Abstract
We recently reported that RabGEF1 is a negative regulator of high-affinity Fc receptor for IgE (FcεRI)-dependent mast cell activation and that mice lacking RabGEF1 develop severe skin inflammation and increased numbers of dermal mast cells. To better understand how RabGEF1 can regulate signaling events and biological responses in mast cells, we examined the responses of bone marrow-derived cultured mast cells (BMCMCs) from wild-type (+/+) and Rabgef1 knockout (−/−) mice after stimulation with the c-Kit ligand, stem cell factor (SCF), an important regulator of mast cell development, survival, proliferation, and activation. We found that RabGEF1-deficient mast cells exhibited enhanced and prolonged activation of Ras and extracellular regulated kinase, and significantly elevated IL-6 secretion, after stimulation with SCF. SCF-induced activation of c-Jun N-terminal kinase was increased in Rabgef1−/− BMCMCs, but without corresponding significant increases in SCF-induced migration or adhesion. SCF-mediated activation of the survival-enhancing kinase, Akt, also was increased in Rabgef1−/− BMCMCs, and these cells had a survival advantage over their +/+ counterparts in vitro. Despite enhanced Ras activation in the absence of RabGEF1, SCF-induced proliferation was lower in Rabgef1−/− BMCMCs compared with their +/+ counterparts. Finally, we found that c-Kit internalization was delayed in the absence of RabGEF1, probably reflecting a positive role for RabGEF1 in the regulation of endocytic events, and that infection of Rabgef1−/− BMCMCs with a wild-type RabGEF1 lentiviral construct normalized c-Kit internalization to the levels seen in +/+ BMCMCs. Thus, RabGEF1 plays a critical role in the regulation of SCF/c-Kit-mediated signaling events and biological responses in mast cells.
Keywords: endocytosis, proliferation, Rab5, Rabex-5, survival
Mast cells are critical effector cells in IgE-associated immediate hypersensitivity and other allergic disorders and also can contribute to T cell-dependent immune responses and certain innate immune responses (1–7). Activated mast cells can secrete three major classes of mediators: (i) preformed mediators (e.g., histamine, tryptase) stored in cytoplasmic granules, by a process called degranulation; (ii) newly synthesized proinflammatory lipid mediators (e.g., leukotrienes, prostaglandins); and (iii) numerous growth factors, cytokines, and chemokines (1, 2, 4–7). Although aggregation of high-affinity IgE receptors (FcεRI) expressed on the mast cell surface induces the release of all three classes of mediators, the type, kinetics, and amounts of particular mediators released depends on the nature of individual activating stimuli and on genetic and microenvironmental factors (6).
By using bone marrow-derived cultured mast cells (BMCMCs) from WT (+/+) and Rabgef1 knockout (−/−) mice, we showed that RabGEF1 (Rab guanine nucleotide exchange factor 1; also known as Rabex-5) can function as a potent negative regulator of IgE plus antigen-induced mast cell degranulation, lipid mediator release, and cytokine production and that RabGEF1 can bind to Ras and can negatively regulate FcεRI-dependent Ras activation in mast cells (8). Interestingly, RabGEF1 was identified by another group as an activator of Rab5, a small GTPase in the Ras superfamily that plays a crucial role in early endosome trafficking (9). Rabgef1−/− mice exhibit increased perinatal mortality, and all Rabgef1−/− mice that survive to adulthood develop severe skin inflammation (8). Dense infiltrates of inflammatory cells, and increased numbers of mast cells, are present in lesional skin; however, skin from nonlesional areas also has significantly elevated numbers of mast cells.
The c-Kit ligand, stem cell factor (SCF) (also known as mast cell growth factor, steel factor, or kit ligand), is a critical factor for the development, survival, proliferation, and functional activation of mast cells (reviewed in ref. 10). SCF can induce mast cell migration, adhesion to extracellular matrix components, and secretion of mediators (11–13). SCF initiates these responses by binding to c-Kit, a tyrosine kinase-containing receptor expressed on the mast cell surface.
We investigated potential roles of RabGEF1 downstream of a receptor tyrosine kinase by comparing SCF-induced signaling events and functional activation in Rabgef1−/− vs. +/+ BMCMCs. We also report methods for the successful infection of mouse BMCMCs with lentiviral expression vectors and demonstrate that infection of Rabgef1−/− BMCMCs with a lentiviral expression vector encoding WT RabGEF1 can normalize RabGEF1 expression levels and c-Kit internalization in Rabgef1−/− BMCMCs.
Results and Discussion
Our attempts to identify differentially expressed mRNA transcripts after FcεRI-dependent mast cell activation revealed the rapid up-regulation of the transcript encoding RabGEF1 (8). RabGEF1 mRNA expression also was up-regulated at 30 and 60 min in BMCMCs after stimulation with SCF but returned to baseline by 2 h (see Fig. 5, which is published as supporting information on the PNAS web site).
Because SCF is important in the induction of several biological responses in mast cells, we compared SCF-mediated signaling events in primary mast cell populations derived from Rabgef1−/− and +/+ mice. Rabgef1−/− and +/+ BMCMCs [>99% FcεRI and c-Kit positive (data not shown)] expressed comparable levels of the SCF receptor, c-Kit (Fig. 1A; see also Fig. 6, which is published as supporting information on the PNAS web site). Although certain populations of −/− BMCMCs showed a slightly broader peak of c-Kit mean fluorescence by flow cytometry (Fig. 1A), the majority of cells expressed approximately equal levels of c-Kit but lower levels of FcεRI (8) compared with +/+ cells.
Fig. 1.
SCF-mediated activation of Ras, Erk, and JNK is enhanced and prolonged, and cytokine production is enhanced, in Rabgef1−/− BMCMCs. (A) Cell surface expression levels of c-Kit in BMCMCs derived from Rabgef1−/− and +/+ mice were analyzed by flow cytometry. (B) Rabgef1−/− and +/+ BMCMCs were starved for 16 h in DMEM plus 10% FCS and then stimulated with 100 ng/ml SCF for the indicated times. Activated (Upper) and total (Lower) Ras levels were analyzed by Western blot using anti-Ras Abs. (C and D) Rabgef1−/− and +/+ BMCMCs were starved and stimulated as in B. Total cell lysates were subjected to Western blot analysis using anti-phospho-Erk1/2 (C) or anti-phospho-JNK (D) Abs (Left). The blots were reprobed with anti-Erk1/2 (C) or anti-GAPDH (D) Abs to show loading. (B–D Right) Signals were quantified by densitometric scanning and corrected for loading. Results in A–D are representative of results obtained in four or more separate experiments. (E) Rabgef1−/− and +/+ BMCMCs were stimulated as in B for 6 h, and IL-6 protein levels in the supernatants were quantified by ELISA. Data are mean ± SEM of the averages of duplicate determinations pooled from six (0 and 50 ng/ml SCF) or five (100 ng/ml SCF) separate experiments. ∗, P < 0.05; ∗∗, P < 0.01 vs. +/+; +, P < 0.05; +++, P < 0.001 vs. corresponding baseline (0 SCF) value. n.s., not significant.
Enhanced Signaling and Functional Activation in Rabgef1−/− Mast Cells.
We previously reported that RabGEF1 both binds to Ras and negatively regulates FcεRI-dependent Ras activation in mast cells (8). Similarly, RabGEF1-deficient BMCMCs exhibited elevated Ras activation, as assessed by GTP-loading, both at baseline and after stimulation with SCF (Fig. 1B). The conversion of inactive GDP-bound Ras to the active GTP-bound form, a reaction catalyzed by GEFs (e.g., Sos), is required to initiate downstream signaling cascades (14, 15). A diverse group of Ras effectors has been identified [e.g., phosphatidyl inositol-3′-kinase (PI3K) and Ras-interaction-interference 1 (Rin1)] (8, 14, 16); however, the canonical Ras pathway is the Ras/Raf1/extracellular regulated kinase 1/2 (Erk1/2) protein kinase cascade (14, 15). Consistent with the elevated Ras activation observed in response to SCF, Erk1/2 phosphorylation was enhanced and prolonged in the absence of RabGEF1 (Fig. 1C). Also, Rabgef1−/− BMCMCs released significantly more IL-6 in response to SCF than did +/+ BMCMCs (Fig. 1E), which is consistent with the view that the Ras/Raf1/Erk1/2 cascade can regulate the synthesis and release of cytokines in mast cells (17–19).
These results indicate that RabGEF1 can act as a negative regulator of SCF-induced Ras signaling. Although the underlying mechanism responsible for increased Ras activation in the absence of RabGEF1 is unknown, the Ras effector Rin1, which also can act as a GEF for Rab5 (20), was originally identified by its ability to inhibit activated Ras by directly competing with Raf1 for binding to GTP-bound Ras (16). Clearly, it will be important to define the mechanism by which RabGEF1 negatively regulates Ras activation in mast cells.
All experiments in Figs. 1–4 used BMCMCs from Rabgef1−/− mice generated on a 129/SvEv background or a mixed 129/SvEv and C57BL/6 background, in which exon 3 of Rabgef1 was deleted (8). However, as previously reported for IgE plus antigen stimulation (8), fetal liver-derived cultured mast cells from Rabgef1−/− mice generated on a WB/Re × C57BL/6 mixed background, in which the entire exon 2 was targeted, yielded similar results (i.e., enhanced and prolonged Erk1/2 phosphorylation and enhanced IL-6 secretion) after stimulation with SCF (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
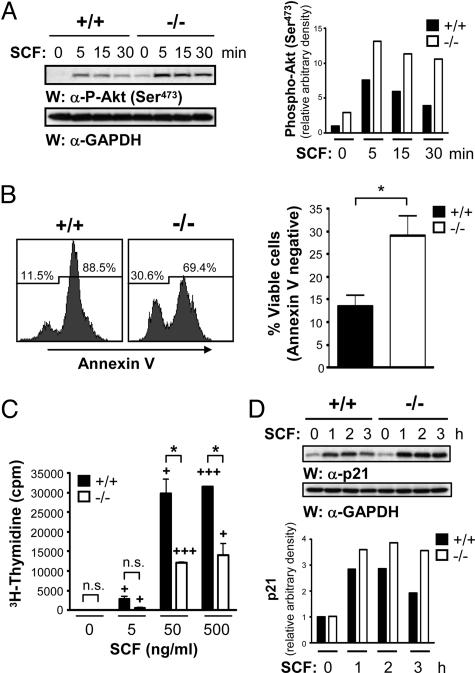
SCF-induced phosphorylation of Akt is enhanced and prolonged in the absence of RabGEF1, and −/− BMCMCs exhibit a survival advantage, but reduced proliferation in response to SCF, compared with +/+ counterparts. (A) Rabgef1−/− and +/+ BMCMCs were starved for 16 h in DMEM plus 10% FCS and then stimulated with 100 ng/ml SCF for the indicated times. (Left) Total cell lysates were subjected to Western blot analysis using anti-phospho-Akt (Ser-473) Abs. The blot was reprobed with anti-GAPDH Abs to show loading. (Right) Signals were quantified by densitometric scanning and corrected for loading. (B) Rabgef1−/− and +/+ BMCMCs were cultured for 72 h in DMEM plus 10% FCS and then stained with Annexin V and analyzed by flow cytometry (Left). (Right) The bar graph represents mean ± SEM of pooled data from five separate experiments. (C) Rabgef1−/− and +/+ BMCMCs were starved for 16 h in DMEM plus 10% FCS, treated with the indicated concentrations of SCF for 48 h, and then labeled with 3H-thymidine for 6 h. Data represent mean ± SEM of triplicate determinations and are representative of results obtained in nine separate experiments (see text). (B and C) ∗, P < 0.05 vs. +/+; +, P < 0.05; +++, P < 0.001 vs. corresponding baseline (0 SCF) value. n.s., not significant. (D) Rabgef1−/− and +/+ BMCMCs were starved and stimulated as in A. (Upper) Total cell lysates were subjected to Western blot analysis using anti-p21 Abs, and the blot was reprobed with anti-GAPDH Abs to show loading. (Lower) Signals were quantified by densitometric scanning and corrected for loading. A and D are representative of results obtained in four and three separate experiments, respectively.
Fig. 3.
SCF-induced c-Kit internalization is delayed in the absence of RabGEF1. (A) Rabgef1−/− and +/+ BMCMCs were starved for 16 h in DMEM plus 10% FCS, then stimulated with 100 ng/ml SCF for the indicated times, and surface c-Kit expression levels were analyzed by flow cytometry. Gray, unstained control. (B) The bar graph represents the mean ± SEM of percent c-Kit internalization determinations from 6 (1 h) or 10 (3 h) separate experiments. Percent c-Kit internalization was calculated by subtracting mean fluorescence intensity at 1 or 3 h from mean fluorescence intensity at 0 h and dividing this number by mean fluorescence intensity at 0 h (×100%). ∗∗∗, P < 0.001 vs. +/+; +, P < 0.05 vs. corresponding 1-h value.
Fig. 4.
Expression of WT RabGEF1 in Rabgef1−/− BMCMCs normalizes c-Kit internalization to the levels seen in +/+ BMCMCs. (A) Rabgef1−/− and +/+ BMCMCs were infected with viral supernatants from 293T cells transfected with control lentiviral vector (empty) or the lentiviral vector containing WT RabGEF1 cDNA, then sorted by GFP expression. RabGEF1 protein levels were analyzed by Western blot. Blots were reprobed with anti-GAPDH Abs to show loading. (B) Infected BMCMCs generated as in A were starved for 16 h in DMEM plus 10% FCS, then stimulated with 100 ng/ml SCF for the indicated times, and surface c-Kit expression was analyzed by flow cytometry. Gray, isotype control. Results in A and B are representative of those obtained in five separate experiments. (C) The bar graph represents the mean ± SEM of percent c-Kit internalization determinations from five separate batches of BMCMCs infected with lentiviral vectors. Percent c-Kit internalization was calculated by subtracting mean fluorescence intensity at 1 or 3 h from mean fluorescence intensity at 0 h and dividing this number by mean fluorescence intensity at 0 h (×100%). ∗, P < 0.05 vs. the indicated population. n.s., not significant.
We also examined SCF-mediated activation of the mitogen-activated protein kinase family member, c-Jun N-terminal kinase (JNK). As with Erk1/2 phosphorylation (Fig. 1C), SCF-induced phosphorylation of JNK was slightly enhanced and prolonged in the absence of RabGEF1 (Fig. 1D). Increased JNK activation may contribute to the elevated IL-6 secretion observed in Rabgef1−/− BMCMCs, because others have shown that JNK activity can regulate cytokine production in mast cells (21, 22). Notably, we did not observe enhanced JNK activation in Rabgef1−/− BMCMCs after stimulation with IgE plus antigen (8). These findings suggest that JNK activation may be differentially regulated downstream of c-Kit and FcεRI, as reported in ref. 22. Although many different signaling pathways can influence JNK activity, the Rho family GTPases (e.g., Rac) that coordinate changes in the actin cytoskeleton are known to be major regulators of JNK activity in mast cells (4, 6, 19, 21, 23).
SCF-Mediated Migration and Adhesion.
Next, we assessed whether RabGEF1 can influence SCF-mediated chemotaxis or adhesion, processes that may contribute to the regulation of mast cell numbers in the skin of Rabgef1−/− mice (6, 12, 24–26). Surprisingly, we found that SCF-induced migration was comparable in Rabgef1−/− and +/+ BMCMCs, as assessed by in vitro transwell chemotaxis (see Fig. 8A, which is published as supporting information on the PNAS web site). However, we observed a significant increase in the basal migration of Rabgef1−/− vs. +/+ BMCMCs in the absence of SCF (Fig. 8A).
These migration assays were performed by using transwells coated with fibronectin, a standard approach in such studies (27, 28). However, we found decreased levels of β1 integrin on the surface of Rabgef1−/− BMCMCs vs. +/+ cells (Fig. 8B). Because α5β1 is the major integrin involved in BMCMC adhesion to fibronectin (12, 29), the migration assay results may simply reflect differences in β1 levels between Rabgef1−/− and +/+ BMCMCs. Notably, despite decreased β1 expression, SCF-mediated adhesion to fibronectin was comparable in Rabgef1−/− and +/+ BMCMCs at lower doses of SCF, but was slightly reduced in the −/− cells at higher concentrations of SCF (Fig. 8C).
Enhanced Akt Phosphorylation and Survival in Rabgef1−/− BMCMCs.
In addition to Erk1/2 and JNK phosphorylation (Fig. 1 C and D), several SCF-induced tyrosine phosphorylation events were enhanced and prolonged in the absence of RabGEF1 (see Fig. 9, which is published as supporting information on the PNAS web site). We therefore looked at other key signaling pathways activated downstream of c-Kit. SCF-induced phosphorylation of Akt (Ser-473) was enhanced and prolonged in RabGEF1-deficient BMCMCs (Fig. 2A) and fetal liver-derived cultured mast cells (Fig. 7A). Because Akt is a survival-enhancing serine/threonine kinase (30, 31), we compared the survival of Rabgef1−/− vs. +/+ BMCMCs. By Annexin V staining, Rabgef1−/− BMCMCs had a survival advantage over their +/+ counterparts upon growth factor withdrawal [pooled data from five independent experiments yielded 13.6 ± 2.3% vs. 29.0 ± 4.5% viability (P < 0.05) in +/+ vs. −/− BMCMCs, respectively] (Fig. 2B). If such a survival advantage also occurs in Rabgef1−/− mast cells in vivo, this survival advantage may contribute to the increased numbers of mast cells observed in the skin of Rabgef1−/− mice. We did not observe enhanced survival of Rabgef1−/− BMCMCs in response to SCF in vitro, as assessed by Annexin V staining (see Fig. 10, which is published as supporting information on the PNAS web site). However, interpretation of these results is confounded by the decreased proliferation observed in BMCMCs in the absence of RabGEF1, as described below.
Decreased Proliferation in Rabgef1−/− BMCMCs.
In addition to its role in survival, Akt plays an important role in the regulation of cell-cycle events (32–35), and Ras activation is known to regulate a wide spectrum of cellular responses, including proliferation (14, 15, 36). Surprisingly, despite increased activation of Ras (Fig. 1B) and enhanced phosphorylation of Akt (Fig. 2A) in the absence of RabGEF1, SCF-induced proliferation was lower in Rabgef1−/− vs. +/+ BMCMCs, as assessed by 3H-thymidine incorporation (Fig. 2C) or carboxyfluorescein diacetate-succinimidyl ester dilution (see Fig. 11A, which is published as supporting information on the PNAS web site). Fig. 2C shows a representative 3H-thymidine incorporation assay from one batch of BMCMCs. In eight other experiments, each with triplicate determinations, we observed a 19–70% reduction (mean ± SEM = 46.5 ± 5.9%) in the proliferation of −/− BMCMCs compared with +/+ BMCMCs (P was <0.12 for results from −/− vs. +/+ BMCMCs in one experiment, but P ranged from <0.05 to <0.01 in the other seven experiments). Cell-cycle analysis indicated that cell-cycle progression was impaired in the Rabgef1−/− BMCMCs (Fig. 11B). Because our BMCMCs were generated in medium containing IL-3, we also compared IL-3-induced proliferation in Rabgef1−/− and +/+ BMCMCs and found that, like SCF-induced proliferation, IL-3-induced proliferation was reduced in the absence of RabGEF1 (see Fig. 12, which is published as supporting information on the PNAS web site).
This unexpected decrease in SCF- or IL-3-induced proliferation in Rabgef1−/− BMCMC may be due, in part, to growth factor-induced up-regulation of the cell-cycle inhibitor, p21 (35,37–39). Microarray analysis indicated that p21 mRNA levels were substantially higher in Rabgef1−/− vs. +/+ BMCMCs after a 2-h stimulation with 100 ng/ml SCF (J.K. and S.J.G., unpublished data). Similarly, SCF stimulation induced higher levels of p21 protein expression in Rabgef1−/− BMCMCs compared with their +/+ counterparts (Fig. 2D). Although it is well established that Ras can act as a dominant transforming oncogene, several studies found that high levels of Ras activity can cause growth arrest in primary fibroblasts and in tumor-derived cell lines (35,37–41). Moreover, various mechanisms to explain the antiproliferative effects of Ras have been proposed, including the Ras/Raf/Erk1/2-mediated induction of p21 (37–39).
Notably, when we tested medium containing both SCF and IL-3, the somewhat reduced proliferation observed in Rabgef1−/− vs. +/+ BMCMCs did not achieve statistical significance (see Fig. 13, which is published as supporting information on the PNAS web site). Although it could be argued that exposing BMCMCs to both SCF and IL-3 may better reflect growth conditions in vivo, the decreased proliferation observed in the Rabgef1−/− BMCMCs may simply reflect other aspects of the in vitro culture conditions. Indeed, under our usual conditions for generating BMCMCs in IL-3-containing medium, we consistently obtained fewer BMCMCs from Rabgef1−/− vs. +/+ bone marrow cells after 4–6 weeks in culture. Thus, the role of RabGEF1 in regulating mouse mast cell survival and proliferation appears to be complex, and none of the conditions examined in vitro may directly reflect the situation in vivo.
Delayed c-Kit Internalization in RabGEF1-Deficient BMCMCs.
Although receptor-mediated endocytosis and signal transduction are tightly coupled, the functional relationship between these two processes is not fully understood (42–44). As indicated above, RabGEF1 was initially isolated as a GEF for Rab5, a positive regulator of early endosome fusion in vitro (9). The GEF activity of RabGEF1 is encoded by its central Vps9 (vacuolar protein sorting 9) domain, which displays high homology to the yeast protein implicated in endosome-to-vacuole transport, Vps9p (9, 45). Although Rab5 has been shown to be an important regulator of ligand-induced receptor endocytosis within intact cells (46, 47), all studies examining the role of RabGEF1 in endosome fusion events to date have relied on in vitro fusion assays (9, 48). Thus, we were interested in evaluating ligand-induced receptor endocytosis in Rabgef1−/− vs. +/+ BMCMCs.
Upon SCF-induced dimerization, c-Kit is internalized by clathrin-mediated endocytosis and then degraded (49). Restoration of surface c-Kit expression is thought to depend primarily on new protein synthesis (50). To evaluate ligand-induced receptor endocytosis in BMCMCs, we assessed by flow cytometry the SCF-induced loss of surface c-Kit staining. SCF-induced internalization of c-Kit was delayed in Rabgef1−/− vs. +/+ BMCMCs (Fig. 3A). Pooled data revealed significant suppression of c-Kit internalization in the absence of RabGEF1: percent c-Kit internalization was 68.6 ± 5.2% vs. 30.5 ± 6.2% (n = 6; P < 0.001) at 1 h and 79.0 ± 4.2% vs. 51.4 ± 4.1% (n = 10; P < 0.001) at 3 h after stimulation with SCF in +/+ vs. −/− BMCMCs, respectively (Fig. 3B).
These data provide evidence that RabGEF1 plays a key role in the regulation of endocytic events in the context of an intact cell. Furthermore, the delayed c-Kit internalization in Rabgef1−/− mast cells suggests that Rab5 activity is compromised in the absence of RabGEF1. Although no studies to date have investigated the dependence of c-Kit internalization on Rab5, the internalization and trafficking of other receptor tyrosine kinases, such as the epidermal growth factor receptor (EGFR), are known to be Rab5-dependent (46). In addition to RabGEF1, several GEFs for Rab5 have been identified, including Rin1 (20), Rin2 (51), Rin3 (52), and ALS2 (53); however, the redundancy for Rab5 activation has not yet been investigated. Our findings indicate that RabGEF1 plays a significant and nonredundant role in regulating endocytosis in BMCMCs.
Receptor endocytosis is traditionally thought to attenuate ligand-induced signaling events and cellular responses (43, 54). Therefore, it is reasonable to propose that delayed c-Kit internalization (Fig. 3) may contribute to the prolonged SCF-induced signaling events observed in Rabgef1−/− BMCMCs. The findings that these cells demonstrate elevated and prolonged signaling after stimulation with either SCF (Figs. 1 and 2) or IgE plus antigen (8) suggest the possibility of a similar defect; however, it will be important to investigate the role of RabGEF1 in the internalization of other receptor families (e.g., FcεRI, IL-3 receptor, TLRs, etc.). Furthermore, a number of signaling molecules activated downstream of c-Kit have been reported to play an important role in the regulation of receptor internalization [e.g., Ras (20, 55), Akt (55), and Src family kinases (54)]; thus, it will be important to study the effects of these signaling molecules on SCF-induced endocytosis in the presence and absence of RabGEF1.
Complicating matters further, ligand-induced endocytosis and normal cellular trafficking of various receptors may be necessary to activate the full spectrum of intracellular signaling pathways downstream of these receptors (42–44, 54, 56). Therefore, it also will be important to evaluate whether additional Rab5-dependent mechanisms, such as endosome fusion and trafficking, are altered in the absence of RabGEF1.
Infection of Rabgef1−/− BMCMCs with a WT RabGEF1 Lentiviral Construct Normalizes c-Kit Internalization.
To better understand the aberrations observed in Rabgef1−/− BMCMCs, we wished to restore RabGEF1 protein expression in these cells. Although it is difficult to introduce foreign DNA into mast cells, successful approaches employing retroviral infection of BMCMCs have been reported (57). For our studies, we attempted to develop a lentiviral vector system for infection of mouse BMCMCs. Lentiviral vectors are actively imported to the nucleus; thus, they are able to integrate and induce stable cDNA expression in nondividing or slowly dividing cells (58). WT RabGEF1 cDNA was cloned into a lentiviral vector containing an internal ribosomal entry site (IRES) controlling GFP expression (58). BMCMCs were infected with viral supernatants from 293T cells transfected with control lentiviral vector (empty) or the lentiviral vector containing WT RabGEF1 cDNA, then sorted by GFP expression. After infection of Rabgef1−/− BMCMCs with the WT construct, Western blot analysis revealed that RabGEF1 expression levels were comparable with the endogenous levels in +/+ cells (Fig. 4A).
To test the function of the reconstituted RabGEF1, we examined c-Kit internalization in Rabgef1−/− BMCMCs infected with the empty or WT RabGEF1 lentiviral constructs. Expression of WT RabGEF1 in Rabgef1−/− BMCMCs virtually normalized the ability of these cells to internalize c-Kit after stimulation with SCF (Fig. 4 B and C; see also Fig. 14, which is published as supporting information on the PNAS web site). These findings provide direct evidence that the abnormality in c-Kit internalization observed in Rabgef1−/− BMCMCs indeed reflects an important role for RabGEF1 in the pathways that regulate c-Kit receptor internalization. Moreover, these findings show that we can use lentiviral approaches to infect BMCMCs directly and thereby alter expression levels of the mutant or WT protein of interest after mast cell differentiation has occurred.
Mast cells have been implicated in many diseases and in host defense. Accordingly, a better understanding of the positive and negative regulation of the signaling pathways that control mast cell function will advance our understanding of the biology of these cells and may suggest novel therapeutic approaches to modulate their function in health and disease. Our results indicate that RabGEF1 plays rather complex, but important, roles in the regulation of c-Kit-dependent signaling and biological activation in mast cells. The approaches described herein, particularly the lentiviral infection of BMCMCs, will facilitate examination of the consequences of specific mutations of RabGEF1 on mast cell function in response to SCF, IgE plus antigen, or other stimuli. Such approaches also will be helpful in attempts to study the role of RabGEF1 and other factors that may link the processes of endocytosis and signaling in mast cells.
Materials and Methods
Cell Culture.
Femoral and tibial bone marrow from 4- to 8-week-old Rabgef1−/− mice generated on a 129/SvEv or 129/SvEv and C57BL/6 mixed background (8) or their +/+ littermates was cultured in DMEM plus IL-3 containing medium for 4–8 weeks to generate populations of BMCMCs that were >95% pure by flow cytometry (described below).
BMCMC Stimulation and Western Blot Analysis.
BMCMCs starved for 16 h in DMEM plus 10% FCS were stimulated with various concentrations of recombinant SCF (PeproTech, Rocky Hill, NJ, or a generous gift from Amgen, Thousand Oaks, CA). Cells were solubilized by boiling in Laemlli sample buffer (pH 6.8). Cell lysates were separated by SDS/PAGE, electroblotted onto Invitrolon poly(vinylidene difluoride) membranes (Invitrogen), then probed with antibodies (Abs) against phospho-Erk1/2 (E10), phospho-Akt (Ser-473), phospho-JNK, or Erk1/2 (Cell Signaling Technology, Beverly, MA), GAPDH (Research Diagnostics, Flanders, NJ), phosphotyrosine (clone 4G10, Upstate Biotechnology, Waltham, MA), p21 (C-19; Santa Cruz Biotechnology), or RabGEF1 (Rabex-5) (BD Biosciences, San Jose, CA).
Measurement of IL-6 Release.
IL-6 ELISAs (BD Biosciences) were performed according to the manufacturer’s instructions.
Flow Cytometry.
BMCMCs were incubated for 5 min with monoclonal Abs to CD16/32 (clone 93) to block nonspecific binding. Cells stained for 30 min with allophycocyanin-conjugated anti-c-Kit (2B8; eBioscience, San Diego), phycoerythrin-conjugated anti-FcεR1α (MAR-1; eBioscience), or biotinylated isotype control (IgG2b κ chain; Pharmingen) Abs were analyzed on a FACSCalibur flow cytometer (Becton Dickinson). Dead cells were excluded by propidium iodide gating, and data were analyzed with flowjo software (Tree Star, Ashland, OR).
c-Kit Internalization.
BMCMCs starved for 16 h in DMEM plus 10% FCS were resuspended in DMEM plus 0.1% BSA and 100 ng/ml SCF. Aliquots were removed at 0, 1, or 3 h, and surface expression levels of c-Kit were analyzed by flow cytometry as described above.
Annexin V Staining.
Annexin V staining and analysis was performed according to the manufacturer’s instructions (BD ApoAlert; BD Biosciences) on a FACSCalibur.
Assessment of Ras Activation.
Ras activation was assayed with the EZ-Detect Ras Activation Kit (Pierce). A GST fusion protein containing the Ras-binding domain of Raf-1 was used to precipitate active GTP-bound Ras.
Chemotaxis Assays.
In vitro transwell chemotaxis assays were performed as described in ref. 59 by using 5-μm pore Transwell inserts (Costar) coated with 50 μg/ml fibronectin (Sigma).
Cell Proliferation Assays.
3H-Thymidine incorporation assays were performed as described in ref. 60. 3H-thymidine incorporated over 6 h was expressed as cpm.
Lentiviral Vector Production.
The entire coding region of RabGEF1 cDNA (8) was PCR amplified by using the 5′ primer 5′-CCGTCCGGAATGTACCCATACGATGTTCCAGATTACGCTATGAGCCTGAAGTCCGAACG-3′ (BspE1 site in bold, HA epitope DNA sequence in italics), and the 3′ primer 5′-CCGGCTAGCTCACCCTGCGTACACCTGAGG-3′ (Nhel site in bold) and subcloned into pRRLsin-18.PPT.PGK.MCS.IRES.GFP.pre (58). Active viral stocks were created and concentrated as described in ref. 58. Briefly, 293T cells were transfected with the transfer vector plasmid pRRLsin-18.PPT.PGK.MCS.IRES.GFP.pre (empty) or pRRLsin-18.PPT.PGK.RabGEF1.IRES.GFP.pre (WT), the VSV-G envelope-encoding plasmid pMD.G, and the packaging plasmid CMVΔR8.74 (58) by using the calcium phosphate method. The supernatants were harvested 48 and 72 h after transfection, pooled, passed through a 0.45-μm filter, ultracentrifuged for 2 h at 100,000 × g in a SW28 rotor, resuspended in 100 μl of 0.1% BSA in PBS, and stored at −80°C.
Lentiviral Infection.
Two- to five-week-old BMCMCs were infected with viral supernatants from 293T cells transfected with control lentiviral vector (empty) or the lentiviral vector containing WT RabGEF1 cDNA as described above. Then, 72–96 h after infection, BMCMCs were sorted by GFP expression using FACS Vantage SE/DiVa (Becton Dickinson) and cultured in Iscove’s modified Dulbecco’s medium plus 10 ng/ml IL-3 (PeproTech).
Statistics.
Unless otherwise specified, all data are expressed as mean ± SEM and were examined for significance by unpaired Student’s t test, 2-tailed.
Supplementary Material
Acknowledgments
We thank members of the S.J.G. laboratory for helpful discussions and M. Liebersbach for animal husbandry. This work was supported by National Institutes of Health Grants AI23990, CA72074, and HL67674 (to S.J.G.); a Natural Sciences and Engineering Research Council of Canada Fellowship (to J.K.); and Stanford Medical Scientist Training Program Grant 5-732-GM07365 (to E.J.R.).
Abbreviations
- BMCMC
bone marrow-derived cultured mast cell
- Erk
extracellular regulated kinase
- FcεRI
high-affinity Fc receptor for IgE
- JNK
c-Jun N-terminal kinase
- SCF
stem cell factor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Metzger H. Immunol. Rev. 1992;125:37–48. doi: 10.1111/j.1600-065x.1992.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 2.Metcalfe D. D., Baram D., Mekori Y. A. Physiol. Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 3.Mekori Y. A., Metcalfe D. D. Immunol. Rev. 2000;173:131–140. doi: 10.1034/j.1600-065x.2000.917305.x. [DOI] [PubMed] [Google Scholar]
- 4.Rivera J. Curr. Opin. Immunol. 2002;14:688–693. doi: 10.1016/s0952-7915(02)00396-5. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami T., Galli S. J. Nat. Rev. Immunol. 2002;2:773–786. doi: 10.1038/nri914. [DOI] [PubMed] [Google Scholar]
- 6.Galli S. J., Kalesnikoff J., Grimbaldeston M. A., Piliponsky A. M., Williams C. M., Tsai M. Annu. Rev. Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 7.Kinet J. P. Annu. Rev. Immunol. 1999;17:931–972. doi: 10.1146/annurev.immunol.17.1.931. [DOI] [PubMed] [Google Scholar]
- 8.Tam S. Y., Tsai M., Snouwaert J. N., Kalesnikoff J., Scherrer D., Nakae S., Chatterjea D., Bouley D. M., Galli S. J. Nat. Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi H., Lippe R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 10.Galli S. J., Zsebo K. M., Geissler E. N. Adv. Immunol. 1994;55:1–96. doi: 10.1016/s0065-2776(08)60508-8. [DOI] [PubMed] [Google Scholar]
- 11.Vosseller K., Stella G., Yee N. S., Besmer P. Mol. Biol. Cell. 1997;8:909–922. doi: 10.1091/mbc.8.5.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam V., Kalesnikoff J., Lee C. W., Hernandez-Hansen V., Wilson B. S., Oliver J. M., Krystal G. Blood. 2003;102:1405–1413. doi: 10.1182/blood-2002-10-3176. [DOI] [PubMed] [Google Scholar]
- 13.Gagari E., Tsai M., Lantz C. S., Fox L. G., Galli S. J. Blood. 1997;89:2654–2663. [PubMed] [Google Scholar]
- 14.Katz M. E., McCormick F. Curr. Opin. Genet. Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 15.Wennerberg K., Rossman K. L., Der C. J. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 16.Han L., Colicelli J. Mol. Cell. Biol. 1995;15:1318–1323. doi: 10.1128/mcb.15.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalesnikoff J., Baur N., Leitges M., Hughes M. R., Damen J. E., Huber M., Krystal G. J. Immunol. 2002;168:4737–4746. doi: 10.4049/jimmunol.168.9.4737. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami Y., Kitaura J., Yao L., McHenry R. W., Kawakami Y., Newton A. C., Kang S., Kato R. M., Leitges M., Rawlings D. J., Kawakami T. Proc. Natl. Acad. Sci. USA. 2003;100:9470–9475. doi: 10.1073/pnas.1633695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siraganian R. P. Curr. Opin. Immunol. 2003;15:639–646. doi: 10.1016/j.coi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Tall G. G., Barbieri M. A., Stahl P. D., Horazdovsky B. F. Dev. Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 21.Song J. S., Haleem-Smith H., Arudchandran R., Gomez J., Scott P. M., Mill J. F., Tan T. H., Rivera J. J. Immunol. 1999;163:802–810. [PubMed] [Google Scholar]
- 22.Ishizuka T., Chayama K., Takeda K., Hamelmann E., Terada N., Keller G. M., Johnson G. L., Gelfand E. W. J. Immunol. 1999;162:2087–2094. [PubMed] [Google Scholar]
- 23.Gu Y., Byrne M. C., Paranavitana N. C., Aronow B., Siefring J. E., D’Souza M., Horton H. F., Quilliam L. A., Williams D. A. Mol. Cell. Biol. 2002;22:7645–7657. doi: 10.1128/MCB.22.21.7645-7657.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamawy M. M., Mergenhagen S. E., Siraganian R. P. Immunol. Today. 1994;15:62–66. doi: 10.1016/0167-5699(94)90135-X. [DOI] [PubMed] [Google Scholar]
- 25.Okayama Y. Clin. Exp. Allergy. 2000;30:455–457. doi: 10.1046/j.1365-2222.2000.00824.x. [DOI] [PubMed] [Google Scholar]
- 26.Maurer M., Galli S. J. Lab. Invest. 2004;84:1593–1602. doi: 10.1038/labinvest.3700196. [DOI] [PubMed] [Google Scholar]
- 27.Sundstrom M., Alfredsson J., Olsson N., Nilsson G. Exp. Cell Res. 2001;267:144–151. doi: 10.1006/excr.2001.5239. [DOI] [PubMed] [Google Scholar]
- 28.Meininger C. J., Yano H., Rottapel R., Bernstein A., Zsebo K. M., Zetter B. R. Blood. 1992;79:958–963. [PubMed] [Google Scholar]
- 29.Houtman R., Koster A. S., Nijkamp F. P. Clin. Exp. Allergy. 2001;31:817–822. doi: 10.1046/j.1365-2222.2001.01104.x. [DOI] [PubMed] [Google Scholar]
- 30.Franke T. F., Cantley L. C. Nature. 1997;390:116–117. doi: 10.1038/36442. [DOI] [PubMed] [Google Scholar]
- 31.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 32.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. Mol. Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 33.Diehl J. A., Cheng M., Roussel M. F., Sherr C. J. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou B. P., Liao Y., Xia W., Spohn B., Lee M. H., Hung M. C. Nat. Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 35.Crespo P., Leon J. Cell Mol. Life Sci. 2000;57:1613–1636. doi: 10.1007/PL00000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shields J. M., Pruitt K., McFall A., Shaub A., Der C. J. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 37.Westbrook T. F., Nguyen D. X., Thrash B. R., McCance D. J. Mol. Cell. Biol. 2002;22:7041–7052. doi: 10.1128/MCB.22.20.7041-7052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewing A., Wiseman B., Lloyd A. C., Land H. Mol. Cell. Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods D., Parry D., Cherwinski H., Bosch E., Lees E., McMahon M. Mol. Cell. Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin A. W., Barradas M., Stone J. C., van Aelst L., Serrano M., Lowe S. W. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 42.Di Fiore P. P., De Camilli P. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 43.Le Roy C., Wrana J. L. Nat. Rev. Mol. Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 44.Miaczynska M., Pelkmans L., Zerial M. Curr. Opin. Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Esters H., Alexandrov K., Iakovenko A., Ivanova T., Thoma N., Rybin V., Zerial M., Scheidig A. J., Goody R. S. J. Mol. Biol. 2001;310:141–156. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- 46.Barbieri M. A., Roberts R. L., Gumusboga A., Highfield H., Alvarez-Dominguez C., Wells A., Stahl P. D. J. Cell Biol. 2000;151:539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galperin E., Sorkin A. J. Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- 48.Lippe R., Miaczynska M., Rybin V., Runge A., Zerial M. Mol. Biol. Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yee N. S., Hsiau C. W., Serve H., Vosseller K., Besmer P. J. Biol. Chem. 1994;269:31991–31998. [PubMed] [Google Scholar]
- 50.Shimizu Y., Ashman L. K., Du Z., Schwartz L. B. J. Immunol. 1996;156:3443–3449. [PubMed] [Google Scholar]
- 51.Saito K., Murai J., Kajiho H., Kontani K., Kurosu H., Katada T. J. Biol. Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 52.Kajiho H., Saito K., Tsujita K., Kontani K., Araki Y., Kurosu H., Katada T. J. Cell Sci. 2003;116:4159–4168. doi: 10.1242/jcs.00718. [DOI] [PubMed] [Google Scholar]
- 53.Topp J. D., Gray N. W., Gerard R. D., Horazdovsky B. F. J. Biol. Chem. 2004;279:24612–24623. doi: 10.1074/jbc.M313504200. [DOI] [PubMed] [Google Scholar]
- 54.Broudy V. C., Lin N. L., Liles W. C., Corey S. J., O’Laughlin B., Mou S., Linnekin D. Blood. 1999;94:1979–1986. [PubMed] [Google Scholar]
- 55.Barbieri M. A., Kohn A. D., Roth R. A., Stahl P. D. J. Biol. Chem. 1998;273:19367–19370. doi: 10.1074/jbc.273.31.19367. [DOI] [PubMed] [Google Scholar]
- 56.Vieira A. V., Lamaze C., Schmid S. L. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 57.Hata D., Kawakami Y., Inagaki N., Lantz C. S., Kitamura T., Khan W. N., Maeda-Yamamoto M., Miura T., Han W., Hartman S. E., et al. J. Exp. Med. 1998;187:1235–1247. doi: 10.1084/jem.187.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ailles L., Schmidt M., Santoni de Sio F. R., Glimm H., Cavalieri S., Bruno S., Piacibello W., Von Kalle C., Naldini L. Mol. Ther. 2002;6:615–626. [PubMed] [Google Scholar]
- 59.Zabel B. A., Allen S. J., Kulig P., Allen J. A., Cichy J., Handel T. M., Butcher E. C. J. Biol. Chem. 2005;280:34661–34666. doi: 10.1074/jbc.M504868200. [DOI] [PubMed] [Google Scholar]
- 60.Tsai M., Takeishi T., Thompson H., Langley K. E., Zsebo K. M., Metcalfe D. D., Geissler E. N., Galli S. J. Proc. Natl. Acad. Sci. USA. 1991;88:6382–6386. doi: 10.1073/pnas.88.14.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.