Abstract
The transcription factor Myc plays a central role in the control of cellular proliferation. Myc expression is induced by growth factors in a pathway mediated by cellular Src (c-Src), but it is not clear whether Myc induction or activity is required for malignant transformation by activated Src. We introduced v-Src into a c-myc−/− derivative of Rat-1 fibroblasts and into 3T9 mouse fibroblasts harboring a conditionally excisable c-myc allele. Expression of activated viral Src in Myc-deficient cells led to loss of actin stress fibers and surface fibronectin, indicating that Myc is dispensable for v-Src-induced morphological transformation. However, v-Src failed to rescue the proliferative defect resulting from the loss of Myc. In Myc-deficient cells, despite its inability to overcome this proliferation block, v-Src was able to regulate the expression of certain Myc transcriptional targets and induce the expression of active cyclin D/Cdk4 and Cdk6 complexes; it also induced the phosphorylation of Rb, albeit at reduced levels. In contrast, however, in the absence of Myc, the level of Cdk2 kinase activity was drastically reduced. This reduction in Cdk2 activity was associated with a decrease in the expression of Cdk7, Cdc25A, and cyclin A. Coexpression of Cdk2 plus cyclin E and/or cyclin A rescued the G1/S block and allowed the cells to enter mitosis. These results indicate that in the absence of Myc, v-Src can activate early G1 cell cycle regulators but fails to activate regulators of the late G1/S transition.
Keywords: cell cycle, transformation, Myc transcriptional targets
Several lines of evidence suggest that cellular Src (c-Src) is required for the mitogenic activity of growth factors such as platelet-derived growth factor, epidermal growth factor (EGF) and colony stimulating factor 1 (1). This mitogenic function of c-Src is mediated, at least in part, by the induction of the cell cycle regulator and transcription factor c-Myc (hereinafter referred to as Myc) (1, 2). In susceptible avian or mammalian cells expression of v-Src, a mutationally activated derivative of c-Src leads to transformation in vitro and the acquisition of malignancy in vivo. However, the role of Myc in transformation by v-Src is controversial, with some reports suggesting that Myc function or induction is required for transformation by v-Src (3), whereas others suggest that it is not (4, 5).
Myc is a basic helix-loop-helix leucine zipper transcription factor and forms obligate heterodimers with Max that are required for transcriptional activation. Induced loss of Myc activity causes cells to enter a quiescent G0 phase (6). Myc is required for several cycle transitions, including passage through G1 and activation of cyclin D-dependent kinases, as well as entry into S and activation of cyclin E/Cdk2 and cyclin A/Cdk2 (7). Myc has been shown to increase the expression levels of cyclins E and A and to repress the expression of cyclin D1 (8). Myc represses the transcription of the Cdk inhibitors p27 and p21 (9, 10) and increases the rate of p27 degradation (11). Myc also activates Cdk2 by inducing the expression of the Cdk-activating kinase Cdk7 and the Cdk-activating phosphatase Cdc25A (12).
To examine the role of Myc in mitogenic signaling by v-Src, we made use of two Myc-deficient rodent cell lines. The first was a Myc-deficient derivative of Rat1 fibroblasts, which retains the capacity for cell proliferation, but at a very reduced rate (13). The second was a 3T9 cell line derived from mice homozygous for a conditional allele of c-myc (6); this latter line undergoes a complete proliferation arrest upon excision of c-myc. We show here that in Myc-deficient cells, v-Src can induce morphological transformation but does not induce DNA synthesis or bypass the Myc requirement for cell proliferation. We further show that in the absence of Myc, v-Src can regulate a variety of Myc transcriptional targets such as Gas1, Gadd45, and p27 and can activate the G1 cyclin-dependent kinases Cdk4 and Cdk6. However, in the absence of Myc, v-Src does not activate the G1/S cyclin-dependent kinase Cdk2 or induce the expression of cyclin A. The G1/S block could be rescued by coexpression of Cdk2 plus cyclin E and/or cyclin A, allowing the cells to synthesize DNA and enter mitosis. Our findings suggest that the role of Myc in mitogenic signaling by Src is to mediate the activation of cyclin E- and cyclin A-dependent kinases.
Results
Myc Deficiency Does Not Affect the Activity of v-Src or Its Ability to Induce Morphological Transformation.
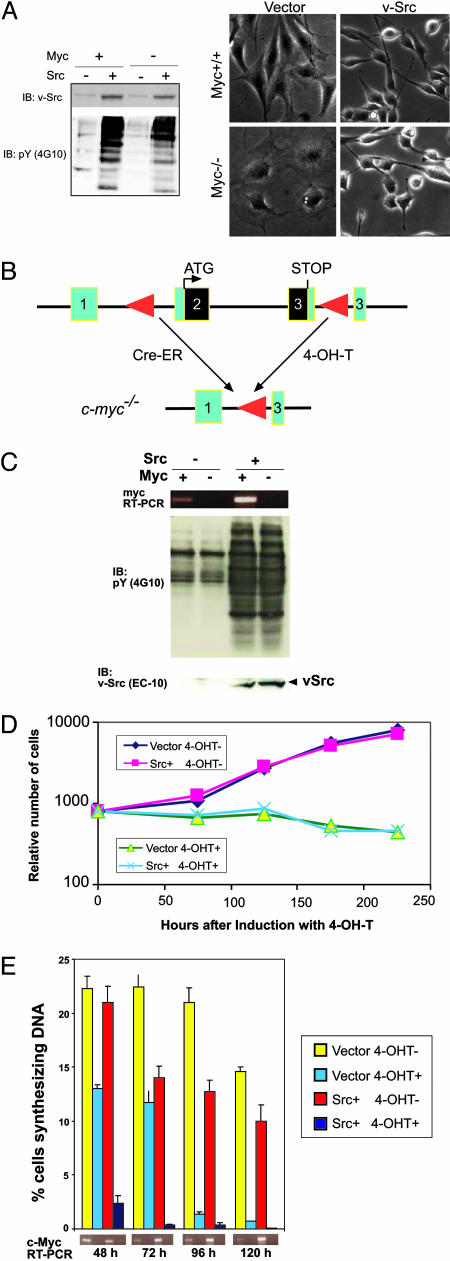
We initially introduced v-Src into a Rat-1 derived line TGR-1, and a Myc-deficient cell line derived from it, HO15.19 (13). HO15.19 cells proliferate, albeit slowly, with a population doubling time of ≈50 h (7). c-myc+/+ and c-myc−/− clones expressing v-Src at equal levels (Fig. 1A Left) were selected for further study. As previously described for the original lines (13), control c-myc−/− cells expressing empty vector were flatter, more spread out, and less refractile than corresponding c-myc+/+ cells. Both c-myc+/+ and c-myc−/− cells expressing v-Src appeared morphologically transformed: Most cells in the population were highly refractile and rounded or spindle-shaped (Fig. 1A Right).
Fig. 1.
Effects of Myc deficiency on transformation of Rat1 and 3T9 cells by v-Src. (A) Morphological transformation of Myc-deficient Rat1 cells by v-Src. (A Left) Lysates of Rat1 myc+/+ and myc−/− cells expressing either empty vector or v-Src were resolved by SDS/PAGE and immunoblotted with anti-Src mAb 2–17 or 4G10. (A Right) Phase contrast micrographs of same cells expressing either empty vector or v-Src. (B) Schematic showing the generation of Myc-deficient 3T9 cells by Cre-mediated myc excision. (C) Absence of Myc expression does not affect Src levels and activity. 3T9mycflox/flox cells stably expressing Cre-ER and either empty vector or v-Src were induced with 4-OH-T to excise c-myc. Total RNA isolated from uninduced (Myc+) and 4-OH-T-induced (Myc−) cells was analyzed for myc mRNA levels by RT-PCR. Expression of v-Src and phosphotyrosine activity were analyzed by immunoblotting. (D) Expression of v-Src does not overcome the proliferative defect resulting from the deletion of c-myc. Equal numbers of 3T9mycflox/flox cells stably expressing Cre-ER and either empty vector or v-Src were seeded in triplicate in medium containing 10% serum and induced with 4-OH-T for the indicated periods. Cells were trypsinized and counted with a Coulter counter. The data represent the number of cells relative to the number of cells at the time of plating (mean of three independent experiments). (E) v-Src expression does not rescue the defect in DNA synthesis resulting from the absence of Myc. Equal numbers of 3T9mycflox/flox cells stably expressing Cre-ER and either empty vector or v-Src were induced with 4-OH-T for the indicated periods, and BrdUrd incorporation was assayed to determine the percentage of cells synthesizing DNA. Data represent the mean of three independent experiments ± SD. Myc levels were analyzed by RT-PCR.
To confirm these conclusions, we made use of a 3T9 line, 3T9-mycflox/flox, derived from mice in which the endogenous c-myc locus had been replaced by an allele with loxP sites in intron 1 and the 3′-untranslated region of c-myc (6). A construct expressing a fusion of Cre-recombinase and estrogen receptor (Cre-ER) was introduced into this cell line (Fig. 1B). In the resulting cell line, c-myc can be excised by inducing Cre activity with 4-hydroxytamoxifen (4-OH-T) (Fig. 1B), leading to a complete block in cell proliferation (see below). We then generated derivatives of these cells stably expressing v-Src. Treatment of these cells with 4-OH-T led to c-myc excision, which was confirmed by RT-PCR and Northern blot hybridization for c-myc (Fig. 1C and data not shown). Immunoblotting with anti-v-Src and anti-phosphotyrosine antibodies indicated that v-Src expression levels and activity are maintained after c-myc excision (Fig. 1C). Although the Myc-deficient cells became larger in size, they retained the spindly and refractile morphology characteristic of v-Src-transformed fibroblasts (data not shown).
Morphological transformation is a complex process involving the loss of actin stress fibers and focal adhesions, together with a decrease in extracellular matrix deposition. In c-myc−/− Rat-1 and 3T9 cells, expression of v-Src led to the loss of stress fibers and surface fibronectin that are hallmarks of v-Src transformation (Fig. 6, which is published as supporting information on the PNAS web site, and data not shown). This result indicates that Myc function is not required for the initiation or maintenance of these molecular correlates of morphological transformation.
Src Expression Does Not Bypass the Requirement for Myc Function in Cell Proliferation.
To determine whether Src could bypass the requirement for Myc function for cell proliferation, we examined the proliferation of c-myc−/− Rat-1 cells expressing v-Src and of 3T9-mycflox/flox cells expressing v-Src after c-myc excision. c-myc−/− Rat-1 cells expressing v-Src proliferated at the same rate as cells expressing empty vector, both in 10% serum and in 0.5% serum (Fig. 7, which is published as supporting information on the PNAS web site). Moreover, within 72 h after Cre-ER activation with 4-OH-T, all of the 3T9-mycflox/flox cells expressing v-Src had ceased to proliferate (Fig. 1D) and failed to incorporate BrdUrd into DNA (Fig. 1E). These results indicate that expression of v-Src is not able to overcome the proliferation defect induced by deficiency of Myc. It should be noted, however, that the cells studied here are immortalized and likely to be defective in the p19ARF-p53 pathway; whether the same results would be obtained in primary cells remains to be determined.
v-Src Expression Regulates Many Myc Targets in the Absence of Myc.
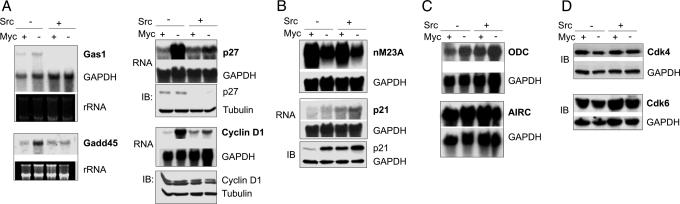
To examine the effect of Src expression on the regulation of Myc targets in Myc-deficient cells, we focused primarily on the effects of Src expression in 3T9 c-myc−/− cells. In the absence of Myc, v-Src was able to strongly repress two Myc targets that are inhibitors of cell proliferation: Gas-1, a growth arrest-specific gene, and the cdk inhibitor p27Kip1 (Fig. 2A). Both Gas1 and p27Kip1 have been shown to be repressed by both Myc and Src at the transcriptional level (14–16). Consistent with these observations, loss of c-myc led to induction of Gas1 and p27 mRNAs. The expression of Gas1 and p27 mRNAs was inhibited in Src-transformed cells, both in the presence and absence of Myc (Fig. 2A); the same result was also obtained in the Rat-1 system (data not shown). The expression of p27 protein was strongly repressed by v-Src, again irrespective of the status of Myc (Fig. 2A). Because Gas-1 and p27 are both repressed by Src in the absence of Myc, we conclude that failure to repress Gas-1 or p27 cannot account for the inability of Myc-deficient cells expressing v-Src to proliferate.
Fig. 2.
Effects of Myc deficiency on the expression of Myc targets. Total RNA isolated from uninduced and 4-OH-T induced cells was analyzed by agarose gel electrophoresis and Northern hybridization, and RNA loading was monitored either by visualizing ribosomal RNAs or by Northern hybridization with a probe for GAPDH. Cell lysates were resolved by SDS/PAGE and protein expression monitored by immunoblotting, with either tubulin or GAPDH as loading control. (A) Expression of Gas1, Gadd45, p27, and cyclin D1. (B) Expression of nM23A and p21. (C) Expression of ODC and AIRC. (D) Expression of Cdk4 and Cdk6.
The expression of a number of other Myc targets also failed to correlate with the proliferation status of the cells. Examples include the following:
Gadd45.
The expression of Gadd45 (growth arrest and DNA damage-inducible protein) was inhibited in Src-transformed cells irrespective of the status of Myc (ref. 17; Fig. 2A).
Cyclin D1.
Cyclin D1 is repressed by Myc (8), but under certain growth conditions, is up-regulated by v-Src (18). We observed that cyclin D1 mRNA is indeed up-regulated in c-myc−/− cells (Fig. 2A). A modest induction of cyclin D1 mRNA was observed in Src-transformed cells, both in the presence and absence of Myc expression.
In summary, the failure of v-Src to induce c-myc−/− cells to proliferate cannot be accounted for by the effects of Myc deficiency on the expression of p27, cyclin D1, Gas-1, or Gadd45.
v-Src Fails to Regulate Some Myc Targets in the Absence of Myc.
In contrast to the results described above, v-Src failed to regulate some Myc targets such as nM23A (19) in the presence and absence of endogenous Myc (Fig. 2B).
Finally it should be noted that under the conditions used here, Myc deficiency did not appear to affect the expression of two well characterized Myc targets, ODC and AIRC (Fig. 2C; refs. 19 and 20). Previous studies have shown that an effect of Myc deficiency on the expression of certain targets such as ODC is only observed as a transient response during serum stimulation (19).
v-Src Induces the Activity of Cyclin D-Dependent Kinases in the Absence of Myc.
Cyclin D-associated Cdks are largely responsible for the phosphorylation and inactivation of the retinoblastoma family of proteins in the G1 phase of the cell cycle (21). Myc promotes the induction of cyclin D2 and the activation of the cyclin D-associated kinases Cdk4 and Cdk6 during the reentry of cells into the cell cycle (7).
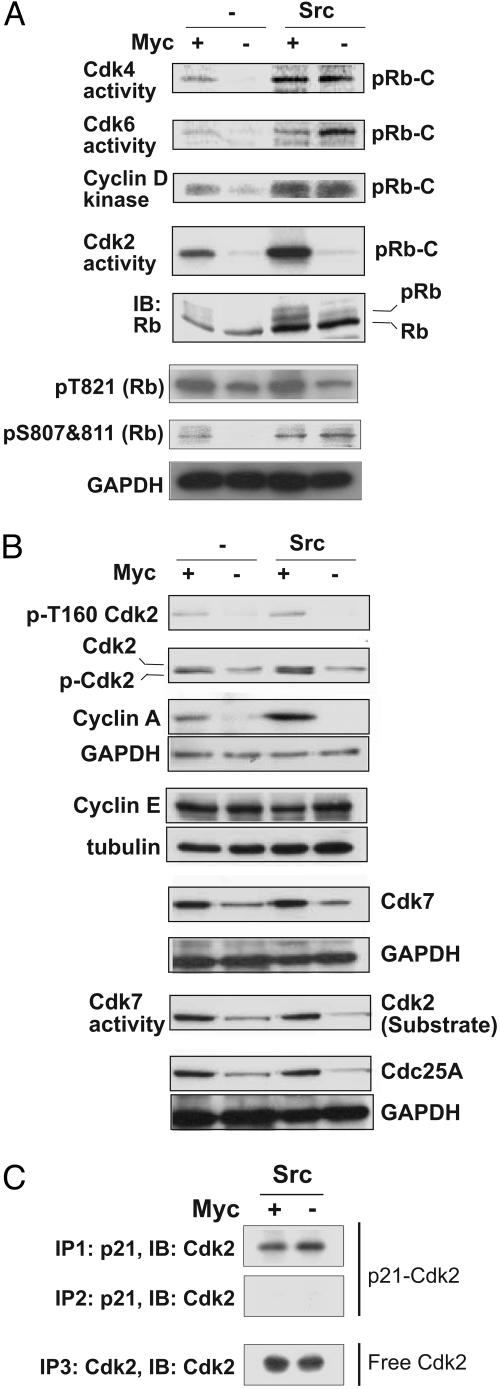
The overall levels of Cdk4 and Cdk6 did not appear to be significantly affected by Myc deficiency (Fig. 2D). As expected, cyclin D-associated Cdk activity was impaired in Myc-deficient cells expressing only empty vector. In striking contrast, v-Src induced cyclin D-associated Cdk activity even in the absence of Myc (Fig. 3A). Furthermore v-Src induced the phosphorylation of Rb at the cyclin D-Cdk4/6 site Ser 807/811, both in the presence and absence of Myc (Fig. 3A). Thus, failure to induce cyclin D-associated Cdk activity cannot account for the inability of v-Src to induce the proliferation of Myc-deficient cells.
Fig. 3.
Effects of Myc deficiency on activation of cell cycle regulators by v-Src. (A) In the absence of Myc, v-Src induces cyclin D-dependent kinases but fails to activate Cdk2. In the top four blots, complexes were immunoprecipitated from extracts with antibodies indicated and kinase activities were assayed by using carboxyterminal Rb as substrate. In blots 5–7, cell lysates were analyzed by immunoblotting with anti-Rb antibody and phospho-specific Rb antibodies: p-T821 and p-S807 plus S811. (B) Failure of v-Src to induce active Cdk2 in c-myc−/− cells results from reduced expression of Cdk2, cyclin A, Cdk7, and Cdc25A. Equal amounts of lysates were analyzed by immunoblotting with antibodies directed against phospho T160-Cdk2, Cdk2, cyclins A and E, Cdk7, and Cdc25A. Cdk-activating enzyme/Cdk7 activity was measured by an immune complex kinase assay by using anti-Cdk7 antibody with recombinant Cdk2 protein as substrate. (C) The fraction of Cdk2 complexed to p21 is increased in the absence of Myc. p21 was immunodepleted from cell lysates by two successive rounds of immunoprecipitation with anti-p21 antibody, and coimmunoprecipitation of Cdk2 was monitored by immunoblotting. The supernatants obtained from the second round of p21 immunoprecipitation were again immunoprecipitated with anti-Cdk2 antibody to determine the levels of Cdk2 not bound to p21 (Bottom).
In the Absence of Myc, v-Src Fails to Activate Cdk2.
We then examined the effects of myc excision on Cdk2 activity. Cdk2 activity was strongly inhibited in the absence of Myc, even in cells expressing v-Src (Fig. 3A). This failure to induce Cdk2 activity appears to be responsible for the inability of v-Src to induce cell proliferation in the absence of Myc (see below). Cyclin D–Cdk4/6 complexes and cyclin E/Cdk2 complexes collaborate in the phosphorylation of the Rb family of proteins, triggering their release from the repressive Rb family/E2F transcriptional complex (22, 23). Consistent with the reduction in Cdk2 activity in Myc-deficient cells expressing v-Src, both the overall level of Rb phosphorylation and the level of phosphorylation of Rb at the Cdk2 site Thr-821 were significantly reduced (Fig. 3A).
Cdk2 activity is subject to complex regulatory mechanisms (24), including association with cyclins E and A, association with negative regulatory subunits, and phosphorylation (25). Cdk2 is activated by phosphorylation on Thr-160 in the activation loop, a phosphorylation that is carried out by Cdk-activating enzymes (25). Cdk2 is also subject to inactivation by phosphorylation in the nucleotide-binding site (Thr-14 and Tyr-15); these latter phosphorylations can be reversed by the dual-specific phosphatase Cdc25A, a Myc target (12).
The level of Cdk2 protein was reduced in Myc-deficient cells expressing v-Src (Fig. 3B). Thr-160 phosphorylation was strongly reduced, both as judged by immunoblotting with an anti-pThr160 antibody and as judged by the absence of the faster migrating band of Cdk2 that represents pThr160-Cdk2 (ref. 26; Fig. 3B). The expression and total kinase activity of Cdk7, the catalytic subunit of Cdk-activating enzyme (25), were also reduced in the absence of Myc, consistent with the down-regulation of pThr160-Cdk2 (Fig. 3B). In addition, the level of cyclin A was reduced, whereas cyclin E expression levels were not altered, irrespective of the status of Src and Myc (Fig. 3B). Finally, the expression of Cdc25A was also down-regulated in the absence of Myc, irrespective of the expression of v-Src (Fig. 3B Bottom).
Overexpression of c-Myc reduces the levels of the Cdk inhibitor p21WAF1/CIP1 (10, 27). In contrast, in mouse fibroblasts stably transformed by v-Src, expression levels of p21 are elevated, and the p21 promoter is induced by v-Src (28). As can be seen from Fig. 2B, p21 expression is induced in c-myc−/− cells, whereas v-Src expression induced p21 expression, consistent with ref. 28. The highest levels of p21 expression were observed in c-myc−/− cells expressing Src.
To determine whether induction of p21 and formation of p21/Cdk2 complexes contributes to the inactivation of Cdk2, we immunodepleted p21 from cell lysates by two successive rounds of immunoprecipitation with anti-p21 antibody and monitored the distribution of Cdk2 by immunoblotting. The fraction of Cdk2 coimmunoprecipitating with p21 in v-Src-expressing cells was increased by Myc deficiency (Fig. 3C), suggesting that increased expression of p21 and the formation of p21/Cdk2 complexes contributes to the G1/S block. However, ≈50% of the Cdk2 remained unbound, consistent with the idea that additional factors contribute to the inactivation of Cdk2.
We conclude that the reduced activity of Cdk2 is due, at least in part, to decreased expression of Cdk7, Cdc25A, and cyclin A and increased complex formation with p21. There may well be additional Cdk2 regulators that cannot be regulated by Src in the absence of Myc.
Cooverexpression of Cdk2 and Cyclins E and A Overcomes the G1/S Block.
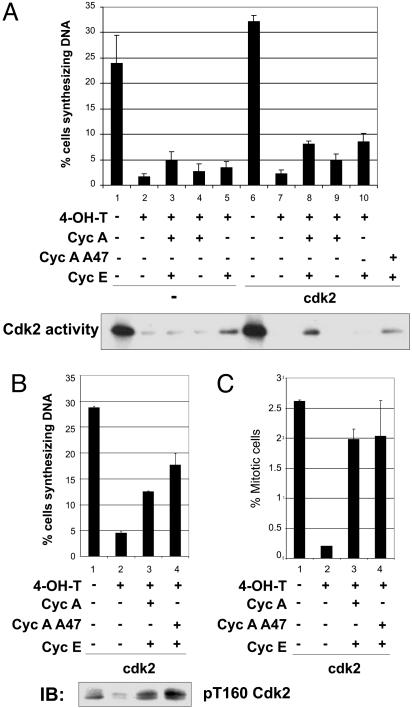
To determine whether the failure of v-Src to drive c-myc−/− cells into S phase results from the reduced activation of cyclin E/Cdk2 and/or cyclin A/Cdk2, Cdk2 was stably overexpressed in the 3T9-mycflox/flox cells expressing Cre-ER and v-Src. The cells were then transiently transfected with expression constructs encoding either cyclin E, cyclin A, a stabilized mutant of cyclin A (A47), or both cyclin E and A constructs, and myc excision was induced by 4-OH-T. Overexpression of Cdk2, cyclin E, and cyclin A (Fig. 8, which is published as supporting information on the PNAS web site) led to the restoration of Thr-160-phosphorylated Cdk2 (Fig. 4B Lower) and also rescued Cdk2 activity as assayed in an immune complex kinase assay (Fig. 4A Lower). The rescue of Cdk2 activity was inefficient, presumably because Cdc25A and other Cdk2 regulators were not also coexpressed. Despite the low level of rescue of Cdk2 activity, overexpression of Cdk2, cyclin E, and cyclin A increased the level of BrdUrd incorporation in the Myc− cells (Fig. 4 A and B). Under optimal conditions (coexpression of Cdk2, cyclin E, and cyclin A (A47), the fraction of cells incorporating BrdUrd was ≈50% of the level observed in the Myc+ cells. Because the transfection efficiency was ≈50% in these experiments, this result indicates that cooverexpression of Cdk2 and cyclins E and A efficiently overcomes the replication block.
Fig. 4.
Cooverexpression of cyclins E and A and Cdk2 in v-Src transformed cells is able to rescue the G1/S block resulting from lack of Myc. 3T9mycflox/flox Cre-ER v-Src-positive cells constitutively overexpressing Cdk2 were generated and then transiently transfected with expression plasmids as indicated: cyclin E, or cyclin A, both cyclin E and A, or cyclin E and a stabilized mutant of cyclin A (A47). Cells were induced with 4-OH-T to excise c-myc and BrdUrd incorporation was assayed to determine the percentage of cells synthesizing DNA. A and B represent two separate sets of experiments; for each set, the data represent the mean of three independent experiments ± SE. (A Lower) Cdk2 activity assayed as described in Fig. 3A. (B Lower) Immunoblotting with anti-phospho-T160-Cdk2 antibody. (C) Percentage of mitotic cells, obtained by anti-phospho-histone H3 staining.
To determine whether overexpression of Cdk2/cyclin A and cyclin E also enabled the cells to enter mitosis, we stained the cells with antibody against phospho-histone H3, a marker of mitotic cells (29). Interestingly, overexpression of cyclin A and E also rescued the block in mitosis (Fig. 4C).
Discussion
Our findings indicate that in the absence of Myc, v-Src is able to regulate early G1 cell cycle regulators such as p27, gadd45, and Cdk4 but is not able to activate late G1 regulators such as cyclin E/Cdk2 and cyclin A/Cdk2 (Fig. 5). These findings raise two questions. First, why do early and late G1 cyclin/Cdk complexes in Myc-deficient cells respond differently to v-Src expression? Second, is the defect in Cdk2 activity sufficient to account for the inability of v-Src to drive cell proliferation in the absence of Myc?
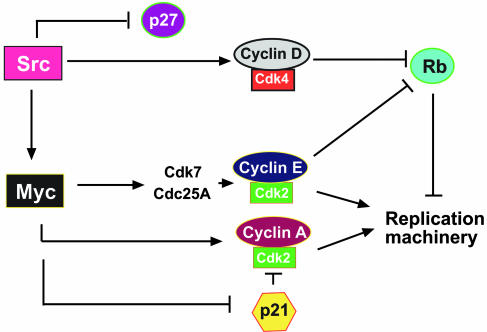
Fig. 5.
Schematic model depicting the significance of Myc in v-Src transformation. In the absence of Myc, v-Src regulates early cell cycle regulators such as p27 and cyclin D-Cdk4 but fails to induce the G1/S transition because of a failure to activate cyclin E/Cdk2 and cyclin A/Cdk complexes.
A variety of observations have converged on a model in which Myc does not promote gene expression by itself, but rather sensitizes loci to other mitogenic signals (19, 30). Myc-dependent recruitment of histone acetyltransferase-containing complexes and acetylation of chromatin proteins sensitizes gene expression to transcription factors activated or recruited to the locus as a consequence of mitogenic signaling. One could therefore postulate that in response to a strong mitogenic stimulus, such as that elicited by v-Src, certain loci are expressed independent of this sensitizing function of Myc, whereas others remain dependent on Myc function. Although it appears that Myc functions at multiple independent cell cycle transitions (7), one could speculate that Myc function may be more critical for the G1/S transition than for passage through the G1 restriction point, and that the promoters of loci involved in regulating Cdk2 activity may be more tightly dependent on Myc than promoters of loci involved in regulating cyclin D-Cdk4/6 activity.
The hypothesis that failure to induce Cdk2 activity is sufficient to account for the inability of Myc-deficient cells expressing v-Src to enter the cell cycle is contradicted by recent reports that cells genetically deficient in Cdk2 or cyclin E are capable of proliferation (reviewed in ref. 31). However, it seems likely that in these genetically deficient cells, other cyclins or cyclin-dependent kinases may complement the deficiencies. Thus, cyclin A may compensate for a deficiency in cyclin E, whereas Cdk1 (Cdc2) has been shown to be able to compensate for a deficiency in Cdk2 (32). Interestingly, cyclin A2 is essential for early embryonic development, indicating that deficiency in this cyclin cannot be compensated by other cyclins (33). In the present studies, the Myc-deficient cells contained very low levels of both cyclin A protein and Cdk2 kinase activity. Furthermore, cooverexpression of Cdk2, cyclin E, and/or cyclin A could overcome the block to G1/S entry and even allowed entry into mitosis, suggesting that in cells expressing v-Src, active cyclin E-Cdk2 and/or cyclin A-Cdk2 are sufficient to drive entry into S in the absence of Myc. Consistent with our observations, both cyclins E and A were earlier shown to be likely targets of Src for platelet-derived growth factor-induced mitogenesis (34).
In summary, our findings demonstrate that whereas Myc is required for activation of cyclin D-dependent kinases in normal cells, it is not required for activation of these kinases in Src-transformed cells. However, expression of v-Src does not abrogate the requirement for Myc induction of cyclin E/Cdk2 and/or cyclin A/Cdk2 for entry into S phase. These findings suggest that the requirement for a particular cell cycle regulator may change according to cellular context. This cell context dependence may have significant implications for the possible use of inhibitors of these regulators as anticancer therapeutics.
Materials and Methods
Cells.
The Rat-1 fibroblast-derived cell line, TGR-1, and a Myc-deficient (c-myc−/−) cell line derived from it, HO15.19, were a generous gift from John Sedivy (Brown University, Providence, RI) (13). Conditionally Myc-deficient cells, 3T9-mycflox/flox cells, have been described in ref. 6. 3T9-mycflox/flox cells stably expressing Cre-ERT2 (a fusion of Cre recombinase to a modified human estrogen receptor binding domain that is responsive to 4-OH-T; ref. 35) were generated by infecting the cells with a mouse stem cell virus expressing a bicistronic message encoding a Cre-ERT2 cDNA and a puromycin resistance cassette separated by an internal ribosome entry site. Derivatives of Rat1 and 3T9-mycflox/flox stably expressing v-Src were generated by retroviral infection and antibiotic selection. Rat1 cells were cultured in F10-DMEM (2:1) supplemented with 10% calf serum. 3T9 cells were cultured in DMEM supplemented with 10% FBS.
Transfections for the rescue experiments were carried out by using LIPOFECTAMINE PLUS (Invitrogen) as per the manufacturer's protocol. Cells incubated with 4-OH-T for 24 h were transfected with either pCDLSRa296-Cyclin A or CSMT-Cyclin A47 and pCS2-Cyclin E, and were analyzed 72 h after transfection.
RT-PCR and Northern Blots.
RNA was prepared from cultured cells by using the RNAeasy kit (Qiagen, Valencia, CA). Myc RT-PCR was carried out by using c-myc-specific primers as described in ref. 6. cDNA probes for mRNAs encoding various Myc targets were generated by RT-PCR by using gene-specific primers. Probe hybridization and washing were performed according to procedures provided by Amersham Pharmacia.
Immunoblotting.
Lysis of cells and immunoblotting were carried out as described in ref. 36. mAbs 2–17 and EC10 were used to detect Src, and mAb4G10 (Upstate Biotechnology, Lake Placid, NY) was used to detect cellular phosphotyrosyl proteins. Antibody against GAPDH was from Abcam, Inc. (Cambridge, MA). All other antibodies were from Santa Cruz Biotechnology.
BrdUrd Incorporation Assays.
The BrdUrd incorporation and flow cytometry analysis was carried out by using a BrdUrd assay kit from Roche Diagnostics (Indianapolis). Cells were labeled with BrdUrd for 1 h.
Immunoprecipitation and Cdk Activity Assays.
Immunoprecipitation and kinase assays were performed as described in ref. 37. Equal amounts of protein (1 mg) were immunoprecipitated with anti-p21WAF1/CIP1 antibody (Abcam, Inc.). Cdk2 was immunoprecipitated from p21-depleted supernatants by incubation with anti-Cdk2 antibody (Santa Cruz Biotechnology) and adsorption to protein-A agarose.
Supplementary Material
Acknowledgments
We thank John Sedivy for providing Rat1 myc+/+ and myc−/− cells; Bob Eisenman, David Morgan, Gary Firestone, Pierre Chambon, and Taku Chibazakura for expression constructs; and Ramadevi Prathapam, Derek Sun, and other members of Martin Laboratory for their help and discussions. The work has been supported by National Institutes of Health Grant CA17542 (to G.S.M.) and by grants from the Swiss National Science Foundation, the Swiss Cancer League, and the UBS Optimus Foundation (to A.T.). T.P. was supported by a Philip Morris External Research Program Fellowship, and S.T. was supported by National Research Service Award Training Grants GM07232 and CA09041.
Abbreviations
- 4-OH-T
4-hydroxytamoxifen
- Cre-ER
fusion of Cre-recombinase and estrogen receptor
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Abram C. L., Courtneidge S. A. Exp. Cell Res. 2000;254:1–13. doi: 10.1006/excr.1999.4732. [DOI] [PubMed] [Google Scholar]
- 2.Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. Mol. Cell. Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman T., Broome M. A., Sinibaldi D., Wharton W., Pledger W. J., Sedivy J. M., Irby R., Yeatman T., Courtneidge S. A., Jove R. Proc. Natl. Acad. Sci. USA. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welham M. J., Wyke J. A., Lang A., Wyke A. W. Oncogene. 1990;5:161–169. [PubMed] [Google Scholar]
- 5.Berg T., Cohen S. B., Desharnais J., Sonderegger C., Maslyar D. J., Goldberg J., Boger D. L., Vogt P. K. Proc. Natl. Acad. Sci. USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., Bishop J. M. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 7.Mateyak M. K., Obaya A. J., Sedivy J. M. Mol. Cell. Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen-Durr P., Meichle A., Steiner P., Pagano M., Finke K., Botz J., Wessbecher J., Draetta G., Eilers M. Proc. Natl. Acad. Sci. USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W., Shen J., Wu M., Arsura M., FitzGerald M., Suldan Z., Kim D. W., Hofmann C. S., Pianetti S., Romieu-Mourez R., et al. Oncogene. 2001;20:1688–1702. doi: 10.1038/sj.onc.1204245. [DOI] [PubMed] [Google Scholar]
- 10.Wu S., Cetinkaya C., Munoz-Alonso M. J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L. G. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 11.O’Hagan R. C., Ohh M., David G., de Alboran I. M., Alt F. W., Kaelin W. G., Jr, DePinho R. A. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galaktionov K., Chen X., Beach D. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 13.Mateyak M. K., Obaya A. J., Adachi S., Sedivy J. M. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 14.Lee T. C., Li L., Philipson L., Ziff E. B. Proc. Natl. Acad. Sci. USA. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossi M., La Rocca S. A., Pierluigi G., Vannucchi S., Ruaro E. M., Schneider C., Tato F. Oncogene. 1998;17:1629–1638. doi: 10.1038/sj.onc.1202090. [DOI] [PubMed] [Google Scholar]
- 16.Johnson D., Frame M. C., Wyke J. A. Oncogene. 1998;16:2017–2028. doi: 10.1038/sj.onc.1201727. [DOI] [PubMed] [Google Scholar]
- 17.Marhin W. W., Chen S., Facchini L. M., Fornace A. J., Jr, Penn L. Z. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 18.Riley D., Carragher N. O., Frame M. C., Wyke J. A. Oncogene. 2001;20:5941–5950. doi: 10.1038/sj.onc.1204826. [DOI] [PubMed] [Google Scholar]
- 19.Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bello-Fernandez C., Packham G., Cleveland J. L. Proc. Natl. Acad. Sci. USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beijersbergen R. L., Bernards R. Biochim. Biophys. Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 22.Lundberg A. S., Weinberg R. A. Mol. Cell. Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black A. R., Azizkhan-Clifford J. Gene. 1999;237:281–302. doi: 10.1016/s0378-1119(99)00305-4. [DOI] [PubMed] [Google Scholar]
- 24.Morgan D. O. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 25.Harper J. W., Elledge S. J. Genes Dev. 1998;12:285–289. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 26.Sheaff R. J. Methods Enzymol. 1997;283:173–193. doi: 10.1016/s0076-6879(97)83015-7. [DOI] [PubMed] [Google Scholar]
- 27.van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., et al. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 28.Sinibaldi D., Wharton W., Turkson J., Bowman T., Pledger W. J., Jove R. Oncogene. 2000;19:5419–5427. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 29.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 30.Amati B., Frank S. R., Donjerkovic D., Taubert S. Biochim. Biophys. Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- 31.Gladden A. B., Diehl J. A. Cancer Cell. 2003;4:160–162. doi: 10.1016/s1535-6108(03)00217-4. [DOI] [PubMed] [Google Scholar]
- 32.Aleem E., Kiyokawa H., Kaldis P. Nat. Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 33.Murphy M., Stinnakre M. G., Senamaud-Beaufort C., Winston N. J., Sweeney C., Kubelka M., Carrington M., Brechot C., Sobczak-Thepot J. Nat. Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- 34.Furstoss O., Manes G., Roche S. FEBS Lett. 2002;526:82–86. doi: 10.1016/s0014-5793(02)03120-4. [DOI] [PubMed] [Google Scholar]
- 35.Indra A. K., Warot X., Brocard J., Bornert J. M., Xiao J. H., Chambon P., Metzger D. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prathapam T., Kuhne C., Banks L. Oncogene. 2001;20:7677–7685. doi: 10.1038/sj.onc.1204960. [DOI] [PubMed] [Google Scholar]
- 37.Cover C. M., Hsieh S. J., Cram E. J., Hong C., Riby J. E., Bjeldanes L. F., Firestone G. L. Cancer Res. 1999;59:1244–1251. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.