Abstract
Mycobacterium tuberculosis remains a major cause of morbidity and mortality worldwide. Studies have reported human pathogens to have geographically structured population genetics, some of which have been linked to ancient human migrations. However, no study has addressed the potential evolutionary consequences of such longstanding human–pathogen associations. Here, we demonstrate that the global population structure of M. tuberculosis is defined by six phylogeographical lineages, each associated with specific, sympatric human populations. In an urban cosmopolitan environment, mycobacterial lineages were much more likely to spread in sympatric than in allopatric patient populations. Tuberculosis cases that did occur in allopatric hosts disproportionately involved high-risk individuals with impaired host resistance. These observations suggest that mycobacterial lineages are adapted to particular human populations. If confirmed, our findings have important implications for tuberculosis control and vaccine development.
Keywords: coevolution, deletions, lineage, polymorphism, population
Several studies have reported geographically structured populations in human pathogens (1–4). Recently, the genetic population structure of Helicobacter pylori and Mycobacterium leprae have been linked to ancient human migrations (1, 4, 5). Such long-standing host–pathogen associations could lead to adaptive genetic changes between interacting host and pathogen populations. Studies in invertebrate model systems have shown that pathogens can adapt to specific host species (6). However, no example of host-specific pathogen adaptation has yet been documented in pathogens affecting different human populations. The observation of geographically structured populations of human pathogens implies that particular strains and their corresponding patient populations can be classified as sympatric or allopatric (6). Compatibility, defined as the ability of a given pathogen to infect a particular host, often differs in sympatric versus allopatric host–pathogen combinations, with sympatric combinations usually displaying a greater compatibility (6).
Mycobacterium tuberculosis occurs world-wide and is still killing 2–3 million people each year (7). New tools for tuberculosis control are urgently needed, including a more effective vaccine (8). A series of genotyping tools for M. tuberculosis have been developed (9). Most of these make use of mobile genetic elements or repetitive DNA. Even though these tools have been invaluable for detecting ongoing tuberculosis transmission, the markers upon which they are based change relatively rapidly, making it difficult to define deep phylogenetic relationships (4). In contrast, large sequence polymorphisms (LSPs) represent unique event polymorphisms that can be used to construct robust phylogenies for M. tuberculosis (10). An additional advantage is that, once LSPs have been identified (e.g., by comparative whole-genome hybridization), simple PCR can be used to screen large numbers of strains in a high-throughput fashion.
In this study, we used comparative genomic and molecular epidemiological tools to define the global population structure of M. tuberculosis and to investigate its influence on the transmission dynamics of M. tuberculosis in San Francisco during an 11-year period.
Results and Discussion
To define the global population structure of M. tuberculosis, we performed genomic deletion analysis on a global sample of 875 strains originating from 80 countries (Table 1 and Table 3, which is published as supporting information on the PNAS web site). This sample included strains isolated from foreign-born tuberculosis patients in San Francisco who contracted the infection in their country of origin and was complemented with geographically representative strains from other reference collections. We analyzed a subset of 111 strains by comparative whole-genome hybridization (11). The results of 74 of these experiments were published earlier (11–13), and 37 are reported herein. Overall, we identified 19 phylogenetically informative and lineage-specific LSPs (Fig. 1a and Table 4, which is published as supporting information on the PNAS web site). These LSPs were confirmed by sequencing, validated by PCR and sequencing in 72 additional strains, and used to screen the remaining 692 strains by PCR or multiplex real-time PCR (Table 4 and Table 5, which is published as supporting information on the PNAS web site). We used as additional phylogenetic markers the previously reported regions of difference (RD) TbD1 and RD9 (ref. 14), the 7-bp deletion in the pks15/1 locus (15), and the katG463 ctg to cgg substitution (16).
Table 1.
Assignment of 875 strains of M. tuberculosis from 80 countries to six main phylogenetic lineages in eleven geographic regions
| Geographic region (no. of countries) | Total strains | Indo-Oceanic lineage | East-Asian lineage | East-African-Indian lineage | Euro-American lineage | West-African lineage 1 | West-African lineage 2 |
|---|---|---|---|---|---|---|---|
| Americas (18) | 207 | 6 | 7 | 194 | |||
| Europe (14) | 35 | 1 | 4 | 30 | |||
| North Africa/Middle East (6) | 10 | 10 | |||||
| West Africa (8) | 28 | 12 | 9 | 7 | |||
| Central Africa (5) | 15 | 1 | 14 | ||||
| South Africa (1) | 5 | 2 | 3 | ||||
| East Africa (6) | 20 | 3 | 8 | 9 | |||
| Indian Subcontinent (4) | 17 | 3 | 1 | 10 | 3 | ||
| Southeast Asia (9) | 272 | 169 | 73 | 1 | 29 | ||
| East Asia (7) | 262 | 17 | 190 | 55 | |||
| Pacific Islands (2) | 4 | 4 | |||||
| Total (80) | 875 | 199 | 277 | 20 | 363 | 9 | 7 |
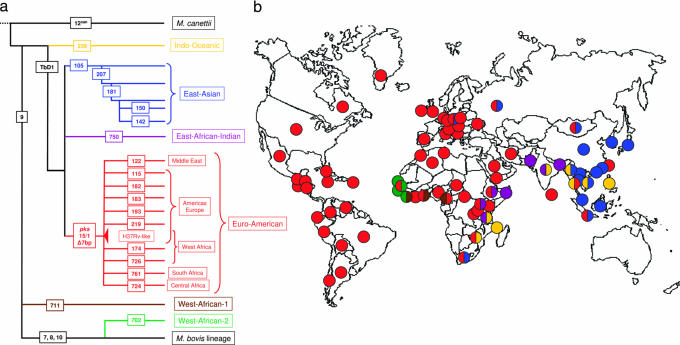
Fig. 1.
The global population structure and geographical distribution of M. tuberculosis. (a) LSPs define a global phylogeny for M. tuberculosis. The names of the lineage-defining LSPs or regions of difference are shown in rectangles. The geographic regions associated with specific lineages are indicated. (b) The six main lineages of M. tuberculosis are geographically structured. Each dot corresponds to 1 of 80 countries represented in the global strain collection. The colors of the dots relate to the six main lineages defined in Fig. 1a and indicate the dominant lineage(s) in the respective countries.
The analysis of our global sample of 875 strains revealed six main lineages and 15 sublineages of M. tuberculosis (Fig. 1a, Tables 1 and 3, and Table 6, which is published as supporting information on the PNAS web site). Some of these lineages correspond to strain groupings that have previously been reported. For example, the Indo-Oceanic lineage includes a group of strains that have been referred to as “ancestral” due to the fact that they conserve the TbD1 genomic region, which is deleted in “modern” strains of M. tuberculosis (14). The East-Asian lineage includes, but is not limited to, the Beijing family of strains (13). The West-African lineages 1 and 2 correspond to strains that have traditionally been named Mycobacterium africanum (12), and the Euro-American lineage regroups strains that have generally been described as principal genetic groups 2 and 3 (15–17).
Besides confirming some of the mycobacterial groupings that have been described previously, our analysis of an extended global strain collection revealed that the population genetics of M. tuberculosis is highly geographically structured. Each of the six main lineages was associated with particular geographical areas, and the lineage names reflect these geographical associations (Fig. 1b and Table 1). For example, the East-Asian lineage is dominant in many countries of the Far East, and the Indo-Oceanic lineage occurs all around the Indian Ocean. The Euro-American lineage is clearly the most frequent lineage in Europe and the Americas, but specific sublineages within the Euro-American lineage predominate also in different regions of Africa and the Middle East (Fig. 1). Although we did observe such geographical substructuring within the Euro-American lineage, no other sublineage was associated with any specific geographical area (results not shown).
Although most other areas were associated with only one or two lineages, all six main lineages were represented in Africa (Fig. 1b). These lineages included the two West-African lineages that did not occur elsewhere, as well as the Indo-Oceanic lineage, the most ancestral of the six lineages, which was associated with East Africa. A recent study suggests that ancestral mycobacteria may have already affected early hominids in East Africa around 3 million years ago (18). Taken together, these findings are consistent with a scenario for the origin and evolution of human tuberculosis in which M. tuberculosis expanded and diversified during its spread out of East Africa. This speculative scenario suggests that M. tuberculosis might be significantly older than previously estimated (16). As a consequence, different M. tuberculosis lineages may have adapted to different human host populations.
Taking advantage of the cosmopolitan setting of San Francisco, with its diverse tuberculosis patient and bacterial populations, we investigated the effects of host–pathogen mixing on the occurrence of secondary cases of tuberculosis. We used multiplex real-time PCR to screen for the main lineages of M. tuberculosis in a stratified random sample of 1,321 isolates, corresponding to 71% of all tuberculosis cases reported in San Francisco between 1991 and 2001 who were born in the United States (U.S.), China, The Philippines, Vietnam, or Central America. These patients represent the five largest tuberculosis patient populations in San Francisco. This sample included all of the restriction fragment length polymorphism (RFLP) clustered cases belonging to any of these five populations as well as a random sample of unique cases. The clustered cases were considered part of chains of relatively recent tuberculosis transmission in San Francisco, and the unique cases were considered to have developed tuberculosis as a consequence of reactivation of latent infection (19).
Our results showed that 99.6% of all isolates in San Francisco belonged to three of the six main lineages. Twenty-six percent of the 1,321 isolates belonged to the Indo-Oceanic lineage, 26% to the East-Asian lineage, and 48% to the Euro-American lineage. When we stratified the bacterial lineage data by the five patient populations, a strong association was evident (Fig. 2a; Pearson χ28 = 1295, P < 0.0001). In four of the five patient populations, one specific lineage accounted for at least 72% of all tuberculosis cases.
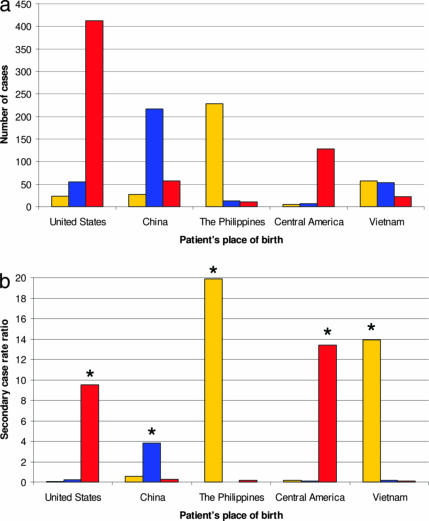
Fig. 2.
Lineage-specific prevalence and transmission of M. tuberculosis in San Francisco (1991 to 2001). (a) Prevalent strains in San Francisco are strongly associated with sympatric patient populations. The sums of both unique and clustered cases are shown. (b) The propensity of specific mycobacterial lineages to transmit is significantly higher in sympatric compared with allopatric patient populations. ∗, comparison with the other two lineages combined, P < 0.0001; yellow, Indo-Oceanic lineage; blue, East-Asian lineage; red, Euro-American lineage.
We explored whether the association between lineage of M. tuberculosis and human population reflects host-specific differential transmission of mycobacterial lineages using RFLP clustering as a proxy for transmission (20). We hypothesized that lineages that are rare in a specific human population are not adapted to transmit and cause secondary cases in this specific human population. We first calculated the secondary case-rate ratios of the three M. tuberculosis lineages irrespective of the patient’s place of birth. All three lineages had statistically different secondary case rate ratios (Table 7, which is published as supporting information on the PNAS web site). In San Francisco, patients infected with the Euro-American lineage were three times more likely to generate a secondary case during the 11-year study period than patients infected with any other strain. The Indo-Oceanic lineage had a significantly lower secondary case-rate ratio and the East-Asian lineage the lowest. When we calculated the lineage-specific secondary case-rate ratios stratified by human population, we found that, in every instance, the secondary case-rate ratios of sympatric lineages were significantly greater in comparison with that of allopatric lineages in the same population (Fig. 2b and Table 7). Taken together, these observations suggest that particular lineages of M. tuberculosis might be adapted to specific human populations and maladapted to others.
Given that some tuberculosis cases were caused by allopatric lineages, we investigated the characteristics of patients with disease caused by allopatric lineages. We chose to look at the U.S.-born population because it represents the largest patient population in San Francisco (Fig. 2a) and because sociological determinants of transmission are well documented. The characteristics of the U.S.-born patients, stratified by the three lineages of M. tuberculosis, are presented in Table 8, which is published as supporting information on the PNAS web site. Significant variables were selected for multivariate logistic regression modeling. The multivariate analysis revealed that U.S.-born patients of self-defined Chinese and Filipino ethnicity tend to harbor the same strains as patients born in China and the Philippines, respectively (Table 2). Because self-defined ethnicity is a good predictor of human genetic ancestry (21), these findings provide significant additional support for the importance of this host–pathogen association.
Table 2.
Risk factors independently associated with one of three M. tuberculosis lineages in 490 U.S.-born patients from San Francisco
| M. tuberculosis lineage | Risk factor | Adjusted odds ratio | (95% CI) | P value |
|---|---|---|---|---|
| East-Asian | Chinese ethnicity | 19.8 | (4.6–84.2) | <0.001 |
| Homelessness | 3.0 | (1.4–6.2) | 0.004 | |
| Indo-Oceanic | Filipino ethnicity | 43.2 | (5.6–335) | <0.001 |
| Age ≥45 years | 3.9 | (1.5–10.1) | 0.004 | |
| HIV positive | 3.4 | (1.3–8.7) | 0.01 | |
| Euro-American | Chinese ethnicity | 0.18 | (0.06–0.6) | 0.004 |
CI, confidence interval.
Another study has reported similar associations between M. tuberculosis strain families and human populations (17). Such host–pathogen associations, although indicative, do not by themselves provide proof that specific host–pathogen adaptations occur. They can also be explained by sociological and epidemiological factors (6). For example, social mixing is nonrandom among ethnic groups in San Francisco, which certainly impacts transmission of M. tuberculosis between U.S.- and foreign-born individuals in the short term (22, 23). However, some foreign strains, for example those associated with Chinese immigrants, must have been introduced into San Francisco repeatedly since the beginning of the Gold Rush in the 1800s (http://en.wikipedia.org/wiki/Chinatown/%2C_San_Francisco). We propose that over such large time frames, there has been ample opportunity for the spread of foreign strains into the U.S.-born population.
We cannot exclude the possibility that social factors contribute or even drive our observation of lineage-specific association with particular human populations. However, further results from our multivariate analysis support biological causality for the observed host–pathogen association. U.S.-born tuberculosis patients of non-Chinese and non-Filipino ethnicity infected with allopatric strains (i.e., belonging to the Indo-Oceanic or East-Asian lineages) were more likely to be HIV-positive or homeless (Table 2). This finding suggests that, although these lineages are less adapted to transmit and cause disease in fully competent members of allopatric human populations, they can do so in the context of impaired host resistance. Such differences in host–pathogen compatibility or local adaptation have been associated with host-specific pathogen adaptation and have been demonstrated in several invertebrate host–pathogen model systems (6).
Conclusions
Overall, our findings demonstrate a global genetic population structure for M. tuberculosis and support the notion that this pathogen has adapted to specific human populations. These results have implications for tuberculosis control efforts, especially for the development of new vaccines. The importance of strain genetic variation for vaccine escape has been documented in several bacterial species (24–26). In bacillus Calmette-Guérin (BCG), the currently available tuberculosis vaccine, significant geographical variation in protective efficacy has been observed (8). Environmental factors and differences in vaccine strain have been invoked (8, 27–29), but our findings suggest that regional differences in host–pathogen interactions could be partially responsible. Although recent progress has been made in the development of new tuberculosis vaccines (30), the global population structure of M. tuberculosis and host-specific pathogen adaptation may need to be considered when engineering and evaluating new vaccine candidates.
Materials and Methods
Molecular Epidemiology in San Francisco.
In an ongoing population-based molecular epidemiological study in San Francisco, CA (19), 2,807 tuberculosis patients were enrolled between January 1991 and December 2001. Of these patients, 2,382 (84.9%) had M. tuberculosis isolated in culture. Demographic and epidemiological data, including place of birth and self-defined ethnicity, were recorded for each patient, and IS6110 RFLP genotyping was performed on 2,141 (89.9%) of the bacterial isolates following standardized methods (19). Isolates with fewer than six IS6110copies were further genotyped by polymorphic GC-rich sequence (PGRS) RFLP (9). Isolates with matching (clustered) RFLP patterns were considered part of a chain of relatively recent tuberculosis transmission. The protocols and the procedures for the protection of human subjects were approved by Stanford University and the University of California, San Francisco.
Global Sample of M. tuberculosis.
Fifty of the strains included in the global sample had unique RFLP patterns and were isolated from U.S.- and foreign-born patients from San Francisco. We previously reported that these patients represented cases of reactivation of infections acquired in their respective country of origin and that the genomic deletion profiles of these strains were associated with the respective patient’s place of birth (10). Therefore, the unique foreign-born cases from San Francisco could be used to sample the diversity of M. tuberculosis. We validated this approach in 108 reference strains obtained from several additional strain collections representative of specific geographic areas (Table 3). These reference strains were selected because they represented the most common genotypes in the corresponding geographic areas based on our previous molecular epidemiological studies (9, 31) (B.C.d.J., S. Narayanan, M.N., S. Niemann, and M.H., unpublished results). We then screened an additional 709 unique strains isolated from U.S.- and foreign-born patients from San Francisco. Eight strains from different patient clusters comprising only U.S.-born individuals were also included.
Identification of Large Sequence Polymorphisms.
Comparative whole-genome hybridization was performed by using an Affymetrix (Santa Clara, CA) DNA chip, following procedures described previously (11). Genomic regions putatively deleted in the test strains compared with the sequenced reference strain H37Rv were identified by using delscan software (AbaSci, San Pablo, CA). Putative deletions were confirmed by PCR and sequencing (11).
Lineage Determination by PCR and Mutliplex Real-Time PCR.
We used the phylogenetically informative LSPs to screen by PCR and/or TaqMan multiplex real-time PCR (Applied Biosystems) in the unique strains and one isolate representative of each of the 184 (97.7% of all) patient clusters that occurred in San Francisco between 1991 and 2001. The screening results from the clustered isolates were extrapolated to the remaining isolates of the respective clusters. The primer and probe sequences used in this study are shown in Tables 4 and 5. The Euro-American lineage was defined based on a characteristic 7-bp deletion in pks15/1(ref. 15) or the ctg to cgg substitution at codon 463 of katG(ref. 16), which are known to be equivalent markers (15, 17).
Lineage-Specific Transmission in San Francisco.
Of 2,141 patients with available RFLP data, 1,849 (86.4%) were born in the U.S., China, the Philippines, Central America including Mexico, and Vietnam. This set of 1,849 patients represented our sampling frame. We classified these patients as follows: all clustered patients, all cases with drug resistance, all patients born in Vietnam or in Central America for which DNA was available, and a random selection of strains with unique RFLP patterns recovered from patients born in the U.S., China, or the Philippines (the three largest patient populations in San Francisco). Overall, 71.4% of eligible patients (1,321/1,849 patients) and their isolates were included in this part of the study, comprising 66.6% (493/740) of patients born in the U.S., 67.3% (301/447) of patients born in China, 69.5% (251/361) of patients born in the Philippines, 89.7% (140/156) of patients born in Central America, and 93.8% (136/145) of patients born in Vietnam.
Statistical Analysis.
The number of secondary cases in each lineage was determined by subtracting the number of RFLP clusters from the total number of clustered cases (20). Because prevalent bacteria have a greater opportunity to transmit, we translated the number of secondary cases in each lineage into lineage-specific secondary case rates by dividing the number of secondary cases in a lineage by the sum of all index cases (the number of clusters plus all of the unique cases) belonging to the same lineage. To compare transmission rates between lineages, we then transformed the lineage-specific secondary case rates into secondary case-rate ratios by dividing the secondary case rate of the lineage of interest by the secondary case rate of the other two lineages combined. To calculate the host population-specific secondary case rate, we made the simplifying assumption that any index case in San Francisco, regardless of which host population he or she belonged to, could have infected the secondary case in question. Thus, we used as the denominator the sum of the number of clusters plus unique cases for the whole of San Francisco. A total of 188 patient clusters with 604 secondary cases occurred in San Francisco between 1991 and 2001. For the analysis, there were 184 clusters (97.9%) with their corresponding 596 (98.7% of all) secondary cases with at least one isolate with DNA available for screening. Overall, 1,349 tuberculosis cases with a unique RFLP pattern occurred during the same time period, 754 (55.9%) of which had lineage information available (including 213 cases who were not part of the five main patient populations). To account for the number of unique cases that were not screened for lineage-defining markers, we weighted the denominator of the secondary case rate by multiplying the number of unique cases in each lineage by 1.79 (1,349 total unique cases/754 screened unique cases).
To identify the risk determinants of transmission of allopatric strains in the U.S.-born population, we sought associations between the three lineages and patient characteristics using univariate analyses with a 3 × 2 χ2 test of proportions with two degrees of freedom. Variables with a P value <0.20 in the 3 × 2 comparison were further tested by individual 2 × 2 comparisons by using the regular χ2 test of proportions or Fisher’s two-tailed exact test. All variables with a P value <0.20 in the 2 × 2 univariate analysis and biological plausibility were considered for the multivariate logistic regression model. We performed forward stepwise model construction and compared the log likelihood ratios of successive models until the final, most parsimonious model was identified. We used the Hosmer-Lemeshow goodness-of-fit test to validate the final models (32). Statistical analyses were performed with stata 7e (Stata Corporation, College Station, TX).
Supplementary Material
Acknowledgments
We thank Bruce Levin and James H. Jones for their valuable comments on the manuscript, and Colette Diguimbaye (Laboratoire de Recherches Vétérinaires et Zootechniques de Farcha, N’Djamena, Chad) and Erik Post (German Leprosy and TB Relief Association, Würzburg, Germany) for providing strains. This research was supported by the Swiss National Science Foundation and the Novartis Foundation (S.G.), the National Institutes of Health (K.D., B.C.d.J., and P.C.H.), and the Wellcome Trust (P.M.S.).
Abbreviations
- RFLP
restriction fragment length polymorphism
- LSP
large sequence polymorphism.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Falush D., Wirth T., Linz B., Pritchard J. K., Stephens M., Kidd M., Blaser M. J., Graham D. Y., Vacher S., Perez-Perez G. I., et al. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 2.Musser J. M., Kroll J. S., Granoff D. M., Moxon E. R., Brodeur B. R., Campos J., Dabernat H., Frederiksen W., Hamel J., Hammond G. Rev. Infect. Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 3.Agostini H. T., Yanagihara R., Davis V., Ryschkewitsch C. F., Stoner G. L. Proc. Natl. Acad. Sci. USA. 1997;94:14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monot M., Honore N., Garnier T., Araoz R., Coppee J. Y., Lacroix C., Sow S., Spencer J. S., Truman R. W., Williams D. L., et al. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 5.Wirth T., Wang X., Linz B., Novick R. P., Lum J. K., Blaser M., Morelli G., Falush D., Achtman M. Proc. Natl. Acad. Sci. USA. 2004;101:4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolhouse M. E., Webster J. P., Domingo E., Charlesworth B., Levin B. R. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization . Global Tuberculosis Control: Surveillance, Planning, Financing. Geneva: World Health Organization; 2005. [Google Scholar]
- 8.Andersen P., Doherty T. M. Nat. Rev. Microbiol. 2005;8:656–662. doi: 10.1038/nrmicro1211. [DOI] [PubMed] [Google Scholar]
- 9.Kremer K., van Soolingen D., Frothingham R., Haas W. H., Hermans P. W., Martin C., Palittapongarnpim P., Plikaytis B. B., Riley L. W., Yakrus M. A., et al. J. Clin. Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsh A. E., Tsolaki A. G., DeRiemer K., Feldman M. W., Small P. M. Proc. Natl. Acad. Sci. USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsolaki A. G., Hirsh A. E., DeRiemer K., Enciso J. A., Wong M. Z., Hannan M., Goguet de la Salmoniere Y. O., Aman K., Kato-Maeda M., Small P. M. Proc. Natl. Acad. Sci. USA. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostowy S., Onipede A., Gagneux S., Niemann S., Kremer K., Desmond E. P., Kato-Maeda M., Behr M. J. Clin. Microbiol. 2004;42:3594–3599. doi: 10.1128/JCM.42.8.3594-3599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsolaki A. G., Gagneux S., Pym A. S., Goguet de la Salmoniere Y. O., Kreiswirth B. N., Van Soolingen D., Small P. M. J. Clin. Microbiol. 2005;43:3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brosch R., Gordon S. V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., et al. Proc. Natl. Acad. Sci. USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmiesse M., Brodin P., Buchrieser C., Gutierrez C., Simoes N., Vincent V., Glaser P., Cole S. T., Brosch R. Microbiology. 2004;150:483–496. doi: 10.1099/mic.0.26662-0. [DOI] [PubMed] [Google Scholar]
- 16.Sreevatsan S., Pan X., Stockbauer K. E., Connell N. D., Kreiswirth B. N., Whittam T. S., Musser J. M. Proc. Natl. Acad. Sci. USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker L., Brown T., Maiden M. C., Drobniewski F. Emerg. Infect. Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez C., Brisse S., Brosch R., Fabre M., Omais B., Marmiesse M., Supply P., Vincent V. PLoS Pathogens. 2005;1:1–7. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasmer R. M., Hahn J. A., Small P. M., Daley C. L., Behr M. A., Moss A. R., Creasman J. M., Schecter G. F., Paz E. A., Hopewell P. C. Ann. Intern. Med. 1999;130:971–978. doi: 10.7326/0003-4819-130-12-199906150-00004. [DOI] [PubMed] [Google Scholar]
- 20.Burgos M., DeRiemer K., Small P. M., Hopewell P. C., Daley C. L. J. Infect. Dis. 2003;188:1878–1884. doi: 10.1086/379895. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg N. A., Pritchard J. K., Weber J. L., Cann H. M., Kidd K. K., Zhivotovsky L. A., Feldman M. W. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 22.Chin D. P., DeRiemer K., Small P. M., de Leon A. P., Steinhart R., Schecter G. F., Daley C. L., Moss A. R., Paz E. A., Jasmer R. M., et al. Am. J. Respir. Crit. Care Med. 1998;158:1797–1803. doi: 10.1164/ajrccm.158.6.9804029. [DOI] [PubMed] [Google Scholar]
- 23.Jasmer R. M., Ponce de Leon A., Hopewell P. C., Alarcon R. G., Moss A. R., Paz E. A., Schecter G. F., Small P. M. Int. J. Tuberc. Lung Dis. 1997;1:536–541. [PubMed] [Google Scholar]
- 24.Gagneux S. P., Hodgson A., Smith T. A., Wirth T., Ehrhard I., Morelli G., Genton B., Binka F. N., Achtman M., Pluschke G. J. Infect. Dis. 2002;185:618–626. doi: 10.1086/339010. [DOI] [PubMed] [Google Scholar]
- 25.Van Loo I. H., Mooi F. R. Microbiology. 2002;148:2011–2018. doi: 10.1099/00221287-148-7-2011. [DOI] [PubMed] [Google Scholar]
- 26.Mbelle N., Huebner R. E., Wasas A. D., Kimura A., Chang I., Klugman K. P. J. Infect. Dis. 1999;180:1171–1176. doi: 10.1086/315009. [DOI] [PubMed] [Google Scholar]
- 27.Behr M. A., Wilson M. A., Gill W. P., Salamon H., Schoolnik G. K., Rane S., Small P. M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 28.Rook G. A., Dheda K., Zumla A. Nat. Rev. Immunol. 2005;5:661–667. doi: 10.1038/nri1666. [DOI] [PubMed] [Google Scholar]
- 29.Fine P. E. Lancet. 1995;346:1339–1345. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 30.McShane H., Pathan A. A., Sander C. R., Keating S. M., Gilbert S. C., Huygen K., Fletcher H. A., Hill A. V. Nat. Med. 2004;10:1240–1244. doi: 10.1038/nm1128. [DOI] [PubMed] [Google Scholar]
- 31.Niobe-Eyangoh S. N., Kuaban C., Sorlin P., Cunin P., Thonnon J., Sola C., Rastogi N., Vincent V., Gutierrez M. C. J. Clin. Microbiol. 2003;41:2547–2553. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selvin S. Monographs in Epidemiology and Biostatistics. Vol. 28. New York: Oxford Univ. Press; 1998. pp. 385–391. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.