Abstract
The crystal structure at 1.93-Å resolution is determined for the Ca2+-discharged obelin containing three bound calcium ions as well as the product of the bioluminescence reaction, coelenteramide. This finding extends the series of available spatial structures of the ligand-dependent conformations of the protein to four, the obelin itself, and those after the bioluminescence reaction with or without bound Ca2+ and/or coelenteramide. Among these structures, global conformational changes are small, typical of the class of “calcium signal modulators” within the EF-hand protein superfamily. Nevertheless, in the active site there are significant repositions of two residues. The His-175 imidazole ring flips becoming almost perpendicular to the original orientation corroborating the crucial importance of this residue for triggering bioluminescence. Tyr-138 hydrogen bonded to the coelenterazine N1-atom in unreacted obelin is moved away from the binding cavity after reaction. However, this Tyr is displaced by a water molecule from within the cavity, which now forms a hydrogen bond to the same atom, the amide N of coelenteramide. From this observation, a reaction scheme is proposed that would result in the neutral coelenteramide as the primary excited state product in photoprotein bioluminescence. From such a higher energy state it is now energetically feasible to account for the shorter wavelength bioluminescence spectra obtained from some photoprotein mutants or to populate the lower energy state of the phenolate anion to yield the blue bioluminescence ordinarily observed from native photoproteins.
Keywords: coelenterazine, photoprotein, EF hand, luciferase, aequorin
Among several luciferins that have been identified in luminous organisms, one well characterized type is “coelenterazine,” an imidazopyrazinone derivative. The best studied proteins using this luciferin are the Ca2+-regulated photoproteins such as those responsible for the bioluminescence of marine coelenterates (1), for example, aequorin from the jellyfish Aequorea (2, 3) and obelin from the hydroid Obelia (4, 5). All of the photoproteins characterized to date consist of a single polypeptide chain with high sequence homology and contain three “EF-hand” Ca2+-binding consensus sequences (6–8) like some other Ca2+-binding proteins (9). Their bioluminescence property derives from the fact that they contain in their active site an “activated” coelenterazine, 2-hydroperoxycoelenterazine, tightly but noncovalently bound. Thus, photoproteins can be regarded as luciferases except that they have a high energy reaction intermediate frozen in place (10). The binding of Ca2+ into the loops of the EF-hands apparently leads to some conformational change in the photoprotein that destabilizes this intermediate leading to rapid emission of bioluminescence. This response on the millisecond time scale must have to do with the biological function in the animal, e.g., frightening of predators, although this function is not established.
It is the purpose of our structural studies to determine how the Ca2+ binding triggers the destabilization of the intermediate and how interactions with the residues comprising the active site direct the decarboxylation of the peroxycoelenterazine to efficiently produce the protein bound product, coelenteramide, in its excited (fluorescent) state (11, 12). The blue bioluminescence emission results when this excited coelenteramide relaxes to its ground state. The bioluminescence spectra are broad with maxima in the range 465–495 nm depending on the type of photoprotein (13). Spatial structures recently determined for aequorin and obelin confirm the presence of this 2-hydroperoxycoelenterazine and show that it is situated within a hydrophobic cavity and stabilized there by a hydrogen bond network (14, 15).
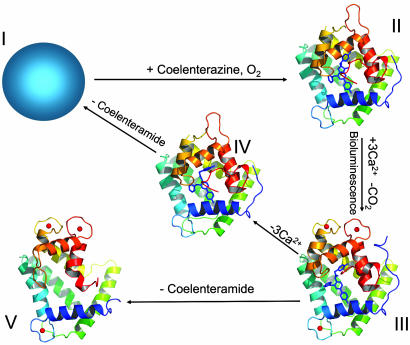
In this paper, we report the crystal structure at 1.93 Å of Ca2+-discharged obelin, i.e., obelin after the addition of Ca2+ to generate bioluminescence with both bound coelenteramide product and three calcium ions (state III, Fig. 1). This result now extends the series of available crystal structures of the ligand-dependent conformations of obelin to four (16) (Fig. 1). This structure shows important differences in how some residues are arranged in the binding cavity compared to the previously determined Ca2+-discharged obelin that lacked the bound Ca2+ (state IV, Fig. 1) and leads to a suggestion for a pathway forming coelenteramide in its neutral excited state as the primary product of this bioluminescence reaction.
Fig. 1.
The three-dimensional structures of conformation states of obelin: II, obelin with 2-hydroperoxycoelenterazine in the absence of Ca2+ (PDB ID code 1EL4) (15); III, Ca2+-discharged obelin with both coelenteramide and bound Ca2+ (present result); IV, Ca2+-discharged obelin with coelenteramide but without Ca2+ (PDB ID code 1S36) (17); V, Ca2+-loaded apo-obelin (PDB ID code 1SL7) (18). The structure of apo-obelin (state I) is not determined. The crystal structures of different obelin conformation states are presented in the same orientation. The 2-hydroperoxycoelenterazine and coelenteramide molecules are displayed by the stick models in the center of the protein; the calcium ions are shown as red balls.
It should be noted that comparable structural insight into photoprotein bioluminescence is not available from the structures of other types of luciferases (19–21) and indeed, there are few other enzymes available with substrate or product able to be visualized within the active site. Many of the EF-hand family of calcium proteins exert their biological function via interactions usually with macromolecular ligands and structural determinations of these complexes is an active area of research (22).
Results
Overall Structure.
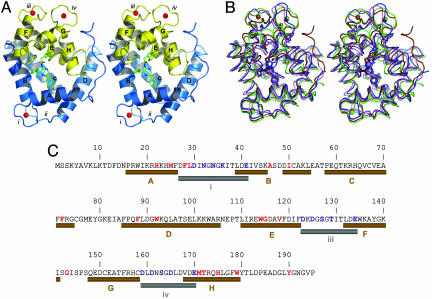
The spatial structure of the protein retains the same overall scaffold with its characteristic two-domain fold (Fig. 2A), in all conformation states (Fig. 2B and Table 1). The range of overall main chain rms deviations is only from a minimum 1.922 Å (between conformation states II and III) to a maximum 2.077 Å (between states III and V). Additionally, the C-terminal domain undergoes a bigger change than the N-terminal domain. EF-hand ii in the N-terminal domain does not have the loop consensus sequence for binding Ca2+ and therefore shows no significant change in any of the conformation states. However, rms deviations of side chain atoms are much bigger (Table 1). The largest repositioning of atoms is observed again in the C-terminal domain and EF-hand iv. Also, the side chain atoms of loop iv undergo the biggest repositioning even among loops capable of binding Ca2+. A calcium atom is found at each of the expected Ca2+-binding sites, EF-hand loops i, iii, and iv, as also observed in state V, the Ca2+-loaded apo-obelin (18).
Fig. 2.
The spatial structure of Ca2+-discharged obelin and its comparison with other ligand-dependent confirmation states shown in Fig. 1. (A) Stereoview of the overall crystal structure of Ca2+-discharged obelin. The stick model in the center of the protein is the coelenteramide molecule. The helices are marked by capital letters A–H. Numbers i–iv designate the loops. Calcium ions are shown as red balls. (B) Stereoview of the superimposition of different obelin conformation states: II, blue; III, brown; IV, pink; and V, green. The 2-hydroperoxycoelenterazine and coelenteramide molecules are displayed by the stick models in the center of the protein; the calcium ions are shown as balls. The 2-hydroperoxycoelenterazine, coelenteramide, and calcium are colored according to the conformation state. (C) Sequence of the obelin from O. longissima (45). The residues comprising the coelenteramide binding cavity or participating in calcium binding are marked with red and blue, respectively. The helices and loops involved in the binding of Ca2+ are shown as brown and gray cylinders, respectively, according to the structure of Ca2+-discharged obelin.
Table 1.
rms deviation values of state III vs. different obelin states
| rms deviation* | State II | State IV | State V |
|---|---|---|---|
| Overall† | 1.922/2.745‡ | 1.996/2.783 | 2.077/3.274 |
| N-terminal domain§ | 1.342/2.012 | 1.349/1.979 | 0.958/2.069 |
| C-terminal domain¶ | 1.969/3.164 | 1.978/3.181 | 2.652/4.225 |
| EF-hand i∥ | 1.685/2.283 | 1.701/1.937 | 0.826/1.924 |
| EF-hand ii | 0.463/1.536 | 0.589/1.925 | 0.472/1.767 |
| EF-hand iii | 1.818/3.094 | 1.801/3.015 | 1.524/3.247 |
| EF-hand iv | 1.839/3.350 | 1.876/3.446 | 1.763/3.738 |
| Loop i** | 1.119/1.948 | 0.735/1.371 | 0.180/0.919 |
| Loop ii | 0.455/1.866 | 0.444/1.889 | 0.275/0.488 |
| Loop iii | 1.404/2.574 | —†† | 0.598/0.814 |
| Loop iv | 1.905/4.632 | 1.912/4.586 | 0.721/0.838 |
*rms deviation was calculated using the program LSQKAB(W 1976).
†The conformational flexible residues 1–10 were excluded in the calculation to better reveal the effect of conformation changes caused by calcium binding and coelenterazine decarboxylation.
‡rms deviation values of main chain/side chain atoms.
§Residues 11–106.
¶Residues 107 to C terminus.
∥EF-hands i, ii, iii, and iv are defined as residues 17–54, 58–104, 110–141, and 149–180, respectively.
**Loops i, ii, iii, and iv are defined as residues 30–40, 75–85, 123–133, and 158–168, respectively.
††Loop iii is disordered in this crystal structure.
In obelin (Fig. 2A), as in aequorin, the C terminus caps the substrate cavity providing a solvent-inaccessible and nonpolar environment. This condition apparently optimizes the efficient population of the first electronic excited state of the product coelenteramide and favors a high quantum yield for its fluorescence (23). The inaccessibility of the substrate for solvent occurs as a result of the numerous hydrogen bond interactions of residues situated in helix A with residues from helix H and from others in the C terminus. For instance, in obelin (state II) the Nε2 atoms of His-22 and His-24 (helix A) form hydrogen bonds with carbonyl oxygens of Trp-179 (helix H) and Gly-193 (C terminus); Nη1 and Nη2 atoms of Arg-21 (helix A) are hydrogen bonded with the carbonyl oxygen of Phe-178 (helix H), with the Oδ1 oxygen of Asp-187 (C terminus), and with the oxygen of Pro-195, all situated in the C terminus. All these listed hydrogen bonds persist in the Ca2+-discharged obelin having bound coelenteramide and calcium (state III) and this strengthens the idea that the binding cavity remains inaccessible for solvent, thus providing the environment favoring the efficient fluorescence of the bound coelenteramide in the Ca2+-discharged photoproteins (8, 24).
Ca2+-Binding Loops.
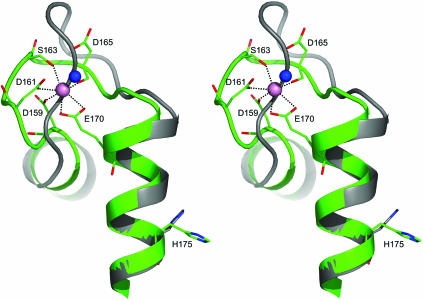
For binding Ca2+ into the EF-hand, the 12 residues of the loop shift their positions to accommodate the Ca2+ in its preferred configuration. Fig. 3 illustrates this movement for the case of loop iv. Loops i and iii have similar but lesser adjustments (Table 1). The typical geometrical arrangement of oxygen atoms in a pentagonal bipyramid is observed with the Ca2+occupying the center of the pyramid. All three Ca2+-binding sites of obelin contribute six oxygen ligands to the metal ion, derived from the carboxylic side groups of Asp and Glu residues, the carbonyl groups of the peptide backbone, or the side chain of Asn, and the hydroxyl group of Ser, all with a coordination distance of ≈2.4 Å. The seventh ligand comes from the oxygen of a water molecule. Based on studies of the EF-hand Ca2+-binding proteins from different sources, it has been observed that high-affinity Ca2+-binding sites have either no water or at most one water ligand (25). The three Ca2-binding sites of the Ca2+-discharged obelin each contain only one water molecule as a ligand, indicating that they all can have high affinity for calcium. Fig. 3 also shows how the loop residue shifts propagate along the chain to reorientate His-175. This displacement of His-175 is postulated as the trigger of the bioluminescence reaction (13).
Fig. 3.
Stereoview of the superposition of Ca2+-binding loop iv of obelin in the Ca2+-discharged state (green) (III) and the native state (gray) (II). The calcium ion and the water molecule are shown as pink and blue balls respectively.
Structure of the Coelenteramide Binding Cavity.
The coelenteramide is situated at the center of this protein structure in the place occupied by the substrate coelenterazine in obelin. The residues comprising the binding cavity, i.e., within 4 Å of the coelenteramide, are indicated in red in Fig. 2C. After the bioluminescence discharge, most residues essentially remain in place compared to their location in the obelin cavity except Ile-142, Ile-111, Phe-119, Ile-142, Ile-144, Trp-135, and Tyr-138, which are displaced, and Phe-28, Gly-143, and Thr-172, which move into the cavity. On average, the residues moving into the cavity are more polar than those that are displaced. As in obelin, there are two water molecules in the cavity of the discharged protein but they are repositioned and for one of them, W2, this appears to have mechanistic significance.
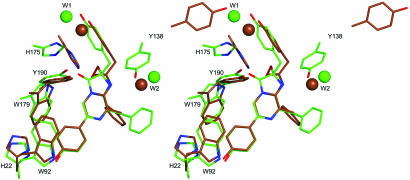
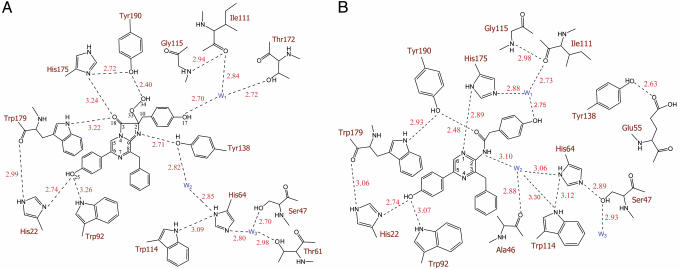
Aside from the loss of CO2 from the chemical decomposition of coelenterazine, the biggest change in its molecular structure is in the reaction center around the C-2 position (C-2, O-33, and C-10) and C-8, resulting in an obvious orientational deviation of the phenol group at the C-10 position and the phenyl group at the C-8 (Fig. 4). Other parts of the molecule also adjust positions a little, but not dramatically. His-22 and Trp-92, which were in hydrogen bond distances with the oxygen of the 6-(p-hydroxyphenyl) group of coelenterazine before reaction (Fig. 5A), are at practically the same distances to coelenteramide in the product cavity. The Nε atoms of these residues are at 2.74 Å and 3.07 Å from the oxygen atom of the 5-(p-hydroxyphenyl) group of coelenteramide, indicative of moderate hydrogen bond interactions (Fig. 5B). In obelin, the Nε atom of Trp-179 is 3.32 Å from the C3-carbonyl oxygen (Fig. 5A) but after reaction, this residue is moved in the direction of Tyr-190 with formation of a new hydrogen bond to it. The Tyr-190 OH group, which apparently stabilizes the 2-hydroperoxy group of coelenterazine in obelin by a hydrogen bond and is also hydrogen bonded to His-175, is also slightly repositioned in the Ca2+-discharged protein. The hydrogen bond to His-175 is lost but a new hydrogen bond develops to the carbonyl of coelenteramide. There is a major reorientation of the His-175 with its imidazole ring now almost perpendicular to the original orientation (Fig. 4). His-175 also forms new hydrogen bonds with the N1 atom of coelenteramide and the water molecule W1 (Fig. 5B). Tyr-138 is moved out of the binding cavity (Fig. 4). The hydrogen bond from Tyr-138 originally to the N1 of coelenterazine now goes to Glu-55 (Fig. 5B), and the Tyr-138 appears to be replaced by that water molecule (W2) originally connecting Tyr-138 to His-64 (Fig. 5A). As a result, the His-64 is also slightly shifted toward coelenteramide. The second water molecule (W1) is also moved, apparently after the change in position of the 2-(p-hydroxybenzyl) group.
Fig. 4.
Stereoview of the superposition of 2-hydroperoxycoelenterazine (green) from PDB ID code 1QV0 and coelenteramide (brown) from present structure, with the key residues facing into the binding cavity. Water molecules are shown as balls.
Fig. 5.
Two-dimensional comparison of the hydrogen bond (dashed lines) network in the binding cavities of obelin (A) and Ca2+-discharged obelin (B). It is seen that with the decomposition of the coelenterazine in the binding site of obelin (A), the Tyr-138 hydrogen bonded to N1 moves away, to be replaced by the water molecule W2.
Thus, only His-175 and Tyr-138 undergo a noticeable repositioning in the binding cavity after the bioluminescence reaction. The importance of the His residue in this position for photoprotein bioluminescence was demonstrated with aequorin (26). Substitution of His-169 (His-175 in obelin) resulted in loss of bioluminescent activity. In contrast, Tyr-138 is not absolutely essential for the bioluminescence. Thus, the change to Y138F obelin preserves practically the same bioluminescence as wild-type obelin, the only differences being a 10-nm longer bioluminescence maximum (λmax = 493 nm), ≈30% lower light yield, and a 10 times slower kinetics.
Discussion
Calcium binding to obelin does not result in large changes in conformation. If this is a general property of Ca2+-regulated photoproteins, it indicates that they belong to the category of Ca2+ signal modulators rather than to the Ca2+ sensors in the EF-hand protein family (27). This categorization probably has to do with function, where for the latter category, the prime example being calmodulin, protein–protein association is involved, whereas the photoproteins have to operate on the millisecond time-scale enabling just a subtle shift to disturb the hydrogen bond network that triggers bioluminescence. This type of shift is exemplified for Ca2+ binding to loop iv (Fig. 3), which propagates to the repositioning of His-175. However, in view of the connectivity among all of the loops which we have described elsewhere (18), it is likely that the triggering of the bioluminescence is a cooperative process. This would lead to quite a complex triggering process, explaining why the interpretation of the bioluminescence kinetics has been controversial.
The most remarkable finding in this present protein structure that has both coelenteramide and the three Ca2+’s bound, is that the water molecule (W2) has positioned itself as a hydrogen bond donor to the amide N (Fig. 5B). In obelin (also in aequorin), the Tyr-138 is hydrogen bonded to this same N of coelenterazine (15); however, in the Ca2+-discharged obelin here, it is moved out of the cavity with its side chain twisted >90° to become solvent exposed and hydrogen bonded to a surface Glu-55. In the structure of the discharged obelin without Ca2+ (state IV), this Tyr-138 is also moved but remains in the binding cavity and water molecule W2 does not move in to fill the vacant position.
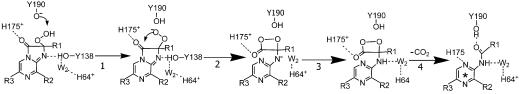
In Scheme 1, we propose a critical function of this water molecule W2 that would rationalize many of the bioluminescence properties. This reaction sequence is along the lines of that derived by Usami and Isobe (28) for the chemiluminescence of a coelenterazine model compound in a protic solvent, a situation appropriate for what might be the condition of the binding cavity of photoproteins. Usami and Isobe observed a chemiluminescence emission after the photooxygenation of the coelenterazine at low temperature (−78°C). They trapped the product of the photooxygenation and established its structure as a dioxetanone derivative using low-temperature NMR. On warming the product, a shorter wavelength chemiluminescence (400 nm) from the neutral dioxetanone intermediate decomposing to the excited state of the neutral coelenteramide was observed at much lower temperature than a longer wavelength (475 nm) chemiluminescence corresponding to the excited state of the coelenteramide model anion, indicating that the dioxetanone anion was more stable. They concluded that there were two pathways available for decarboxylation, one from the neutral dioxetanone, the other from the dioxetanone anion, and that conversion of the anion to the neutral did not take place under the low temperature conditions.
Scheme 1.
It has been proposed (13) that the triggering step in bioluminescence involves transfer of a proton from Tyr-190 to His-175, followed by step 1 in Scheme 1, the deprotonation of hydroperoxide, then step 2 in Scheme 1, the committed formation of the dioxetanone anion. It is assumed that the water molecule has moved into position at this point, but this cannot be verified easily because the protein with dioxetanone and calcium bound would be technically difficult to trap and prepare for crystal structure analysis. However, because Tyr-138 is not essential for the bioluminescence as shown by its substitution to Phe, this assumption is reasonable.
Because the pK of the amide is likely to be higher than the water molecule in this environment, protonation of the anion from the proximate water molecule (Scheme 1, step 3) would be rapid and lead to the neutral dioxetanone. This would be made more energetically feasible by a proton relay from His-64 if that histidine was in its protonated state. We propose that the critical function of this specific water molecule here is to catalyze the decarboxylation reaction by protonating the dioxetanone anion (step 3). Step 4 in Scheme 1 then yields the neutral coelenteramide as the primary excited product in the bioluminescence.
Scheme 1 would rationalize many properties peculiar to coelenterazine bioluminescence in vitro. Both luciferase and photoprotein systems have in vitro bioluminescence spectral maxima in the range of 460–495 nm. A similar range is observed for the fluorescence of coelenteramide analogs in solvents of varying polarity and the presence of strong base and therefore is attributed as originating from the excited state of the coelenteramide anion (29, 30). However, in some mutants of both aequorin and obelin (31–34) two bioluminescence bands can be generated, one similar to the wild type and the other at higher energy with a maximum of ≈400 nm. This latter value corresponds to the fluorescence from the neutral state of coelenteramide. In many studies of these bioluminescence systems, it is stated that the excited coelenteramide amide anion is the primary excited state product. This belief arises from earlier reports such as by McCapra and Chang (35), who investigated the chemiluminescence of an imidazopyrazinone derivative in an aprotic solvent with strong base and provided convincing evidence for a coelenteramide anion as the primary excited state. However, in photoprotein bioluminescence, if the coelenteramide anion were the primary excited state product, it would be energetically infeasible for it to populate the neutral excited state at higher energy. Scheme 1 therefore provides a pathway for formation of the neutral excited state as the primary product. For coelenterazine having the dissociable 6-(p-hydroxyphenyl) group, the longer wavelength bioluminescence bands arise from the excited phenolate after excited state proton transfer to His-22. This process has been described (13).
There is now a sufficient collection of Ca2+-regulated photoprotein structures to conclude that all have a similar cavity structure and should operate by the same mechanism as in Scheme 1. Although there are differences in spectral properties among them, it has been already demonstrated that these are due to different interactions of the coelenteramide with the side chains of residues making up the binding cavity (32, 36). Kinetic properties, Ca2+ sensitivity, thermostability, and so on, are much more difficult to account for, in common with trying to account for enzymatic properties from structural information in general.
Coelenterazine is widely used as a bioluminescent substrate. In addition to Ca2+-regulated photoproteins, this same molecule has been identified as the luciferin for Renilla luciferase from the soft coral Renilla (37) and for the luciferases from copepods (38) as well as for luciferases from some other organisms (the deep-sea shrimp Oplophorus, the scyphozoan medusa Periphylla, etc.). The spatial structures of coelenterazine-type luciferases are not yet available and only a few sequences are in hand, but already these data indicate that luciferase overall structures will differ from the photoproteins. In addition, the luciferases are not Ca2+-dependent proteins, but the bioluminescence properties and the overall chemistry appear to be common with the photoproteins. From the spectral data, we predict a consensus property to be the arrangement of luciferase binding site residues hydrogen bonded to the p-hydroxy group of the 6-substituent of coelenterazine, corresponding to Trp-92 and His-22 in obelin. For the luciferases, there should be a different hydrogen bond network around the C2–C3 atoms of coelenterazine, one that favors the oxygen peroxidation of coelenterazine and then its decarboxylation, rather than stabilization of the hydroperoxide as in the photoproteins.
Materials and Methods
Protein Sample.
The high purity obelin was produced as reported (15). The Ca2+-discharged obelin was prepared by slow addition at room temperature with stirring, of calcium acetate to a diluted obelin solution (≈0.5–1 mg/ml), both in 10 mM Bis·Tris buffer (pH 6.5), to a final Ca2+concentration of 1 mM. During this procedure, a bright blue bioluminescence was observed. The Ca2+-discharged obelin displayed green fluorescence (8), indicating that the coelenteramide remains protein-bound because coelenteramide is not fluorescent in aqueous solution. The protein was concentrated to 26.6 mg/ml with Millipore centrifuge tubes.
Mutagenesis.
Site-directed mutagenesis was done on the template pET19-OL8 expression plasmid (39) carrying the O. longissima wild-type apo-obelin. The Y138F mutation was carried out with the QuikChange site-directed mutagenesis kit (Stratagene) using the 5′-CGAATGGAAAGCTTTTGGAAAAATCTCTG-3′ (forward) and 5′-CAGAGATTTTTCCAAAAGCTTTCCATTCG-3′ (reverse) oligonucleotides. The plasmids harboring mutations were verified by DNA sequencing. The mutant purification protocol was similar to that for the wild-type obelin (15). The bioluminescence spectrum was measured with an AMINCO spectrofluorimeter (Thermo Spectronic) as described (32).
Crystallography.
Initial crystallization screening was carried out by the multiple robotic and other methods, as described (40, 41). The plates were incubated at 4°C for >1 month. One rod-shaped single crystal was grown with 2% polyethylene glycol 400/2.0 M ammonium sulfate/0.1 M Hepes-Na, pH 7.5 (Crystal Screen, Hampton Research). The crystal was cut into three pieces of equal sizes (0.07 × 0.08 × 0.1 mm). Each part was picked up from the crystallization drop using a fiber loop and immersed for 3 s into a 1-μl drop of a 3:1 mixture of well solution and 100% glycerol (42). Still in the loop, it was dragged along the surface of a microscopic cover slide to remove excess liquid and flash-frozen in liquid nitrogen before the data collection process (43).
Data were collected at 0.9725-Å wavelength at beamline 22-ID (SER-CAT) at the Advanced Photon Source (APS) using a MAR 300 charge-coupled deveice detector at a 150-mm crystal-to-detector distance. A single sweep of 360 1.0° oscillation images was recorded with 2-s exposure time per frame. Diffraction data were indexed, integrated, and scaled (Table 2) using the hkl2000 software suite (44). Phases were determined with the molecular replacement program molrep (45) in CCP4 (46, 47) using Ca2+-loaded apo-obelin from O. longissima [Protein Data Bank (PDB) ID code 1SL7] as a search model. There is one molecule per asymmetric unit. Automated tracing was performed by using the initial model phases and the program arp/warp (48). The final model was obtained after several cycles of manual adjustment of the autotraced model using xfit (49), followed by restrained refinement with REFMAC5 (50). The electron density of coelenteramide was clearly visible in the center of the protein molecule. The final refinement statistics are shown in Table 2. The validation was performed with the molprobity online tool (51, 52). The coordinates were deposited in the PDB (53) with an access code 2F8P.
Table 2.
Summary of crystallographic statistics
| Statistics | Value |
|---|---|
| Data processing | |
| Resolution range (Å) | 50.00–1.93 (2.00–1.93) |
| Wavelength, Å | 0.9725 |
| Space group | P43 |
| Cell dimensions, Å | a = 64.26, c = 63.05 |
| Unique reflections | 16,980 (875) |
| Completeness (%) | 86.7 (41.2) |
| I/σ(I) | 26.22 (2.54) |
| Rsym (%) | 7.5 (30.0) |
| Redundancy | 5.2 (1.8) |
| Refinement | |
| Resolution range, Å | 50.00–1.93 |
| Reflections used (free) | 16,136 (844) |
| Rwork (Rfree) | 17.1% (21.3%) |
| Mean B factor (Å2) | 28.5 (27.4) |
| Protein atoms (solvent) | 1561 (139) |
| RMSD bond lengths, Å | 0.016 |
| RMSD bond angles, ° | 1.532 |
Acknowledgments
This work was supported by Russian Foundation for Basic Research Grant 05-04-48271, the “Molecular and Cellular Biology” program of the Russian Academy of Sciences, and the University of Georgia Research Foundation and the Georgia Research Alliance. The data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Abbreviation
- PDB
Protein Data Bank.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The crystal structure reported in this paper has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2F8P).
References
- 1.Morin J. G. In: Coelenterate Biology: Reviews and New Perspectives. Muscatine L., Lenhoff H. M., editors. New York: Academic; 1974. pp. 397–438. [Google Scholar]
- 2.Shimomura O., Johnson F. H., Saiga Y. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 3.Shimomura O. Symp. Soc. Exp. Biol. 1985;39:351–372. [PubMed] [Google Scholar]
- 4.Morin J. G., Hastings J. W. J. Cell. Physiol. 1971;77:305–312. doi: 10.1002/jcp.1040770304. [DOI] [PubMed] [Google Scholar]
- 5.Campbell A. K. Biochem. J. 1974;143:411–418. doi: 10.1042/bj1430411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charbonneau H., Walsh K. A., McCann R. O., Prendergast F. G., Cormier M. J., Vanaman T. C. Biochemistry. 1985;24:6762–6771. doi: 10.1021/bi00345a006. [DOI] [PubMed] [Google Scholar]
- 7.Tsuji F. I., Ohmiya Y., Fagan T. F., Toh H., Inouye S. Photochem. Photobiol. 1995;62:657–661. doi: 10.1111/j.1751-1097.1995.tb08713.x. [DOI] [PubMed] [Google Scholar]
- 8.Markova S. V., Vysotski E. S., Blinks J. R., Burakova L. P., Wang B.-C., Lee J. Biochemistry. 2002;41:2227–2236. doi: 10.1021/bi0117910. [DOI] [PubMed] [Google Scholar]
- 9.Hermann A., Cox J. A. Comp. Biochem. Physiol. B. 1995;111:337–345. doi: 10.1016/0305-0491(94)00218-j. [DOI] [PubMed] [Google Scholar]
- 10.Hastings J. W., Gibson Q. H. J. Biol. Chem. 1963;238:2537–2554. [PubMed] [Google Scholar]
- 11.Shimomura O., Johnson F. H. Biochemistry. 1972;11:1602–1608. doi: 10.1021/bi00759a009. [DOI] [PubMed] [Google Scholar]
- 12.Cormier M. J., Hori K., Karkhanis Y. D., Anderson J. M., Wampler J. E., Morin J. G., Hastings J. W. J. Cell. Physiol. 1973;81:291–297. doi: 10.1002/jcp.1040810218. [DOI] [PubMed] [Google Scholar]
- 13.Vysotski E. S., Lee J. Acc. Chem. Res. 2004;37:405–415. doi: 10.1021/ar0400037. [DOI] [PubMed] [Google Scholar]
- 14.Head J. F., Inouye S., Teranishi K., Shimomura O. Nature. 2000;405:372–376. doi: 10.1038/35012659. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z.-J., Vysotski E. S., Chen C.-J., Rose J., Lee J., Wang B.-C. Protein Sci. 2000;9:2085–2093. doi: 10.1110/ps.9.11.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Glushka J. N., Markova S. V., Vysotski E. S. In: Bioluminescence & Chemiluminescence 2000. Case J. F., Herring P. J., Robison B. H., Haddock S. H. D., Kricka L. J., Stanley P. E., editors. Singapore: World Scientific; 2001. pp. 99–102. [Google Scholar]
- 17.Deng L., Markova S. V., Vysotski E. S., Liu Z.-J., Lee J., Rose. J., Wang B.-C. J. Biol. Chem. 2004;279:33647–33652. doi: 10.1074/jbc.M402427200. [DOI] [PubMed] [Google Scholar]
- 18.Deng L., Vysotski E. S., Markova S. V., Liu Z.-J., Lee J., Rose J., Wang B.-C. Protein Sci. 2005;14:663–675. doi: 10.1110/ps.041142905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher A. J., Raushel F. M., Baldwin T. O., Rayment I. Biochemistry. 1995;34:6581–6586. doi: 10.1021/bi00020a002. [DOI] [PubMed] [Google Scholar]
- 20.Conti E., Franks N. P., Brick P. Structure (London) 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 21.Schultz L. W., Liu L., Cegielski M., Hastings J. W. Proc. Natl. Acad. Sci. USA. 2005;102:1378–1383. doi: 10.1073/pnas.0409335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya S., Bunick C. G., Chazin W. J. Biochim. Biophys. Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Watkins N. J., Campbell A. K. Biochem. J. 1993;292:181–185. doi: 10.1042/bj2930181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimomura O., Johnson J. F. Nature. 1970;227:1356–1357. doi: 10.1038/2271356a0. [DOI] [PubMed] [Google Scholar]
- 25.Strynadka N. C., James M. N. In: Encyclopedia of Inorganic Chemistry. King R. B., editor. New York: Wiley; 1994. pp. 477–507. [Google Scholar]
- 26.Ohmiya Y., Tsuji F. I. FEBS Lett. 1993;320:267–270. doi: 10.1016/0014-5793(93)80600-y. [DOI] [PubMed] [Google Scholar]
- 27.Nelson M. R., Chazin W. J. Biometals. 1998;11:297–318. doi: 10.1023/a:1009253808876. [DOI] [PubMed] [Google Scholar]
- 28.Usami K., Isobe M. Tetrahedron. 1996;52:12061–12090. [Google Scholar]
- 29.Shimomura O., Teranishi K. Luminescence. 2000;15:51–58. doi: 10.1002/(SICI)1522-7243(200001/02)15:1<51::AID-BIO555>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Imai Y., Shibata T., Maki S., Niwa H., Ohashi M., Hirano T. Photochem. Photobiol. A. 2001;146:95–107. [Google Scholar]
- 31.Ohmiya Y., Ohashi M., Tsuji F. I. FEBS Lett. 1992;301:197–201. doi: 10.1016/0014-5793(92)81247-j. [DOI] [PubMed] [Google Scholar]
- 32.Malikova N. P., Stepanyuk G. A., Frank L. A., Markova S. V., Vysotski E. S., Lee J. FEBS Lett. 2003;554:184–188. doi: 10.1016/s0014-5793(03)01166-9. [DOI] [PubMed] [Google Scholar]
- 33.Deng L., Vysotski E. S., Liu Z.-J., Markova S. V., Malikova N. P., Lee J., Rose J., Wang B.-C. FEBS Lett. 2001;506:281–285. doi: 10.1016/s0014-5793(01)02937-4. [DOI] [PubMed] [Google Scholar]
- 34.Vysotski E. S., Liu Z.-J., Markova S. V., Blinks J. R., Deng L., Frank L. A., Herko M., Malikova N. P., Rose J. P., Wang B.-C., Lee J. Biochemistry. 2003;42:6013–6024. doi: 10.1021/bi027258h. [DOI] [PubMed] [Google Scholar]
- 35.McCapra F., Chang Y. C. Chem. Commun. 1967;19:1011–1012. [Google Scholar]
- 36.Stepanyuk G. A., Golz S., Markova S. V., Frank L. A., Lee J., Vysotski E. S. FEBS Lett. 2005;579:1008–1014. doi: 10.1016/j.febslet.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Hori K., Charbonneau H., Hart R. C., Cormier M. J. Proc. Natl. Acad. Sci. USA. 1977;74:4285–4287. doi: 10.1073/pnas.74.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markova S. V., Golz S., Frank L. A., Kalthof B., Vysotski E. S. J. Biol. Chem. 2004;279:3212–3217. doi: 10.1074/jbc.M309639200. [DOI] [PubMed] [Google Scholar]
- 39.Markova S. V., Vysotski E. S., Lee J. In: Bioluminescence & Chemiluminescence 2000. Case J. F., Herring P. J., Robison B. H., Haddock S. H. D., Kricka L. J., Stanley P. E., editors. Singapore: World Scientific; 2001. pp. 115–118. [Google Scholar]
- 40.Liu Z. J., Tempel W., Ng J. D., Lin D., Shah A. K., Chen L., Horanyi P. S., Habel J. E., Kataeva I. A., Xu H., et al. Acta Crystallogr. D. 2005;61:679–684. doi: 10.1107/S0907444905013132. [DOI] [PubMed] [Google Scholar]
- 41.Shah A. K., Liu Z. J., Stewart P. D., Schubot F. D., Rose J. P., Newton M. G., Wang B. C. Acta Crystallogr. D. 2005;61:123–129. doi: 10.1107/S0907444904027064. [DOI] [PubMed] [Google Scholar]
- 42.Teng T. Y. J. Appl. Crystallogr. 1990;23:387–391. [Google Scholar]
- 43.Hope H. Acta Crystallogr. D. 1988;44:22–26. doi: 10.1107/s0108768187008632. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Vagin A., Teplyakov A. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 46.Potterton E., Briggs P., Turkenburg M., Dodson E. Acta Crystallogr. D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 47.Winn M. D. J. Synchrotron Radiation. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- 48.Perrakis A., Morris R., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 49.McRee D. E. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Lovell S. C., Davis I. W., Arendall W. B., III, de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 52.Richardson J. S. Methods Biochem. Anal. 2003;44:305–320. [PubMed] [Google Scholar]
- 53.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissing H., Shindyalov I. N., Bourne P. E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]