Abstract
Xeroderma pigmentosum (XP) is a heritable human disorder characterized by defects in nucleotide excision repair (NER) and the development of skin cancer. Cells from XP group E (XP-E) patients have a defect in the UV-damaged DNA-binding protein complex (UV-DDB), involved in the damage recognition step of NER. UV-DDB comprises two subunits, products of the DDB1 and DDB2 genes, respectively. Mutations in the DDB2 gene account for the underlying defect in XP-E. The UV-DDB complex is a component of the newly identified cullin 4A-based ubiquitin E3 ligase, DDB1-CUL4ADDB2. The E3 ubiquitin ligases recognize specific substrates and mediate their ubiquitination to regulate protein activity or target proteins for degradation by the proteasomal pathway. In this study, we have addressed the role of the UV-DDB-based E3 in NER and sought a physiological substrate. We demonstrate that monoubiquitinated histone H2A in native chromatin coimmunoprecipitates with the endogenous DDB1-CUL4ADDB2 complex in response to UV irradiation. Further, mutations in DDB2 alter the formation and binding activity of the DDB1-CUL4ADDB2 ligase, accompanied by impaired monoubiquitination of H2A after UV treatment of XP-E cells, compared with repair-proficient cells. This finding indicates that DDB2, as the substrate receptor of the DDB1-CUL4A-based ligase, specifically targets histone H2A for monoubiquitination in a photolesion-binding-dependent manner. Given that the loss of monoubiquitinated histone H2A at the sites of UV-damaged DNA is associated with decreased global genome repair in XP-E cells, this study suggests that histone modification, mediated by the XPE factor, facilitates the initiation of NER.
Keywords: E3 ubiquitin ligase, monoubiquitinated histone H2A, nucleotide excision repair
Nucleotide excision repair (NER) is the principal pathway for removal of a broad spectrum of structurally unrelated lesions. In human cells, it is primarily responsible for repair of UV-induced cyclobutane pyrimidine dimers (CPD) and (6-4) photoproducts. Defects in NER result in the UV-sensitive cancer-prone disorder, xeroderma pigmentosum (XP) (1). Seven XP proteins, corresponding to the XP group A (XP-A) through XP-G complementation groups (2), are components of the NER pathway. XP-C and XP-E are specific factors in the global genome NER (GG-NER) subpathway, which processes lesions in nontranscribed DNA and in nontranscribed strands (3). XP-E is the mildest form of this heritable disorder. XP-E pathology is homogeneous both at the clinical (mild dermatologic manifestations, no neurological abnormalities, and late onset of skin tumors) and cellular levels (a modest NER reduction, due to only a partial deficiency in GG-NER; the fibroblasts from XP-E patients are less sensitive to UV than the cells from the other XP complementation groups) (4, 5). At the biochemical level, XP-E is associated with a defective UV-damaged DNA-binding protein complex (UV-DDB) (6), comprising the 48-kDa DDB2 protein and the 127-kDa DDB1 protein, encoded by the DDB2 and DDB1 genes, respectively (7, 8). Mutations in DDB2 cause the XP-E phenotype (5, 9, 10), and microinjection of DDB2 cDNA into XP-E cells restores NER capacity (5).
Although UV-DDB binds avidly to fragments of DNA that contain damage induced by UV irradiation (11–13), it stimulates NER of “naked” DNA only slightly in vitro (14–16). In contrast to NER in XP-E cells, XP-E cell extracts display proficient NER of “naked” DNA in vitro, suggesting that UV-DDB has a role in the repair of DNA in chromatin (17). DDB2 is the first molecule in the activity sequence of XP factors to colocalize with the DNA lesion (18, 19). Additionally, the efficient performance of XP-C in recognizing a distortion in damaged DNA and colocalization of XP-C with UV-induced photolesions, depends on a functional DDB2 (20, 21). These observations have led to the suggestion that UV-DDB has a role in initiating global NER by modifying chromatin in the vicinity of DNA lesions, which may facilitate rapid access of the repair machinery (17, 22, 23).
The finding that UV-DDB is a component of the newly recognized RBX1-CUL4A-based ubiquitin ligase (DDB1-CUL4ADDB2) has shed some light on the complexity of the role of UV-DDB in NER (24). In response to UV irradiation, the COP9 signalosome complex (CSN) (25) regulates the specific activity of the DDB1-CUL4ADDB2 ligase in NER. RNA interference with one of the CSN subunits, CSN5, affects the proficiency of NER in cells so treated (24). Ubiquitin ligases (E3) are multiprotein complexes, which, as a part of the ubiquitin pathway, specifically recognize the substrates and mediate their ubiquitination (26). Attachment of monoubiquitin or a polyubiquitin chain to a target protein activates proteins or marks them for proteasomal degradation, respectively (27). Recently, evidence was presented that proteins involved in the damage-recognition step of GG-NER and transcription-coupled NER are subject to ubiquitination, which implies a novel regulatory mechanism in DNA repair (28–30). The XP-C protein and its yeast counterpart Rad4 are ubiquitinated after UV irradiation of cells (28, 31). The ubiquitination of Rad4 is a signal for degradation by the 26 S proteasome (30, 32). In human cells, XP-C undergoes reversible ubiquitination with no obvious degradation as a consequence. The UV-induced modification of XP-C by ubiquitin and by SUMO1 depends on the presence of functional XP-E and XP-A proteins (33, 34).
To address the role of the XP-E factor in the initiation of GG-NER in a chromatin environment, we sought an in vivo target of the DDB1-CUL4ADDB2 E3 by investigating the UV-induced monoubiquitination of histone H2A in NER-proficient and -deficient XP-E cells.
Results
DDB1-CUL4ADDB2 E3 Colocalizes with UV-Damaged DNA.
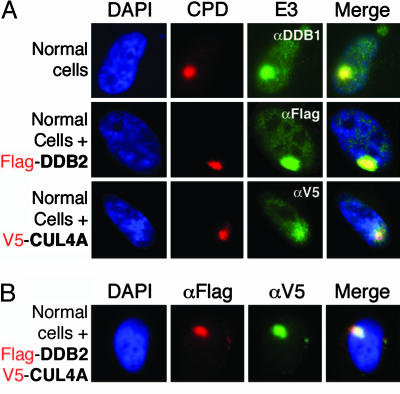
Recent in situ studies confirm that overexpressed DDB2 is recruited to DNA damage sites before other XP proteins colocalize with photolesions (18–20). Given that biochemical studies have demonstrated the binding of the DDB1-CUL4ADDB2 ligase to chromatin after UV treatment (24), this observation poses the question whether CUL4, as a UV-DDB partner, can be detected specifically at sites of DNA damage. Localized cell areas were UV-irradiated through filters, and the initial recruitment of E3 ligase components to CPD lesions was assessed. For these experiments, WI-38 VA13 cells were transiently transfected with CUL4A-V5 and/or DDB2-Flag cDNAs (20, 35). We confirmed, by immunoprecipitation of tagged proteins in lysates of transfected cells, that ectopic CUL4A and DDB2 form a DDB1-CUL4ADDB2 complex with endogenous DDB1 and RBX1 proteins (data not shown).
All three proteins produced a very strong signal in the damaged DNA subnuclear spots, with little background in unirradiated areas of the cell (Fig. 1A). When CUL4A and DDB2 were coexpressed in the WI-38 VA13 cells, both proteins colocalized within irradiated areas (Fig. 1B). Hence, CUL4A as a component of the DDB1-CUL4ADDB2 ubiquitin ligase is concentrated at the sites of UV damage in DNA immediately after irradiation.
Fig. 1.
DDB1-CUL4ADDB2 ubiquitin ligase colocalizes with UV-damaged DNA in vivo. (A) Transformed human fibroblasts (WI-38VA13 cells) were transiently transfected with full length CUL4A- or DDB2-expressing cDNAs. Forty hours after transfection, the cells were irradiated through an 8-μm pore filter with a dose of 60 J/m2. Immediately after treatment, the cells were washed with CSK buffer (100 mM NaCl/300 mM sucrose/10 mM Pipes, pH 7.0/3 mM MgCl2, and protease inhibitors), incubated for 5 min in CSK plus 0.2%Triton X-100, and then fixed. Cells were counterstained with DAPI (blue). UV-irradiated sites were visualized by fluorescent immunostaining by using anti-CPD antibody (red). CUL4A was visualized by an antibody to the V5 epitope present on the transfected Cul4A (green). DDB2 was visualized by an antibody to the Flag epitope present on the transfected DDB2 (green), and endogenous DDB1 was visualized by a DDB1-IgY antibody (green). Merge shows the protein colocalization with damaged DNA. (B) The experiment was performed as described in A, except that the plasmids with CUL4A and DDB2 were cotransfected into WI-38VA13 cells. Merge shows that V5-CUL4A and Flag-DDB2 colocalize in the UV-irradiated subnuclear spot.
Mutations in DDB2 Affect the Formation of DDB1-CUL4ADDB2 E3 in XP-E Cells.
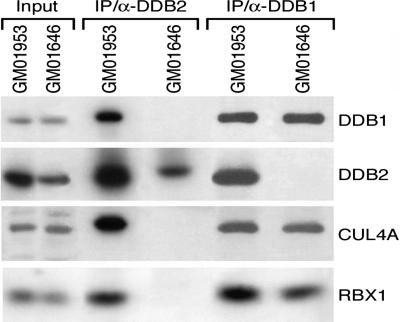
Our previous study of naturally occurring mutations in XP-E patients provides an initial understanding of the functional domains of the DDB2 protein. All substitutions and a small deletion, found in XP2RO, XP25PV, and GM01646 mutants, are responsible for the loss of binding to DDB1 (5). In view of conflicting data obtained on ectopically expressed subunits of the E3 (36, 37), we wondered whether an interaction between UV-DDB and CUL4A exclusively occurs through DDB1, or whether DDB2 could form a complex with CUL4A even though the molecular defects in these mutant lines abolish the interaction with DDB1. Immunoprecipitates (5) with DDB1 or DDB2 antibodies were tested for the presence of endogenous UV-DDB subunits CUL4A and RBX1 in normal and XP-E cells (Fig. 2). Under these conditions, we did not detect CUL4A coimmunoprecipitated with DDB2 protein in an XP-E line (Fig. 2), which suggests that mutations in DDB2 affect the formation of DDB1-CUL4ADDB2 E3 in XP-E cells. The amount of CUL4 pulled down with anti-DDB1 antibody was similar in control and XP-E cells, which confirms that the DDB1 interaction with CUL4A is independent of DDB2. Another E3 complex, formed between DDB1-CUL4A and Cockayne syndrome group A protein (24), was detected in similar amounts in control and XP-E cells (data not shown).
Fig. 2.
A naturally occurring DDB2 mutant fails to interact with CUL4A. The lysates of normal (GM01953) and XP-E (GM01646, with impaired interaction of DDB1 and DDB2 subunits) lymphoblastoid cells were immunoprecipitated with rabbit DDB2 and DDB1-IgY. The coimmunoprecipitates were separated on SDS gels and poly(vinylidene difluoride) membranes probed with DDB1, DDB2, CUL4 (8% gel), and RBX1 (12% gel) antibodies.
XP-E Cells Are Defective in Binding DDB1-CUL4ADDB2 E3 to Chromatin After UV Irradiation.
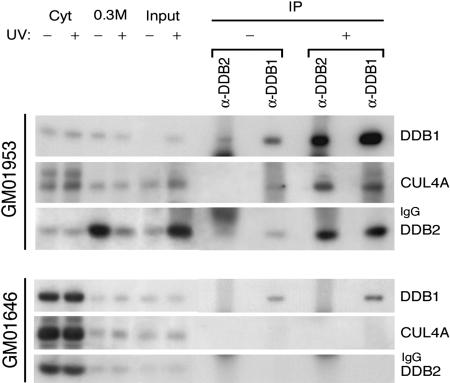
All XP-E cells are deficient in UV-DDB-binding activity (4, 5). Combined with the above findings, this indicates a dysfunctional UV-DDB-based E3 in XP-E cells. We next examined whether the DDB1-CUL4A complex without DDB2 or mutated DDB2 alone can bind to chromatin in XP-E cells 45 min after UV irradiation, when DDB2 is retained in the chromatin-bound fraction of normal cells (Fig. 6, which is published as supporting information on the PNAS web site). The cell fractionation procedure that was applied to UV or mock-treated normal and XP-E cells assured an enrichment in DNA-bound nuclear proteins and also allowed us to monitor the translocation of UV-DDB from nuclear soluble into chromatin-bound fractions upon UV irradiation (Fig. 6 and refs. 22 and 38). Solubilized chromatin fractions, containing mostly mononucleosomes, were immunoprecipitated with DDB1 and DDB2 antibodies. Most DDB2 is translocated to the UV-damaged chromatin of normal cells (compare the inputs of UV and mock-treated cells, Fig. 3) in the form of DDB1-CUL4DDB2 E3. A similar amount of CUL4A and DDB2 was coimmunoprecipitated from UV-damaged chromatin with both antibodies (Fig. 3). The same procedure in XP-E lymphoblastoid cells failed to show any enrichment of CUL4A and DDB2 proteins, either immunoprecipitated with DDB1 or DDB2 antibodies in UV-damaged chromatin (Fig. 3), demonstrating a deficient DDB1-CUL4DDB2 E3. Further, no difference in the subcellular distribution of mutated DDB2 was detected in the fractions before and after irradiation of the XP-E cells.
Fig. 3.
DDB1-CUL4ADDB2 E3 fails to bind to chromatin after UV irradiation of XP-E cells. The solubilized chromatin fractions were immunoprecipitated with rabbit DDB2- and DDB1-IgY antibodies. The fractions (Input) were obtained after MNase complete digestion of the chromatin-bound fractions prepared from 8 × 106 normal and XP-E lymphoblastoid cells that had not been irradiated or had been allowed to recover for 45 min after irradiation (30 J/m2). The coimmunoprecipitates were analyzed by immunoblotting with DDB1, CUL4A, and DDB2sc antibodies to the components of DDB1-CUL4ADDB2 ubiquitin ligase. The cytosolic, 0.3 M nuclear soluble, and solubilized chromatin fractions (see Materials and Methods) were also included in the Western blot to illustrate the difference between the subcellular distribution of mutated and normal DDB2.
Ubiquitination of H2A After UV Irradiation Is Defective in XP-E Cells.
Given that UV-DDB is not essential for NER of purified DNA, but that DDB1-CUL4DDB2 ubiquitin ligase colocalizes with CPD and binds to chromatin after NER-proficient cells have been irradiated with UV (Figs. 1 and 3), we hypothesized that the role of this E3 is to modify chromatin proteins, through ubiquitination, in the vicinity of DNA lesions to which it is bound, so that repair enzymes can access the site of damage. In vivo, ubiquitination of the histone proteins is primarily restricted to histones H2A and H2B, and 10% of H2A is typically ubiquitinated in mammalian cells (39). To assess a possible relation between the DDB1-CUL4DDB2 ubiquitin ligase and monoubiquitination of H2A (uH2A), we investigated the uH2A profile in response to UV irradiation in a panel of NER-proficient and -deficient cells.
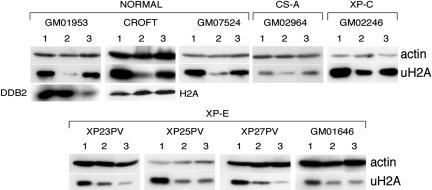
Lymphoblastoid cells from several normal and XP-E cell lines were irradiated with a UV dose of 40 J/m2 and collected immediately or after a recovery period (Fig. 4). Total cell lysates from treated and untreated cells were analyzed by immunoblots to assess the levels of uH2A. Lysates were chosen rather than acid extraction to avoid an extended preparation time and thus possible deubiquitination of H2A. The total cellular uH2A (free and chromatin-imbedded uH2A) was detected with an antibody highly specific for the modified histone (40).
Fig. 4.
Recovery of uH2A in response to UV irradiation differs between XP-E and control cells. The lymphoblastoid cells from control, CS-A, XP-C, and XP-E patients were UV-irradiated with 40 J/m2. The cells were collected either immediately after treatment or after a recovery period of 2 h (1, unirradiated cells; 2, no recovery, and 3, 2-h recovery). After several washes with PBS, cells (5 × 105) were lysed in gel-loading sample buffer and boiled for 15 min; the lysates were analyzed on 12% SDS/PAGE gels. Proteins were transferred and analyzed by immunoblotting by using antibodies against uH2A and actin (loading control). The membrane with the CROFT cell extracts was reprobed with H2A antibody to show that the antibody against uH2A recognizes only the ubiquitinated form of the H2A histone. The level of DDB2 was immunodetected as a marker of the cell response to UV treatment of normal lymphoblastoid cells (GM01953).
Normal cells responded rapidly to UV irradiation, with a dramatic loss of total uH2A (Fig. 4). After 30 min, a recovery of uH2A was detected and by 2 h, the total cellular level of uH2A was restored to the level observed before irradiation (Fig. 7, which is published as supporting information on the PNAS web site, and Fig. 4). In several cases, cells were monitored for up to 15 h without any significant further increase in the uH2A levels (Fig. 7 and data not shown). In contrast, all of the tested XP-E cell lines failed to restore the initial uH2A levels. Moreover, after a 2-h recovery period, there was no increase in uH2A, and in some cases, a further decrease was observed. Nevertheless, these lines also suffered a dramatic loss of uH2A immediately after UV treatment (Figs. 4 and 7). Cockayne syndrome group A and XP-C cells, defective in transcription-coupled NER and GG-NER but with normal DDB2 activity (22), showed responses similar to that of repair-proficient cells (Fig. 4).
Two conclusions can be drawn from these experiments. First, deubiquitination of uH2A as a response to UV irradiation occurs, but independently of the UV-DDB complex. It has been proposed that cleavage of ubiquitin from nucleosomal H2A could be a “nuclear sensor of cellular stress” (41). Second, restoring the levels of uH2A depends on the presence of an intact DDB1-CUL4DDB2 ubiquitin ligase.
DDB1-CUL4ADDB2 E3 Targets Histone H2A for Monoubiquitination at UV-Damaged DNA Sites.
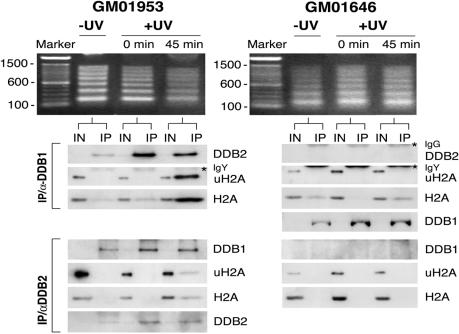
The difference in the uH2A profiles in XP-E cells suggests a role for DDB2 in the recovery of uH2A levels after UV irradiation. To address whether DDB1-CUL4ADDB2 E3 targets nucleosomal H2A for monoubiquitination at UV-damaged sites, immunoprecipitation from native chromatin was used (42). Normal and XP-E lymphoblastoid cells were UV-irradiated and fractionated as in the experiment depicted in Fig. 3. For the purpose of coimmunoprecipitating the UV-DDB E3 complex with chromatin proteins, either with DDB1 or DDB2 antibodies, a solubilized chromatin fraction was prepared by partial micrococcal nuclease (MNase) digestion for 2 min, such that native chromatin was fragmented primarily into penta, tetra-, tri-, di-, and mononucleosomes. Because of the crucial role of DNA fragment size in the outcome of immunoprecipitation of native chromatin with UV-DDB antibodies, the quality of the micrococcal digest was always assayed by agarose gel electrophoresis before the immunoprecipitation of the chromatin-solubilized fraction. The excessive MNase treatment resulted in the dissociation of the UV-DDB E3 complex from chromatin. The complex was found to be intact but was deprived of its contact with chromatin as assayed by the presence of either H2A or uH2A. (compare lanes 2 and 3 in Fig. 8, which is published as supporting information on the PNAS web site).
All solubilized chromatin fractions, obtained from UV or mock-irradiated normal and XP-E cells, contain H2A and uH2A (inputs, Fig. 5). Although UV-DDB moves from the soluble nuclear fraction to the chromatin-bound fractions immediately after UV treatment (Figs. 5 and 6), coimmunoprecipitation failed to show a significant interaction with chromatin, as assayed by H2A levels (Fig. 5). The stable interaction of UV-DDB with H2A and dramatic increase of coimmunoprecipitated uH2A (with both antibodies) were detected only in normal lymphoblastoid cells that were allowed a recovery period of 45 min after irradiation. We also detected uH2A in coimmunoprecipitates with DDB1 antibody, after a recovery period of 25 min (data not shown). During this series of experiments, it became evident that the antibody against DDB2 was less potent than the one against DDB1 in immunoprecipitating target proteins that were still associated with chromatin (Figs. 5 and 8). It is possible that the conformation of the E3 complex with DNA is such that the N terminus of DDB2 is partially shielded and less accessible to the antibody made against an N-terminal peptide. Indeed, the same antibody can perform only a partial supershift of the UV-DDB complex bound to damaged DNA (5).
Fig. 5.
The UV-DDB complex coimmunoprecipitates with uH2A. The chromatin-insoluble fractions from a normal and an XP-E cell line (5 × 106 cells) were digested with MNase to achieve a DNA ladder of mononucleosomes to oligonucleosomes visible in an agarose gel. Histone H2A and uH2A were assayed as coprecipitates of the DDB1-IgY and DDB2 native chromatin immunoprecipitation. Cells were collected at three time points: before UV treatment (−UV), immediately after treatment (0 min), and 45 min after recovering from UV treatment at 37°C (45 min). For each time point, an input lane is included which also serves as a loading control. After SDS/PAGE, proteins were transferred on the appropriate membranes and blotted with antibodies against DDB1, DDB2sc, uH2A, and H2A.
In XP-E lymphoblastoid cells with a deficient E3, there was no interaction between uH2A and UV-DDB protein, as determined by coimmunoprecipitation with DDB1 or DDB2 antibodies (Fig. 5). Although DDB1 is bound to other chromatin targets independent of DDB2 (43) and can associate with nonubiquitinated H2A (Fig. 5), it failed to coimmunoprecipitate with uH2A. It appears that the association of DDB1 and uH2A depends heavily on the presence of an active UV-DDB complex that is tightly bound at the sites of DNA lesions.
Discussion
A major challenge in research on NER is understanding how this process occurs in the nuclear context, and how cells overcome the apparent block to DNA repair occasioned by the surrounding chromatin packaging. Although transcription-coupled NER is transcription-dependent, the initiation of GG-NER seems to depend upon detection of a lesion in chromatin by damage-specific binding proteins (23, 29, 44). In the current study, we demonstrate that DDB1-CUL4ADDB2 E3 colocalizes with CPD and coimmunoprecipitates with uH2A in response to UV irradiation in a manner that depends on DDB2 function. That uH2A was detected in the complex with UV-DDB bound to short polynucleosomes (Fig. 5) suggests that H2A ubiquitination occurs in the vicinity of UV lesions. Despite the fact that DDB1-CUL4ADDB2 E3 is immediately detected at the site of a lesion under the experimental conditions used, uH2A appears on DNA at a later time (between 25 and 45 min), when the stable complex between UV-DDB and histone H2A is achieved. This may indicate that, at an early stage, interactions are fragile and can be disrupted by MNase treatment of native chromatin. However, the appearance of ubiquitinated uH2A in this study correlates with the time when a significant amount of UV-induced ubiquitinated XPC was detected (33). Remarkably, activation of the DDB1-CUL4ADDB2 E3, through dissociation of the COP9 signalosome and neddylation of CUL4A, occurs 30 min after irradiation and gradually decreases thereafter (24). Subsequent reassociation of the COP9 signalosome with the complex (2 h thereafter), leading to inhibition of the E3 (24), correlates with the prominent degradation of the chromatin-bound DDB2 shown in Fig. 6. In XP-E cells, neither DDB1 nor DDB2 colocalizes with uH2A, which is an additional indication that the uH2A detected in normal cells is located at the sites of DDB1-CUL4ADDB2 E3 recruitment and cannot be attributed to DDB1 binding alone. Taken together, our data lead to the conclusion that the DDB1-CUL4ADDB2 E3 in vivo targets histone H2A for ubiquitination at UV-damage DNA sites, where DDB2 serves as the substrate receptor.
The monoubiquitination of lysine 119 of nucleosomal histone H2A is a bulky structural alteration in the vicinity of a binding site for linker histone. It is presumably associated with changes in chromatin structure and does not constitute a degradation signal. This raises the question of the actual role of uH2A. One possibility is that histone ubiquitination shifts chromatin from a compact structure to a relaxed conformation (39). The resulting architectural change of the nucleosome(s) at the UV lesion could facilitate access of the NER machinery (44), further supporting the model of chromatin relaxation monoubiquitinated H2A preferentially localized at decondensed trancriptionally active chromatin, whereas heterochromatin is practically free of ubiquitinated histones (45). Moreover, the H2A monoubiquitination and deubiquitination cycle is a key regulator of chromatin condensation during the cell cycle (46). In mammalian cells, it was shown recently that uH2A marks certain inactive chromatin regions, possibly playing a role in maintaining the inactive chromatin state (47). To complicate matters even more, the in vitro reconstitution of nucleosomes with uH2A or uH2B does not result in any obvious perturbation of the overall structure (48–50).
In addition to a possible role of uH2A in affecting the conformation of the chromatin at the site of a DNA lesion, uH2A could constitute a signal for the assembly of NER factors. A ubiquitin-binding domain (51) of a repair protein could couple uH2A to the rest of the NER machinery. Indeed, the DNA repair protein RAD23B (which forms the damage recognition complex with XPC) has two ubiquitin-associated domains (a class of ubiquitin-binding domain) reported to bind ubiquitin (52). Moreover, UV-induced XP-C translocation within chromatin is compromised in XP-E cells (53). Further, Sugasawa et al. (33) reported that XP-E cells are defective in polyubiquitination of XP-C, and that recombinant UV-DDB-ubiquitin ligase catalyzes this modification of the XP-C protein.
Our results indicate an important link between defective uH2A and defective DNA repair. Impaired ubiquitination of histone H2A after UV treatment of XP-E cells may lead to a chromatin structure with restricted access for the NER machinery or to defective recruitment of DNA repair proteins.
Materials and Methods
Cell Lines.
Normal human lymphoblastoid (GM01953, CROFT, and GM07524), XP-E lymphoblastoid (GM01646, XP23PV, XP25PV, and XP27PV), CS-A (GM02964), and XP-C (GM02246) cells (obtained from the Coriell Cell Repository) were grown in RPMI media 1640 supplemented with 10% FCS and glutamax. WI-38 VA13 transformed primary human fibroblasts (obtained from American Type Culture Collection) were grown in DMEM media supplemented with 10% FCS, essential and nonessential amino acids, and glutamax. All cells were kept at 37°C in a humidified atmosphere with 5% CO2.
Plasmids and Transfections.
The human CUL4A clone (no. 3537176) was obtained from Open Biosystems (Huntsville, AL) in bacterial stocks. The full length CUL4A cDNA (GenBank accession no. AY365124) was PCR-amplified with primers designed to introduce restriction sites at the 5′ and 3′ ends, and a V5 tag was introduced at the C terminus. The PCR products were cloned into the pcDNA 3.1 vector (Invitrogen). The Flag-DDB2 cDNA (5) was subcloned into the pcDNA 3.1 vector. The WI-38 VA13 cells were transiently transfected with cDNA constructs by using Fugene 6 (Roche Applied Science, Indianapolis), according to the manufacturer’s instructions.
Antibodies.
A new antibody [DDB1-immunoglobulin Y (IgY)] was prepared against full-length human DDB1 protein (54) in chickens by Aves Labs (Tigard, OR). The rabbit DDB2 and DDB1 antibodies were previously described (5). Goat cullin 4 and goat DDB2sc (Santa Cruz Biotechnology), mouse Flag: M2 (Sigma), mouse V5 (Invitrogen), rabbit RBX1 (Sigma), mouse hemagglutinin (Covance, Richmond, CA), and H2A as well as uH2A (Upstate Biotechnology, Lake Placid, NY) antibodies were obtained commercially. Mouse anti-CPD (Kamiya Biomedical, Thousand Oaks, CA), goat V5-FITC (Bethyl Laboratories, Montgomery, TX), and rabbit Flag (Sigma) were used for indirect immunofluorescence. Secondary antibodies conjugated to Alexa Fluor 594 or Alexa Fluor 488 were purchased from Molecular Probes.
UV and Indirect Immunofluorescence.
For micropore UV irradiation (35, 55), cells grown on glass coverslips were covered with the filter and irradiated from above with UV-C radiation at a dose rate of 0.5 J/m2 per sec. After in situ detergent extraction, cells were fixed in 2% paraformaldehyde in PBS for 15 min, made permeable with one wash and two 10-min incubations with cold 0.2% Triton X-100 in PBS (Triton buffer) and blocked in 5% BSA for 20 min. DNA was denatured with 0.4 M NaOH for 4 min and washed repeatedly with PBS. Cells were incubated with the primary and secondary antibodies for 1 h each and washed with cold Triton buffer. The coverslips were mounted on a slide with vectashield containing DAPI. The indirect fluorescence images were obtained with an Olympus (Melville, NY) Fluorescence Microscope AX70 Provis. The digital images were captured with a cooled charge-coupled device camera, processed, and superimposed with the help of spot software (Diagnostic Instruments, Sterling Heights, MI).
Preparation of Native Chromatin.
Coimmunoprecipitation from uncrosslinked chromatin was used to test for a physical interaction between histone H2A and DDB1-CUL4ADDB2 E3. The high affinity of UV-DDB for chromatinized DNA in irradiated cells enables it to withstand treatment with MNase (24) during immunoprecipitation of native chromatin (42).
The cell pellet was washed in cold PBS and fractionated into cytosolic and 0.3 M nuclear soluble fractions, as described (5). To obtain a soluble chromatin fraction (42), the remaining pellet (chromatin-bound fraction) was resuspended in CS buffer (20 mM Tris·HCl, pH 7.5/100 mM KCl/2 mM MgCl2/1 mM CaCl2/0.3 M sucrose/0.1% Triton X-100, and complete protease inhibitors). The concentration of MNase and time of incubation at 37°C varied (24, 42): For complete digestion into mononucleosomes, 400 units/ml MNase was added, and samples were incubated at 37°C for 5 min. For partial digestion, MNase was added at 27 units/ml, and the samples were incubated at 37°C for 2.5, 5, 7.5, 10, and 25 min. The reaction was stopped by adding 5 mM EDTA. Samples were centrifuged and the supernatant used as the solubilized chromatin fraction.
Chromatin Immunoprecipitation and Immunoblotting.
For coimmunoprecipitation of free or chromatin-bound UV-DDB subunits, the cells or solubilized chromatin fractions were resuspended in Nonidet P-40 buffer [50 mM Tris·HCl (pH 8.0)/100 mM NaCl/5 mM MgCl2/1% Nonidet P-40, and protease inhibitors). The suspensions were incubated for 2 h with antibody at 4°C followed by 1 h with protein A coupled agarose beads or anti-IgY coupled agarose beads. The coimmunoprecipitated complexes were washed four times with Nonidet P-40 buffer and boiled in gel-loading sample buffer [50 mM Tris (pH 6.8)/4 M Urea/1% SDS/4% 2-mercaptoethanol, and dye) for 10 min. Proteins were separated on 8% and 12% SDS/PAGE gels and transferred to Immobilon-P membranes.
For histone coimmunoprecipitation and immunoblotting, the solubilized chromatin fractions were incubated overnight with DDB1-IgY or DDB2 antibodies at 4°C, followed by the same procedure as above. Proteins were separated on a 12% SDS/PAGE gel and transferred to an Immobilon-P membrane (upper part, including proteins >30 kDa) or a nitrocellulose membrane (lower part, including proteins between 10 and 30 kDa). The membranes were incubated overnight with primary antibodies and for 1 h with the corresponding alkaline phosphatase (AP) or horseradish peroxidase (HRP)-conjugated secondary antibodies. Proteins were detected by a chemiluminescent signal produced by the appropriate AP or HRP substrate. The chemiluminescent signal was captured by BioMax-Light film (Kodak) and a Chemi-Doc System (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Dr. Richard Wood for helpful comments on the manuscript; Dr. Birgitte Wittschieben for critical reading of the manuscript and help in expression of DDB1 in the insect cell system; Drs. Sanford Leuba and Miroslav Tomschik for assistance with chromatin preparation; and Drs. Miria Stefanini (Consiglio Nazionale delle Ricerche Instituto di Genetica Molecolare, Pavia, Italy) and Olivera Finn (University of Pittsburgh) for providing CROFT and XP23PV, XP25PV, and XP27PV lymphoblastoid cells, respectively. This work was supported by a University of Pittsburgh Medical School startup fund and a grant from The Pittsburgh Foundation (to V.R.-O.).
Abbreviations
- CPD
cyclobutane pyrimidine dimers
- NER
nucleotide excision repair
- XP
xeroderma pigmentosum
- XP-A–G
XP groups A–G
- UV-DDB
UV-damaged DNA-binding protein
- uH2A
monoubiquitinated histone H2A
- GG-NER
global genome NER
- MNase
micrococcal nuclease
- IgY
immunoglobulin Y.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.de Boer J., Hoeijmakers J. H. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 2.Wood R. D., Mitchell M., Sgouros J., Lindahl T. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt P. C. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 4.Tang J., Chu G. DNA Rep. 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapic-Otrin V., Navazza V., Nardo T., Botta E., McLenigan M., Bisi D. C., Levine A. S., Stefanini M. Hum. Mol. Genet. 2003;12:1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- 6.Chu G., Chang E. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 7.Keeney S., Chang G. J., Linn S. J. Biol. Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 8.Dualan R., Brody T., Keeney S., Nichols A. F., Admon A., Linn S. Genomics. 1995;29:62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 9.Itoh T., Mori T., Ohkubo H., Yamaizumi M. J. Invest Dermatol. 1999;113:251–257. doi: 10.1046/j.1523-1747.1999.00652.x. [DOI] [PubMed] [Google Scholar]
- 10.Nichols A. F., Ong P., Linn S. J. Biol. Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 11.Reardon J. T., Nichols A. F., Keeney S., Smith C. A., Taylor J. S., Linn S., Sancar A. J. Biol. Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 12.Fujiwara Y., Masutani C., Mizukoshi T., Kondo J., Hanaoka F., Iwai S. J. Biol. Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- 13.Abramic M., Levine A. S., Protic M. J. Biol. Chem. 1991;266:22493–22500. [PubMed] [Google Scholar]
- 14.Aboussekhra A., Biggerstaff M., Shivji M. K., Vilpo J. A., Moncollin V., Podust V. N., Protic M., Hubscher U., Egly J. M., Wood R. D. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 15.Mu D., Park C. H., Matsunaga T., Hsu D. S., Reardon J. T., Sancar A. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 16.Wakasugi M., Shimizu M., Morioka H., Linn S., Nikaido O., Matsunaga T. J. Biol. Chem. 2001;276:15434–15440. doi: 10.1074/jbc.M011177200. [DOI] [PubMed] [Google Scholar]
- 17.Rapic-Otrin V., Kuraoka I., Nardo T., McLenigan M., Eker A. P., Stefanini M., Levine A. S., Wood R. D. Mol. Cell. Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakasugi M., Kawashima A., Morioka H., Linn S., Sancar A., Mori T., Nikaido O., Matsunaga T. J. Biol. Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 19.Green C. M., Almouzni G. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitch M. E., Nakajima S., Yasui A., Ford J. M. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 21.Moser J., Volker M., Kool H., Alekseev S., Vrieling H., Yasui A., van Zeeland A. A., Mullenders L. H. DNA Rep. 2005;4:571–582. doi: 10.1016/j.dnarep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Rapic-Otrin V., McLenigan M. P., Bisi D. C., Gonzalez M., Levine A. S. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed S. H. DNA Rep. 2005;4:909–918. doi: 10.1016/j.dnarep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A. F., Tanaka K., Nakatani Y. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 25.Cope G. A., Deshaies R. J. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 26.Petroski M. D., Deshaies R. J. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 27.Finley D., Ciechanover A., Varshavsky A. Cell. 2004;116:S29–S32. doi: 10.1016/s0092-8674(03)00971-1. [DOI] [PubMed] [Google Scholar]
- 28.Sweder K., Madura K. J. Biomed. Biotechnol. 2002;2:94–105. doi: 10.1155/S1110724302205033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svejstrup J. Q. J. Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- 30.Lommel L., Ortolan T., Chen L., Madura K., Sweder K. S. Curr. Genet. 2002;42:9–20. doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- 31.Ng J. M., Vermeulen W., van der Horst G. T., Bergink S., Sugasawa K., Vrieling H., Hoeijmakers J. H. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsey K. L., Smith J. J., Dasgupta A., Maqani N., Grant P., Auble D. T. Mol. Cell. Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Hanaoka F. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q. E., Zhu Q., Wani G., El-Mahdy M. A., Li J., Wani A. A. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volker M., Mone M. J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J. H., van Driel R., van Zeeland A. A., Mullenders L. H. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 36.Chen X., Zhang Y., Douglas L., Zhou P. J. Biol. Chem. 2001;276:48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 37.Nag A., Bondar T., Shiv S., Raychaudhuri P. Mol. Cell. Biol. 2001;21:6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otrin V. R., McLenigan M., Takao M., Levine A. S., Protic M. J. Cell Sci. 1997;110:1159–1168. doi: 10.1242/jcs.110.10.1159. [DOI] [PubMed] [Google Scholar]
- 39.Jason L. J., Moore S. C., Lewis J. D., Lindsey G., Ausio J. BioEssays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- 40.Vassilev A. P., Rasmussen H. H., Christensen E. I., Nielsen S., Celis J. E. J. Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- 41.Mimnaugh E. G., Kayastha G., McGovern N. B., Hwang S. G., Marcu M. G., Trepel J., Cai S. Y., Marchesi V. T., Neckers L. Cell Death Differ. 2001;8:1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- 42.O’Neill L. P., Turner B. M. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 43.Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 44.Gong F., Kwon Y., Smerdon M. J. DNA Rep. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Davie J. R., Murphy L. C. Biochemistry. 1990;29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 46.Bradbury E. M. BioEssays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 47.Baarends W. M., Wassenaar E., van der Laan R., Hoogerbrugge J., Sleddens-Linkels E., Hoeijmakers J. H., de Boer P., Grootegoed J. A. Mol. Cell Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleinschmidt A. M., Martinson H. G. Nucleic Acids Res. 1981;9:2423–2431. doi: 10.1093/nar/9.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies N., Lindsey G. G. Biochim. Biophys. Acta. 1994;1218:187–193. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 50.Jason L. J., Finn R. M., Lindsey G., Ausio J. J. Biol. Chem. 2005;280:4975–4982. doi: 10.1074/jbc.M410203200. [DOI] [PubMed] [Google Scholar]
- 51.Hicke L., Schubert H. L., Hill C. P. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 52.Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. J. Mol. Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q. E., Zhu Q., Wani G., Chen J., Wani A. A. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- 54.Wittschieben B., Iwai S., Wood R. D. J. Biol. Chem. 2005;280:39982–39989. doi: 10.1074/jbc.M507854200. [DOI] [PubMed] [Google Scholar]
- 55.Katsumi S., Kobayashi N., Imoto K., Nakagawa A., Yamashina Y., Muramatsu T., Shirai T., Miyagawa S., Sugiura S., Hanaoka F., et al. J. Invest. Dermatol. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.