Abstract
Bacterial type IV secretion systems (T4SS) translocate DNA and/or proteins to recipient cells, thus providing a mechanism for conjugative transfer of genetic material and bacterial pathogenesis. Here we describe the first structure of a core component from the archetypal Agrobacterium tumefaciens T4SS: the 2.2-Å resolution crystal structure of the VirB8 periplasmic domain (pVirB8AT). VirB8 forms a dimer in the crystal, and we identify residues likely important for stabilization of the dimer interface. Structural comparison of pVirB8AT with Brucella suis VirB8 confirms that the monomers have a similar fold. In addition, the pVirB8AT dimer superimposes very closely on the B. suis VirB8 dimer, supporting the proposal that dimer formation in the crystal reflects self-interactions that are biologically significant. The evolutionary conservation level for each residue was obtained from a data set of 84 VirB8 homologs and projected onto the protein structure to indicate conserved surface patches that likely contact other T4SS proteins.
Keywords: bacterial protein export, conjugation, type IV secretion
Type IV secretion systems (T4SS) function as conjugation systems, DNA uptake/release systems, and effector protein translocator systems (1) and are required for virulence of several human pathogens including Bartonella henselae, Bordetella pertussis, Brucella suis, Helicobacter pylori, and Legionella pneumophila, as well as the plant pathogen Agrobacterium tumefaciens (2). The A. tumefaciens T4SS is composed of 12 proteins, VirB1–11 and VirD4. The known properties of these components are well reviewed (1), and each contributes to one or more functional subgroups. The A. tumefaciens system genetically transforms plant cells with a specific DNA element, called the T-DNA. VirB4, VirB11, and VirD4 are NTPases located at the inner membrane that may provide energy for DNA transfer or assembly of the secretion system. VirB6–VirB10 likely form a “core” complex that spans the inner and outer membranes. VirB2, VirB5, and VirB7 assemble the pilus. VirB1 is a lytic transglycosidase thought to promote T4SS assembly by lysis of the peptidoglycan layer. Crystal structures of several T4SS components have been published, including homologs of hexameric NTPases VirB11 and VirD4 (3, 4), VirB5 (5), VirB8, and VirB10 (6). A hexameric structure for A. tumefaciens VirB4 has been recently proposed (7).
A. tumefaciens VirB8 is a bitopic membrane protein with a short N-terminal cytoplasmically exposed domain, a transmembrane helix, and a large C-terminal periplasmic domain. A T-DNA immunoprecipitation assay has identified contacts between the DNA substrate and six of the T4SS components, including VirB8 (8). It was recently proposed that VirB8 is the scaffold for polar assembly of the A. tumefaciens T4SS (9). Yeast two-hybrid interaction studies suggest that the periplasmic portion of VirB8 contacts VirB1, VirB4, VirB9, VirB10, and VirB11 (10, 11). Biochemical studies confirm that VirB8 interacts with VirB9, VirB10, and itself in vitro (11), and that VirB8 influences the subcellular localization of VirB9 and VirB10 in the membrane (12). Affinity-tagging studies of B. suis VirB8 demonstrate interactions with VirB4 (which are necessary for stabilization of VirB8) and with VirB5 (13). Thus, it is suggested that the core T4SS protein complex is linked to the pilus complex via VirB8–VirB5 interactions (13). Random mutagenesis of A. tumefaciens VirB8 has identified five residues, Gly-78, Ser-87, Ala-100, Arg-107, and Thr-192, that, when substituted, lead to loss of VirB8 activity (14).
Here we describe the production, characterization, and crystal structure of the periplasmic domain of A. tumefaciens pVirB8AT, corresponding to residues 92–237. Our structure suggests that VirB8 forms a dimer, in agreement with similar predictions for the recently determined structure of B. suis VirB8 (6). A consurf analysis of sequence conservation (15, 16) predicts surface residues likely to be involved in dimer formation and interactions with other components of the T4SS.
Results
Structure of the pVirB8AT Monomer.
The structure of pVirB8AT has been determined at 2.2-Å resolution by using the structure of the periplasmic domain of B. suis VirB8 (pVirB8BS) (6) as a model for molecular replacement. The final model contains a homodimer plus two molecules of 2-methyl-2-4-pentanediol and 170 water molecules in the asymmetric unit. The structure of each monomer (denoted monomer A and B throughout) comprises VirB8 residues 92–231 and additional N-terminal tag residues 88–91 (Gly-Pro-His-Met). Residues 232–237 are disordered. Residues 184–187 form a flexible loop that is sufficiently ordered to model the main chain into continuous 2Fc − Fo electron density for monomer A but displays discontinuous electron density at the 1ς level in monomer B. A cis-peptide bond was identified in each monomer at Pro-143.
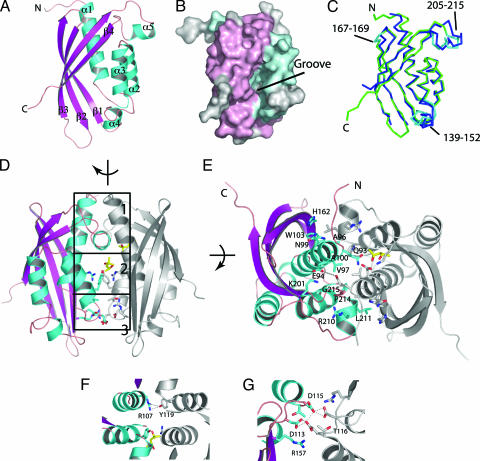
Each monomer is composed of a single domain of approximate dimensions 20 × 30 × 45 Å that contains five helices clustered to one side of a four-strand antiparallel β-sheet (Fig. 1A). The β-sheet extends the length of the molecule and curves around helix α1. Helices α1 to α4 are at the N terminus of the crystallized fragment, whereas helix α5 is inserted in a loop between strands β3 and β4 of the sheet (Fig. 2A). The protein fold is similar to that of the NTF2-like family, including NTF2 (17), scytalone dehydratase (18), ketosteroid isomerases (19), and one domain of CaMKII (20), although the sequence identity is low (4–13%). NTF2 has a cone-shaped structure with a substantial cavity at the base of the cone. pVirB8AT differs by the presence of helix α5 and the absence of two short β-strands. The absence of the β-strands results in a deep groove on one face of pVirB8AT that is flanked by β4 and α3 at the top, extends from midway along the molecule length, and widens at the base (Fig. 1B). The opposite face of pVirB8AT is relatively flat and corresponds with the dimer interface.
Fig. 1.
Structural features of pVirB8AT and comparison with pVirB8BS. Monomer A is colored magenta and cyan and monomer B is gray. (A) Ribbon representation of monomer A with secondary structure elements labeled. (B) Surface rendered image of pVirB8AT in same orientation as A showing the deep groove. The surface is colored according to the secondary structure: cyan for helical segments, pink for β-strands, and gray for loops. (C) Cα trace of superimposed pVirB8AT (green with regions of greatest divergence highlighted in cyan) and pVirB8BS (blue) monomers. (D) Ribbon diagram of the dimer rotated 90° with respect to A. Boxes indicate the three dimer contact regions. The side chains of Arg-107, Asp-113, Asp-115, Thr-116, Tyr-119, and Arg-157 are shown, and two 2-methyl-2-4-pentanediol molecules (yellow) are shown as stick models. (E–G) correspond to boxes D1, D2, and D3 and show details of the three dimer contact regions viewed down the dimer axis, equivalent to a 90° rotation from the view in D. (E) Residues involved in the major region of dimer contact. (F) Interactions across the central channel between the monomers. (G) Four aspartates at the lowest level of the interface.
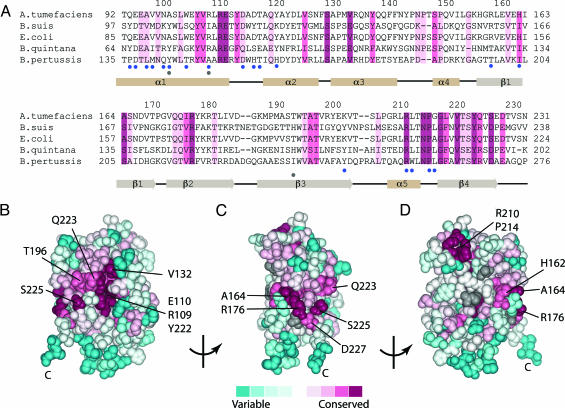
Fig. 2.
consurf analysis of pVirB8AT (residues 92–231 of structural model). (A) Multiple sequence alignment of pVirB8AT and corresponding peptides of selected homologs: Residues in the three most conserved levels are highlighted. The corresponding secondary structural elements are shown. Blue dots indicate residues involved in dimerization. Gray dots indicate residues genetically identified as essential for activity. (B–D) Space filling models of pVirB8AT. Surface residues are shaded according to degree of conservation. Residues in gray have been shown to be essential for VirB8 activity. Surface exposed residues in the two most conserved levels are labeled. (B) The protein is presented in approximately the same orientation as Fig. 1A. (C) Rotated by 90°. (D) Rotated by 180° with the dimer interface presented face on. GenInfo Identifier numbers for the multiple sequence alignment are A. tumefaciens VirB8, 17939307; B. suis VirB8, 23463398; E. coli TraG, 10955150; Bartonella quintana VirB8, 21260578; and Bordatella pertussis VirB8, 420953.
VirB8 Dimer Interface.
The presence of a homodimer of pVirB8AT in the asymmetric unit allows us to describe the dimer interactions in detail. The pVirB8AT monomers forming the dimer can be superimposed with a root-mean-square deviation of 0.175 Å between all 144 Cα atoms. The dimer interface buries a total of 1,720 Å2, corresponding to 10% of the accessible surface area of each monomer, and is the largest contact area between two monomers in the crystal. Dimer interactions are divided into three distinct contact regions (Fig. 1D, see also Fig. 4, which is published as supporting information on the PNAS web site). The largest contact area, shown in detail in Fig. 1E, buries 1,180 Å2 and is dominated by hydrophobic interactions between 13 surface residues. This contact region involves residues from the N terminus of α1, α5, and the residues connecting α5 to β4. There is only one hydrogen bond, between Glu-94 of monomer A and Gln-93 of monomer B. Two residues at the C-terminal end of α5 (Arg-210 and Pro-214) are highly conserved across VirB8 sequences and form a conserved surface patch at the dimer interface (Fig. 2 A and D).
The second and third contact regions are significantly smaller with interactions predominantly between hydrogen bonding partners. The smallest interaction site in the central region of the dimer buries only 200 Å2 and involves the side chains of Arg-107 and Tyr-119, which protrude from the surface of each monomer (Fig. 1 D and F).
At the base of the dimer interface, the third contact point buries 340 Å2 and is mediated by Asp-113, Asp-115, Thr-116, and Arg-157 from each monomer (Fig. 1G). The four aspartates have well defined electron density and contribute five intersubunit hydrogen bonds. Thus, our model implies that the Asp-115 residues are at least partially protonated. The aspartates are surrounded by the positively charged side chains of Arg-157 and Lys-154, albeit the electron density for these side chains is poor and it is not possible to confidently identify specific interactions. Arg-157 has the potential to form a salt bridge with either Asp-113 of the same monomer, or Asp-115 of the 2-fold related monomer, and may play a role in stabilization of this aspartate cluster. The crystals were grown at pH 4.6 and, depending on the pKa of the aspartates, it is unlikely that the Asp–Asp interactions are present at higher pH. Potentially, dimerization is influenced by pH.
Comparison of pVirB8AT and pVirB8BS.
pVirB8AT and pVirB8BS share 30% sequence identity, and as expected, the overall fold of pVirB8AT is very similar to the published structure of pVirB8BS (6). Both structures correspond to the periplasmic fragment of the VirB8 protein, composed of residues 92–231 of the A. tumefaciens protein and 97–234 of the B. suis protein. Superposition of monomer A or B of pVirB8AT on one of the five monomers of pVirB8BS present in the asymmetric unit gives a root-mean-square deviation between 1.33 and 1.42 Å for 125–127 Cα atoms, depending on the monomers superimposed. The biggest differences in regions of superposition, highlighted in Fig. 1C, are seen at the N- and C-termini, and at three surface loop regions, comprising residues 139–152, 167–169, and 205–215.
Only 4 of 19 residues involved in dimer interactions are conserved between pVirB8AT and pVirB8BS (Fig. 2A). Nevertheless, dimer formation, in both VirB8 proteins, involves packing of the two monomers in a very similar fashion and the pVirB8AT dimer superimposes on the pVirB8BS BD and CE dimers with a root-mean-square deviation of 1.52 Å (251 Cα atoms) and 1.65 Å (252 Cα atoms), respectively (Fig. 4A). Several residues that interact at the largest dimer interface (Fig. 1E) correspond to bulkier side chains in the B. suis protein (A. tumefaciens Gln-93, Val-97, Ala-100, and Gly-215 are replaced by B. suis Tyr-98, Met-102, Tyr-105, and Leu-222) and, as a consequence, the N-termini of α1 in the A and B monomers pack closer in pVirB8AT. In the central dimer region, residues Arg-107 and Tyr-119 of pVirB8AT are replaced, in the B. suis protein, by smaller side chains Ile-112 and Lys-124, respectively, with the consequence that the opening observed between the two pVirB8BS monomers is much less pronounced in pVirB8AT (Fig. 4 B and C).
Mapping of Conserved Residues.
The biological importance of a residue often correlates with its level of evolutionary conservation within the protein family (15). We analyzed a broad range of VirB8 sequences and determined highly conserved residues that are potentially necessary for either structural integrity or biological activity. These residues were then mapped onto the structure of pVirB8AT.
The multiple sequence alignment used for the consurf analysis (15) was composed of 84 sequences (Fig. 5, which is published as supporting information on the PNAS web site). Representative sequences illustrate the degree of conservation (Fig. 2A) and this conservation was mapped onto the protein structure (Fig. 2 B–D). The most highly conserved residues are either buried or contribute to one of two clusters exposed at the surface of the protein. Thirteen residues in the two most conserved consurf levels were found to have little or no solvent accessibility at the monomer surface: Tyr-105, Val-106, Tyr-112, Val-124, Ser-128, Tyr-136, Ser-147, Ile-175, Ala-195, Asn-213, Gly-216, Leu-217, and Val-219. Further analysis of these buried conserved residues revealed that they also appear to form clusters that are located adjacent to the conserved surface patches and, thus, are presumably important for maintaining the structural integrity of these surface patches. It seems highly likely that these conserved surface regions are important for maintaining the functionality of VirB8 (16).
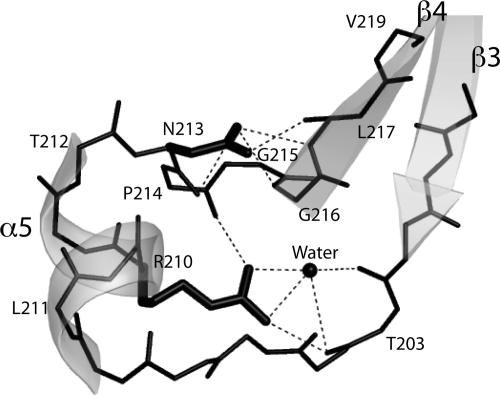
One conserved region, formed by residues 210–219, is defined by the sequence motif RxxNPxGxxV (Fig. 2 A and D) and appears to be important to stabilize and position the loop containing helix α5 between strands β3 and β4. This loop appears to be relatively rigid in our crystal structure, reflected by lower-than-average temperature factors. The local fold is stabilized by extensive hydrogen bonds between the side chain of Arg-210 and main-chain atoms of residues Thr-203 and Pro-214, and the side chain of Asn-213 and main-chain atoms of residues Asn-213, Gly-216, and Leu-217 (Fig. 3). Other stabilizing influences are hydrophobic interactions between residues Leu-217 and Val-219 of this loop, which pack against the N terminus of helix α1. In addition, side-chain and main-chain atoms of residues Arg-210, Leu-211, Pro-214, and Gly-215 interact with the second pVirB8AT monomer at the dimer interface (Fig. 1E). The positioning of this loop is likely essential for maintenance of the dimer interface. The highly conserved residues (Arg-210, Asn-213, Pro-214, Gly-216, and Val-219) of this loop are largely buried in our crystal structure, only Arg-210 and Pro-214 are exposed at the monomer surface, and Pro-214 becomes buried on dimer formation. It is not clear whether Arg-210 would be exposed at the surface of the native protein. In our structure, Arg-210 is predominantly protected from solvent by nonnative residues N-terminal to Thr-92, the first VirB8AT residue of the construct. The exposure of Arg-210, therefore, will depend on the conformation of the native N-terminal residues in solution.
Fig. 3.
Stick diagram of the loop containing helix 5. The highly conserved residues R210, N213, P214, G216, and V219 are part of this loop. Dashed lines represent the hydrogen bonds between the side chains of Arg-210 and Asn-213 and main chain atoms.
The second conserved region is discontinuous along the linear sequence but is composed of a surface patch encompassing the groove opposite to the dimerization interface and extending around the surface of the central region of the β-sheet (Fig. 2 B and C). The groove is situated between β4, at the edge of the β-sheet, and helix α3, with the C-terminal end of helix α1 providing the floor (Fig. 1A). There are several conserved residues inside the groove and on the β4 edge, whereas the α3 side of the groove exhibits more variable residues. The most highly conserved residues, Arg-109, Glu-110, Tyr-222, and Val-132, line the groove (Fig. 2B). The side chains of Arg-109 and Glu-110 extend from the floor and are stabilized by hydrogen bonds to surrounding residues Tyr-222, Tyr-105, and Asn-135. On the edge of the groove, two residues, conserved in most VirB8 homologs as an aspartate and an arginine, form a salt bridge in the pVirB8BS structure (6), but in VirB8AT, these residues, Ser-225 and Gln-223, are not conserved. A highly conserved salt bridge between Arg-176 on β2 and Asp-227 (a glutamate in most VirB8 sequences) at the N-terminal end of β4 serves to pin the position of this C-terminal β-strand to the core of the protein, thus maintaining the integrity of one wall of the groove. Adjacent to the salt bridge, Ala-164 is a highly conserved serine in most VirB8 homologs.
Mapping of Functional Residues.
Kumar and Das (14) introduced random mutations into pTiA6-derived virB8 of A. tumefaciens strain A348 and identified five mutations, G78S, S87L, A100V, R107A, and T192M, that produce stable proteins but fail to complement a virB8-deletion mutant in tumor-forming assays. The S87L mutant exhibited a semidominant phenotype providing early evidence that VirB8 forms a multimer (14). Two mutations, G78S and S87L, are in a region of the polypeptide (residues 60–90) linking the predicted transmembrane helix with the VirB8 periplasmic domain and are not present in the construct we have expressed. This linking peptide is disordered in the crystal structure of pVirB8BS and does not appear to be a structural component of the periplasmic domain, which is stable in its absence. The S87L mutant prevents interaction with VirB9 in the yeast two-hybrid assay and abolishes DNA transfer function (11). Although Ser-87 is not part of our structure, it will be close to the observed N terminus, suggesting that the VirB9 interaction site is close to the top of the VirB8 molecule as viewed in Fig. 1A.
The three remaining mutations, A100V, R107A, and T192M, can be mapped onto the presented structure (Fig. 2 A and D). Although these residues are not highly conserved among VirB8 homologs, it is useful to consider the role of these residues in more detail. All three residues are located on the surface of a VirB8 monomer and are close to the conserved surface patches and likely to be involved in interactions with other proteins. Both Ala-100 and Arg-107 are involved in dimer interactions in the crystal. The side chain of Ala-100 is in close contact with the side chain of Pro-214 (3.7 Å) and the carbonyl oxygen of Leu-211 (3.2 Å) of the opposing monomer. Thus, it is possible that the A100V mutation could disrupt formation of the VirB8 dimer. However, yeast two-hybrid assays show that the mutated protein continues to interact with itself and with VirB9 and VirB10. Yeast two-hybrid assays also show that the R107A mutant has retained the ability for self-interaction but is not able to interact with VirB9 or VirB10. In our structure, Arg-107 is hydrogen-bonded to Tyr-119 from the noncrystallographic symmetry-related monomer (Fig. 1F), and replacement with an alanine is unlikely to disrupt dimer formation. VirB8 may not always be in the dimer state in vivo, allowing interaction with other T4SS components such as VirB9 or VirB10. The apparent conflict between yeast two-hybrid and tumorigenesis data may be explained by innate differences in each of the assays. Tumor formation requires VirB8 activity and may involve dynamic interactions with many T4SS components and T4SS substrates. In contrast, two-hybrid interactions that are compromised by single amino acid changes may be stabilized by other contacts along the length of bait and prey peptides.
A T192M mutant of VirB8 is unable to complement a VirB8 deletion mutant but does not affect VirB8 interactions with itself or with VirB9 and VirB10 (14). Thr-192 is in close contact with the highly conserved salt bridge between Arg-176 and Asp-227 and with Ile-163 and Lys-178. Possibly mutation to a methionine could disrupt the local tightly fitted structure or remove required flexibility in residues surrounding the groove.
Solution Characterization of VirB8.
The apparent molecular mass of pVirB8AT was 23 kDa by SDS/PAGE and 70 kDa by native PAGE, suggesting dimer or trimer formation. To resolve the discrepancy, pVirB8AT was subjected to gel filtration at pH 8.0 and pH 4.5. At both pH values, pVirB8AT was eluted as a monomer with a molecular mass of 20 kDa. Similarly, x-ray scattering experiments provided results that are consistent with a monomer in solution with a radius of gyration of 18.8 ± 0.2 Å and maximum dimension 63 ± 3 Å. Furthermore, simulations based on the monomer and dimer taken from the crystal structure of pVirB8AT suggest the monomer best fits the scattering curve, yielding a goodness-of-fit value (χ) of 2.33 (in contrast to χ = 8.72 for the dimer, data not shown). Thus it appears that the protein is monomeric in solution and runs anomalously on native PAGE.
Discussion
VirB8 is a core component of the A. tumefaciens T4SS. Our structure suggests specific residues involved in dimer formation and allows us to predict surface features that could direct its interaction with other VirB proteins. We believe that the observed dimer is physiologically significant. Yeast two-hybrid studies have shown that VirB8 is able to interact with itself (11) and mutation of residues at the dimer interface, A100V and R107A, reduces VirB8 function (14). Significantly, the crystal structure of pVirB8BS also indicates a nearly identical dimeric structure (Fig. 4A); the similarity between quaternary structures would be very unlikely if dimer formation were solely a consequence of crystallization.
Data mining of protein structure databases can indicate correlative properties such as geometric features, chemical features, and residue conservation that distinguish a biological interface from a nonspecific crystal contact (ref. 21 and references therein). Despite wide variation in the values of these properties, the size of the interface and, to a lesser extent, the degree of hydrophobicity or the presence of conserved residues, can aid differentiation of biological interfaces from crystal contacts (21). The pVirB8AT dimer interface buries 1,720 Å2 of accessible surface area, corresponding to 10% of the total monomer accessible surface area. This interface is larger than the other crystal contacts in our structure and larger than the average crystal contact in the Protein Data Bank (PDB), although it is slightly smaller than the average dimer interface (21). In addition, we observe highly conserved residues participating in dimer formation. Assuming VirB8 dimerization is of biological significance, it would be helpful to investigate the conditions that promote dimerization. It is possible that dimer formation in solution is energetically unfavorable for the isolated periplasmic domain, requiring additional interactions formed by the missing N-terminal polypeptide. Alternatively, VirB8 may be required to interact with itself only under certain conditions, for example, in the presence or absence of other T4SS components
We identify two highly conserved surface regions that are likely of functional significance. Although the precise biological role of VirB8 is unknown, there is good evidence that VirB8 interacts with a number of other VirB proteins and with DNA during translocation (11). Therefore, we suggest that the clustered, solvent-accessible, conserved residues correspond to sites of interaction of VirB8, either with itself, with other VirB proteins, or with DNA. The smaller conserved region corresponds to (210-RxxNPxSxxV-219) and likely functions to position the extended loop containing helix α5, Arg-210, and Pro-214 for proper dimer-forming interactions. The VirB9 interaction site may also be close to this conserved region.
The larger conserved region includes surface-exposed residues Arg-109, Glu-110, Val-132, Ala-164, Arg-176, Tyr-222, Gln-223, Ser-225, and Asp-227, which can be seen as dark red and pink conserved patches in two views of VirB8AT (Fig. 2 B and C). The extended nature of this conserved surface patch, and that VirB8 interacts with multiple VirB components, suggests this region may enable specific protein interactions. The valley of the groove flanked by β4 and α3 likely forms one interaction site. Structurally, the groove corresponds to the hydrophobic cavity of NTF2, which comprises the protein interface region of the NTF2–Ran complex (22). Examination of the NTF2-like family also reveals that the central region of the β-sheet serves as a common site for protein–protein interactions. This region contributes to the homodimer interfaces of NTF2, CaMKII, and ketosteroid isomerase, and to the heterodimer interface of the two NTF2-like mRNA export factors Mtr2 and Mex67 (23). Thus, we suggest that the surface patch centered on highly conserved residues Ala-164, Arg-176, and Ser-225 on the face of the β-sheet of pVirB8AT may form a second specific interaction site.
Mapping the level of conservation onto the structure of pVirB8AT has enabled us to distinguish between residues likely essential for structural integrity and surface residues that may be important for VirB8 activity. The discovery of a highly conserved surface groove is especially intriguing, because it may provide a site for interactions with other T4SS components. The identification of conserved residues at the dimer interface and surface of VirB8 provokes the design of a mutagenesis program to further investigate the importance of these interfaces in the structure and function of the T4SS.
Materials and Methods
Cloning and Protein Preparation.
A fragment encoding residues 92–237 of the VirB8 peptide was amplified from pTiC58 and cloned into a modified pET28 vector. The resulting clone, pDW134, was used to transform Escherichia coli strain BL21(DE3). His-tagged pVirB8AT was expressed and purified on a Ni-NTA column, followed by anion exchange chromatography. Purity of pVirB8AT was checked by SDS/PAGE and native PAGE.
Gel Filtration and Electrophoresis.
SDS/PAGE and native PAGE were run on a PHAST system by using standard protocols. Gel filtration experiments used a Superose-75 column with buffer systems 50 mM Tris·HCl pH 8.0/50 mM NaCl or 50 mM sodium citrate, pH 4.5/50 mM NaCl.
Solution X-Ray Scattering.
Scattering data were collected (station 2.1 of the Daresbury Synchrotron Radiation Source) of protein samples (at concentrations between 0.5 and 12 mg/ml) and buffer in the momentum transfer interval 0.03 Å−1 ≤ q ≥0.71 Å−1, where q = 4πsinθ/λ, (2θ is the scattering angle and λ is the x-ray wavelength 1.5 Å) according to published procedures (24). The distance distribution function p(r), and the radius of gyration were evaluated with the program gnom (25), which also provides a reliable estimation of the overall particle size. Scattering pattern simulations based on crystal structure information were carried out with the program crysol (26). The goodness-of-fit value (χ) is a measure of how well the structure fits the experimental data (a reliable agreement between experiment and simulation is generally obtained for χ < 3).
Crystallization and Data Collection.
Crystals of pVirB8AT (10–13 mg/ml) were obtained at 4°C by vapor diffusion against a reservoir of 0.1 M sodium acetate, pH 4.6/0.2–0.4 M sodium bromide/20% vol/vol 2-methyl-2–4 pentanediol. Crystals belonged to the space group C2 with cell dimensions a = 112.3 Å, b = 72.3 Å, c = 48.8 Å, and β = 110.7 Å and diffracted anisotropically, to 1.8 Å in one direction and to 2.2 Å in the orthogonal direction. Data were collected at the Daresbury Synchrotron Radiation Source and processed by using programs in the ccp4 suite (27) to yield a 2.2 Å native data set. Data collection and processing statistics are presented in Table 1, which is published as supporting information on the PNAS web site.
Structure Determination.
The crystal structure was determined by molecular replacement by using the structure of a periplasmic domain of VirB8 from B. suis (pVirB8BS), PDB entry 2BHM (6), as a search model. The positions of two monomers, related by a noncrystallographic dyad axis, were identified. Maps were improved by using two-fold noncrystallographic symmetry averaging, solvent flattening, and extensive model building. Refinement using refmac5 (28) converged to a model with R and Rfree factors of 23.5% and 29.8% with good stereochemistry, and refinement statistics are given in Table 1. Noncrystallographic symmetry restraints were applied throughout the entire refinement process.
Sequence Comparisons.
Several methodologies were followed for comparison of homologous sequences and identification of conserved sites on the surface of VirB8 and all methodologies yielded similar results. For the analysis described here, a Hidden Markov Model (HMM) was constructed by using the program hmmbuild on a pairwise structural alignment of A. tumefaciens VirB8 residues 78–231 and B. suis VirB8 residues 77–238. Sequences matching the HMM were selected from the uniprot database and aligned to the HMM by using the hmmalign program (29). The new alignment was cropped to the relevant region, edited to remove redundancy, and used as the basis for building another HMM. This process was repeated three times, resulting in a final multiple sequence alignment that included 84 sequences. The multiple sequence alignment was used as input for analysis by using the consurf software (15, 16) and the evolutionary trace program tracesuite ii (30).
Supplementary Material
Acknowledgments
This work was supported by a Laboratory Directed Research and Development (LDRD) grant from the Lawrence Berkeley National Laboratory (to S.B.) and by the Council for the Central Laboratory of the Research Councils Daresbury Laboratory. D.W. was supported by a Novartis grant (to P.C.Z.) and by the LDRD grant (to S.B.). R.M. and P.C.Z. were supported by National Science Foundation Grant 0343566.
Abbreviations
- HMM
hidden Markov model
- T4SS
Type IV secretion system.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2cc3).
References
- 1.Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. Annu. Rev. Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagai H., Roy C. R. Cell Microbiol. 2003;5:373–83. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 3.Yeo H.-J., Savvides S. N., Herr A. B., Lanka E., Waksman G. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 4.Gomis-Rüth F. X., Moncalián G., Pérez-Luque R., González A., Cabezón E., de la Cruz F., Coll M. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 5.Yeo H.-J., Yuan Q., Beck M. R., Baron C., Waksman G. Proc. Natl. Acad. Sci. USA. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradot L., Bayliss R., Oomen C., Leonard G. A., Baron C., Waksman G. Proc. Natl. Acad. Sci. USA. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middleton R., Sjölander K., Krishnamurthy N., Foley J., Zambryski P. Proc. Natl. Acad. Sci. USA. 2005;102:1685–1690. doi: 10.1073/pnas.0409399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascales E., Christie P. J. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Judd P. K., Kumar R. B., Das A. Proc. Natl. Acad. Sci. USA. 2005;102:11498–11503. doi: 10.1073/pnas.0505290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward D. V., Draper O., Zupan J. R., Zambryski P. C. Proc. Natl. Acad. Sci. USA. 2002;99:11493–11500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A., Xie Y. H. J. Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R. B., Xie Y. H., Das A. Mol. Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Q., Carle A., Gao C., Sivanesan D., Aly K. A., Höppner C., Krall L., Domke N., Baron C. J. Biol. Chem. 2005;280:26349–26359. doi: 10.1074/jbc.M502347200. [DOI] [PubMed] [Google Scholar]
- 14.Kumar R. B., Das A. J. Bacteriol. 2001;183:3636–3641. doi: 10.1128/JB.183.12.3636-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 16.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullock T. L., Clarkson D. W., Kent H. M., Stewart M. J. Mol. Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 18.Lundqvist T., Rice J., Hodge C. N., Basarab G. S., Pierce J., Lindqvist Y. Structure (London) 1994;2:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 19.Cho H. S., Choi G., Choi K. Y., Oh B. H. Biochemistry. 1998;37:8325–8330. doi: 10.1021/bi9801614. [DOI] [PubMed] [Google Scholar]
- 20.Hoelz A., Nairn A. C., Kuriyan J. Mol. Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 21.Ponstingl H., Kabir T., Gorse D., Thornton J. M. Prog. Biophys. Mol. Biol. 2005;89:9–35. doi: 10.1016/j.pbiomolbio.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Stewart M., Kent H. M., McCoy A. J. J. Mol. Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 23.Fribourg S., Conti E. EMBO Rep. 2003;4:699–703. doi: 10.1038/sj.embor.embor883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossmann J. G., Hall J. F., Kanbi L. D., Hasnain S. S. Biochemistry. 2002;41:3613–3619. doi: 10.1021/bi015955o. [DOI] [PubMed] [Google Scholar]
- 25.Semenyuk A. V., Svergun D. I. J. Appl. Crystallogr. 1991;24:537–540. [Google Scholar]
- 26.Svergun D., Barberato C., Koch M. J. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 27.Collaborative Computational Project No. 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Eddy S. R. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 30.Lichtarge O., Bourne H. R., Cohen F. E. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.