fischer von Mollard, H. D. Söling (Max Planck Institute for Biophysical Chemistry), and S. Verrier for the gift of antibodies and plasmids; and Drs. Olga Vites (Max Planck Institute for Biophysical Chemistry) and Dirk Fasshauer for the gift of recombinant proteins. S.O.R. is the recipient of long-term fellowships from the European Molecular Biology Organization and from the Human Frontier Science Program. This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 523, TP B6 (to R.J.), and LA127212-1 (to T.L.).
Abbreviations
- SNAP
soluble NEM-sensitive factor attachment protein
- NEM
N-ethylmaleimide
- PNS
postnuclear supernatant
- SNARE
soluble NEM-sensitive fusion attachment receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Fasshauer D. Biochim. Biophys. Acta. 2003;1641:87–97. doi: 10.1016/s0167-4889(03)00090-9. [DOI] [PubMed] [Google Scholar]
- 2.Rizo J., Südhof T. C. Nat. Rev. Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R., Lang T., Südhof T. C. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 4.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 5.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 6.Antonin W., Fasshauer D., Becker S., Jahn R., Schneider T. R. Nat. Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 7.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock J. B., Matern H. T., Peden A. A., Scheller R. H. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 9.Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 10.McNew J. A., Parlati F., Fukuda R., Johnston R. J., Paz K., Paumet F., Söllner T. H., Rothman J. E. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 11.Parlati F., Varlamov O., Paz K., McNew J. A., Hurtado D., Söllner T. H., Rothman J. E. Proc. Natl. Acad. Sci. USA. 2002;99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paumet F., Rahimian V., Rothman J. E. Proc. Natl. Acad. Sci. USA. 2004;101:3376–3380. doi: 10.1073/pnas.0400271101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerrard S. R., Mecklem A. B., Stevens T. H. Traffic. 2000;1:45–55. doi: 10.1034/j.1600-0854.2000.010108.x. [DOI] [PubMed] [Google Scholar]
- 14.Darsow T., Rieder S. E., Emr S. D. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Barlowe C. Mol. Biol. Cell. 2002;13:3314–3324. doi: 10.1091/mbc.E02-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorngren N., Collins K. M., Fratti R. A., Wickner W., Merz A. J. EMBO J. 2004;23:2765–2776. doi: 10.1038/sj.emboj.7600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sørensen J. B., Nagy G., Varoqueaux F., Nehring R. B., Brose N., Wilson M. C., Neher E. Cell. 2003;114:75–86. doi: 10.1016/s0092-8674(03)00477-x. [DOI] [PubMed] [Google Scholar]
- 18.Fasshauer D., Antonin W., Margittai M., Pabst S., Jahn R. J. Biol. Chem. 1999;274:15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 19.Yang B., Gonzalez L., Jr, Prekeris R., Steegmaier M., Advani R. J., Scheller R. H. J. Biol. Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- 20.Maxfield F. R., McGraw T. E. Nat. Rev. Mol. Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 21.Südhof T. C. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 22.Zerial M., McBride H. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 23.Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 24.Christoforidis S., McBride H. M., Burgoyne R. D., Zerial M. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., Zerial M. J. Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills I. G., Urbe S., Clague M. J. J. Cell Sci. 2001;114:1959–1965. doi: 10.1242/jcs.114.10.1959. [DOI] [PubMed] [Google Scholar]
- 27.McBride H. M., Rybin V., Murphy C., Giner A., Teasdale R., Zerial M. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 28.Kreykenbohm V., Wenzel D., Antonin W., Atlachkine V., Fischer von Mollard G. Eur. J. Cell Biol. 2002;81:273–280. doi: 10.1078/0171-9335-00247. [DOI] [PubMed] [Google Scholar]
- 29.Sun W., Yan Q., Vida T. A., Bean A. J. J. Cell Biol. 2003;162:125–137. doi: 10.1083/jcb.200302083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Link E., McMahon H., Fischer von Mollard G., Yamasaki S., Niemann H., Südhof T. C., Jahn R. J. Biol. Chem. 1993;268:18423–18426. [PubMed] [Google Scholar]
- 31.Gorvel J. P., Chavrier P., Zerial M., Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 32.Gruenberg J., Griffiths G., Howell K. E. J. Cell Biol. 1989;108:1301–1316. doi: 10.1083/jcb.108.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holroyd C., Kistner U., Annaert W., Jahn R. Mol. Biol. Cell. 1999;10:3035–3044. doi: 10.1091/mbc.10.9.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz R., Mayorga L. S., Weidman P. J., Rothman J. E., Stahl P. D. Nature. 1989;339:398–400. doi: 10.1038/339398a0. [DOI] [PubMed] [Google Scholar]
- 35.Barnard R. J., Morgan A., Burgoyne R. D. J. Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trowbridge I. S., Collawn J. F., Hopkins C. R. Annu. Rev. Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 37.Antonin W., Holroyd C., Fasshauer D., Pabst S., Fischer von Mollard G., Jahn R. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 39.Struck D. K., Hoekstra D., Pagano R. E. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 40.Scales S. J., Chen Y. A., Yoo B. Y., Patel S. M., Doung Y. C., Scheller R. H. Neuron. 2000;26:457–464. doi: 10.1016/s0896-6273(00)81177-0. [DOI] [PubMed] [Google Scholar]
- 41.Peng R., Gallwitz D. J. Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonsen A., Gaullier J. M., D’Arrigo A., Stenmark H. J. Biol. Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- 43.Antonin W., Riedel D., Fischer von Mollard G. J. Neurosci. 2000;20:5724–5732. doi: 10.1523/JNEUROSCI.20-15-05724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahn R., Schiebler W., Ouimet C., Greengard P. Proc. Natl. Acad. Sci. USA. 1985;82:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelmann L., Hanson P. I., Chapman E. R., Jahn R. EMBO J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruns D., Engers S., Yang C., Ossig R., Jeromin A., Jahn R. J. Neurosci. 1997;17:1898–1910. doi: 10.1523/JNEUROSCI.17-06-01898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnstable C. J., Hofstein R., Akagawa K. Dev. Brain Res. 1985;20:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- 48.Bock J. B., Lin R. C., Scheller R. H. J. Biol. Chem. 1996;271:17961–17965. doi: 10.1074/jbc.271.30.17961. [DOI] [PubMed] [Google Scholar]
- 49.Vites O., Rhee J. S., Schwarz M., Rosenmund C., Jahn R. J. Biol. Chem. 2004;279:26251–26256. doi: 10.1074/jbc.M404079200. [DOI] [PubMed] [Google Scholar]
- 50.Heumann R., Kachel V., Thoenen H. Exp. Cell Res. 1983;145:179–190. doi: 10.1016/s0014-4827(83)80019-6. [DOI] [PubMed] [Google Scholar]
- 51.Huttner W. B., Schiebler W., Greengard P., De Camilli P. J. Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuette C. G., Hatsuzawa K., Margittai M., Stein A., Riedel D., Kuster P., König M., Seidel C., Jahn R. Proc. Natl. Acad. Sci. USA. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kyhse-Andersen J. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 54.Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 55.Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.