Abstract
Numerous proteins that function in DNA metabolic pathways are known to interact with the proliferating cell nuclear antigen (PCNA). The important function of PCNA in stimulating various cellular activities requires its topological linkage with DNA. Loading of the circular PCNA onto duplex DNA requires the activity of a clamp-loader [replication factor C (RFC)] complex and the energy derived from ATP hydrolysis. The mechanistic and structural details regarding PCNA loading by the RFC complex are still developing. In particular, the positive identification of a long-hypothesized structure of an open clamp–RFC complex as an intermediate in loading has remained elusive. In this study, we capture an open yeast PCNA clamp in a complex with RFC through fluorescence energy transfer experiments. We also follow the topological transitions of PCNA in the various steps of the clamp-loading pathway through both steady-state and stopped-flow fluorescence studies. We find that ATP effectively drives the clamp-loading process to completion with the formation of the closed PCNA bound to DNA, whereas ATPγS cannot. The information derived from this work complements that obtained from previous structural and mechanistic studies and provides a more complete picture of a eukaryotic clamp-loading pathway using yeast as a paradigm.
Keywords: clamp loader, DNA replication
The replisome is a dynamic molecular complex responsible for the duplication of a genomic DNA template at the replication fork. The central components of the replisome are the replicative DNA polymerases that copy both leading- and lagging-strand DNA (reviewed in ref. 1). Processivity of DNA replication is accomplished through a clamp protein that forms a holoenzyme complex with the replicative DNA polymerase (2). The clamp proteins identified in different organisms show a common toroidal architecture with a central channel large enough to encircle duplex DNA (3). Once loaded onto duplex DNA, clamp proteins can freely slide in a 2D space. The eukaryotic clamp proliferating cell nuclear antigen (PCNA) forms a holoenzyme complex with DNA polymerases δ and ε through specific interaction(s) between the polymerase and the PCNA ring (4–6).
The task of loading PCNA onto DNA at the primer-template junction is fulfilled by replication factor C (RFC) in an ATP-driven process. Eukaryotic RFC is a heteropentameric protein complex consisting of Rfc1, -2, -3, -4, and -5 subunits. Each Rfc subunit belongs to the AAA+ protein family that contains characteristic ATP-binding/hydrolysis motifs and converts the chemical energy derived from ATP binding and hydrolysis to mechanic force that is required for clamp loading (7). The RFC complex shares partial similarities to its counterpart in lower organisms like bacteriophage T4 and Escherichia coli. However, the small subunits of eukaryotic RFC are more diverged compared with those in prokaryote and archaeon, which hints for a higher complexity in both structural and functional aspects. Indeed, the yeast RFC showed an interesting departure from its prokaryotic counterparts in terms of a step-wise binding of ATP molecules modulated by its interaction with either DNA or PCNA (8). Therefore, a substantial difference is expected in the mechanistic details for clamp loading as compared with its prokaryotic counterpart. Furthermore, additional RFC-like complexes have been identified that share the Rfc2–5 subunits with RFC but carry out specialized functions in the DNA damage checkpoint and in sister chromatid cohesion (reviewed in ref. 9).
Although the clamp loaders from the three model systems, including yeast, E. coli, and bacteriophage T4, all use ATP for loading the clamp onto DNA, the timing and amount of ATP binding and of its hydrolysis differ. For the E. coli system, ATP binding is sufficient for opening the β clamp (10), and subsequent hydrolysis of two or three molecules of ATP is required to dissociate the γ complex from the loaded clamp (11). In bacteriophage T4, ATP hydrolysis by the clamp loader (gp44/62) is needed to adjust the clamp interface before interaction with DNA (12). Eukaryotic clamp loading has not been as thoroughly studied as bacteriophage T4 and E. coli. The current model proposes that ATP binding, not hydrolysis, is required to open up the PCNA interface (9).
An elegant study carried out by Bowman et al. (13) provided us the structure of the Saccharomyces cerevisiae PCNA–RFC complex. By introducing mutations into the “arginine finger” residues of RFC complex that promote ATP hydrolysis, the crystal structure of S. cerevisiae RFC bound to PCNA was solved in the presence of ATPγS. A surprising observation from the high-resolution crystal structure showed a closed PCNA ring in complex with the RFC–ATPγS complex. However, previous biochemical studies had suggested that ATPγS can substitute for ATP in forming the hypothesized ring-opened PCNA–RFC complex. Therefore, it is not certain whether the introduction of the arginine mutations in RFC resulted in a complex incapable of opening PCNA, or whether ATP binding alone, i.e., with ATPγS as a surrogate cofactor, is incapable of opening the PCNA ring. If so, previous biochemical studies would need to be reevaluated.
To address the structural features of the RFC–PCNA complex in solution, we constructed PCNA mutants that allowed us to introduce a FRET pair across the clamp interface and to follow the PCNA topology changes along different steps of clamp loading. Our results clearly implicate a structure of an open PCNA ring bound to RFC induced upon nucleotide binding. We showed that ATPγS can substitute for ATP in forming the aforementioned complex. Only ATP, however, can complete the clamp-loading process through its hydrolytic cleavage that fosters closure of the PCNA ring onto DNA and the dissociation of RFC to form the PCNA–DNA complex. Presteady-state kinetic studies monitored by stopped-flow FRET provided rate constants of the individual steps for the loading of PCNA onto DNA by RFC. Based on this information and modeling, we propose that an in-plane opening of the PCNA ring is required for loading PCNA onto duplex DNA, whereas an out-of-plane opened PCNA may represent an intermediate in the closing of clamp onto duplex DNA.
Results
Construction of a PCNA with a FRET Pair Across the Subunit Interface.
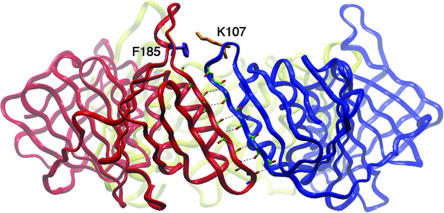
Based on the crystal structure, three identical subunit interfaces are formed in the homotrimeric PCNA ring (5). To develop a FRET pair that would monitor ring opening and closing, we introduced mutations close to, but not at, the interfacial β strand and on the “back” side of PCNA (Fig. 1). These modifications were not anticipated to affect either subunit–subunit interactions or the interaction of PCNA with RFC. First, a unique Trp residue as a FRET donor was introduced by a F185W mutation. Second, across the subunit interface, we chose K107 as the site for introducing a cysteine residue for covalent attachment of the cysteine reactive dye N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine (1,5-IAEDANS). The distance between the backbone carbon atoms of F185 and K107 is 10 Å, based on the crystal structure of yeast PCNA. Finally, to avoid ambiguity in labeling, the four native cysteines (C22/C30/C62/C81) were mutated to serines, ultimately providing the F185W/K107C/C22S/C30S/C62S/C81S mutant protein. We also constructed a second PCNA mutant K107C/C22S/C30S/C62S/C81S as a control protein lacking the FRET donor. These PCNAs will be referred to as PCNA-WC and -C, respectively.
Fig. 1.
The subunit interface of PCNA. The residues located at the interfacial antiparallel β-strands are highlighted. The residues (Phe-185 and Lys-107) selected for introducing the FRET pair are shown as purple and gold, respectively.
Both mutant PCNAs showed identical chromatographic behavior as wild-type protein, suggesting that the mutations had no deleterious effect upon the integrity of the PCNA structure. They were specifically labeled with a 5-[2(acetyl)aminoethyl]aminonaphthalene-1-sulfonate (AEDANS) fluorophore by reacting the unique Cys-107 thio group with N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine (1,5-IAEDANS). To ensure that the labeled PCNA mutants retained normal functions, the activities of AEDANS-labeled and unlabeled PCNA mutants were assayed by determining their ability to stimulate the ATPase activity of yeast RFC. Wild-type yeast RFC (250 nM) alone possesses minimal ATPase activity (basal rate of 2.6 nM s−1), and this ATPase activity is synergistically stimulated to a higher level of 417 nM s−1 by the presence of both DNA and PCNA (see Table 2, which is published as supporting information on the PNAS web site). Importantly, both the unlabeled and AEDANS-labeled PCNA mutants retain full activity in stimulating the RFC ATPase, as compared with wild-type PCNA (see Table 2). These results indicate that neither the mutations nor the site-specific labeling of PCNA with AEDANS disrupted the interactions between PCNA and RFC/DNA.
Determination of the Subunit Interface Distance of S. cerevisiae PCNA Through FRET.
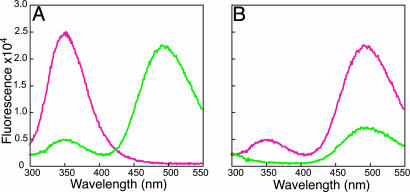
The presence of a FRET donor (W185) and acceptor (C107-AEDANS) pair allowed us to directly measure the distance across the PCNA subunit interface. Steady-state fluorescence spectroscopy comparing the fluorescence of PCNA-WC and -WCad (AEDANS-labeled PCNA-WC) (Fig. 2A) showed that attachment of AEDANS to the C107 position had a large quenching effect on the fluorescence of Trp-185. A concomitant increase in fluorescence centered at 495 nm was also observed upon exciting the W185 at 290 nm, which is a strong indication of fluorescence energy transfer between these two fluorophores. We further confirmed that FRET indeed occurred between donor W185 and C107-AEDANS by determining the fluorescence of PCNA-WCad and -Cad upon excitation at 290 nm (Fig. 2B). PCNA-WCad shows a much stronger fluorescence signal at 495 nm compared with that of PCNA-Cad (the latter is because of moderate direct excitation of AEDANS at 290 nm).
Fig. 2.
Fluorescence spectra of PCNA-WC (magenta) and PCNA-WCad (green) (A) and PCNA-WCad (magenta) and PCNA-Cad (green) (B) with 290 nm excitation.
To calculate the exact distance between W185 and C107-AEDANS, the Förster distance was determined for the Trp–AEDANS FRET pair by using the experimental parameters obtained from steady-state measurements [QD, κ2, FD(λ), εA(λ)] (see Supporting Text). Our calculation of the Förster distance (R0) of 22 Å is consistent with the reported value of 18∼22 Å for the Trp-AEDANS FRET pair (14). The sensitization of AEDANS was used to calculate the FRET energy transfer efficiency (ET) by using Eq. 1 (Supporting Text), and ET was determined to be 0.95. Given the measured values of R0 and ET, the distance between W185 and C107-AEDANS was calculated at 13 Å, according to Eq. 2 (Supporting Text). This FRET-based distance is close to the distance of 10 Å between the backbone carbon atoms of residues 185 and 107 obtained from the crystal structure of trimeric yeast PCNA. It should be noted that, in calculating the ET and spatial distance between the two fluorophores, we assumed that the three subunit interfaces in the closed trimeric PCNA ring are identical. This assumption is validated by both the crystal structure of trimeric yeast PCNA and hydrodynamic studies showing that yeast PCNA exists as a symmetric molecule in solution (see Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site).
The PCNA Subunit Interface Distance Changes Along the Clamp-Loading Pathway.
To accurately interpret FRET signals between W185 and C107-AEDANS across the PCNA subunit interface as a probe for PCNA opening and closing by RFC during the various stages of loading, possible contributions as fluorescence donors by the tryptophan residues in RFC need to be accounted for. Therefore, we carried out parallel experiments with both PCNA-WCad and PCNA-Cad that lack W185, so we could correct for the RFC contribution to the overall signal by a method described before for the T4 clamp (15). The calculated ET between the FRET pair across the subunit interfaces is an averaged value among all three subunit interfaces. To derive distance information for the opened interface during clamp loading, we made the assumption that only one subunit interface is subjected to large conformation changes that result in either an increase or decrease in the interface distances, whereas the remaining two subunit interfaces stay intact (see Discussion).
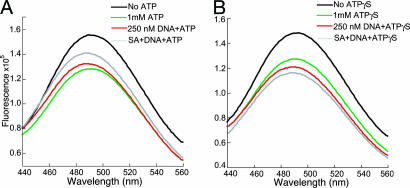
We studied the loading of PCNA onto a model DNA substrate in two steps: (i) binding of labeled PCNA to RFC in the presence of ATP or ATPγS, an analog that is not hydrolyzed by RFC (8); and (ii) interaction of the preformed RFC–PCNA complex with DNA. The DNA substrate is a small forked DNA with a biotin tag at the 5′ end of the template strand, as described before (16). Upon binding of streptavidin to the biotin tag, the loaded PCNA can be effectively retained on DNA. The steady-state fluorescence spectra of labeled PCNA at the different steps of PCNA loading are depicted in Fig. 3. The addition of RFC to PCNA-WCad results in a small fluorescence increase around 495 nm upon excitation at 290 nm because of background fluorescence from excitation of RFC Trps. Once we subtract this background from the total fluorescence, the resulting fluorescence shows little change compared with that of the labeled PCNA alone, which indicates that, in the absence of ATP, the addition of RFC has no effect upon the structure and/or conformation of the PCNA ring. The subsequent addition of ATP to the above solution containing PCNA-WCad and RFC caused a marked decrease in AEDANS fluorescence (Fig. 3A). This decrease in FRET suggested ring opening of PCNA within a RFC–PCNA complex upon ATP binding. Addition of the forked DNA substrate to this ATP–RFC–PCNA complex initiated the clamp-loading reaction. Clearly, the decrease in FRET signal was attenuated by DNA addition (Fig. 3A), suggesting a closure of the PCNA ring around DNA. However, the level of AEDANS fluorescence increase is low compared with the actual fluorescence decrease initially observed upon ATP addition.
Fig. 3.
Steady-state fluorescence spectra of RFC catalyzed PCNA-WCad loading in the presence of ATP (A) and ATPγS (B).
One explanation for this behavior is that the loaded PCNA ring can slide off the duplex DNA from its open end, bind to RFC, and be reopened. As a result, PCNA exists in an equilibrium between the open (in the ATP–RFC–PCNA complex) and the closed forms (in the PCNA–DNA complex), and this equilibrium is maintained through ATP turnover by RFC. By adding a stoichiometric amount of streptavidin to the loading reaction, we blocked the 5′ end of the duplex DNA through streptavidin binding to the biotin tag, thereby inhibiting PCNA sliding off. Indeed, a further increase in AEDANS fluorescence signal was observed, consistent with closing of the PCNA ring around duplex DNA (Fig. 3A).
To obtain quantitative distance information of the PCNA interface at the various steps of PCNA loading, we analyzed the fluorescence ET between Trp-185 and Cys-107-AEDANS, as described before (15). The basic assumptions in this calculation are that (i) two of the three PCNA subunit interfaces remain largely unchanged, with a calculated energy transfer efficiency (EC) of 0.95; and (ii) the total ET is related to the energy transfer efficiency of the open (EO) and closed (EC) subunit interfaces following the equation EO = 3ET − 2EC (15). Then, variation in the distance reported by the FRET pair for the open PCNA interface can be calculated by using Eq. 2 (Supporting Text). These distances for various steps of PCNA loading catalyzed by RFC are listed in Table 1.
Table 1.
Steady-state fluorescence data during PCNA loading
| Loading steps | ATP as cofactor |
ATPγS as cofactor |
||||
|---|---|---|---|---|---|---|
| ET* | EO† | R0, Ň | ET | EO | R0, Å | |
| PCNA | 0.952 ± 0.003 | 0.95 | 13 | 0.952 ± 0.003 | 0.95 | 13 |
| PCNA, RFC, nucleotide | 0.658 ± 0.008 | 0.07 | 34 | 0.71 ± 0.03 | 0.23 | 27 |
| PCNA, RFC, nucleotide, DNA | 0.82 ± 0.01 | 0.56 | 21 | 0.72 ± 0.02 | 0.26 | 26 |
| PCNA, RFC, nucleotide, DNA, SA§ | 0.91 ± 0.01 | 0.83 | 17 | 0.71 ± 0.01 | 0.23 | 27 |
*The averaged energy transfer efficiency of all three subunit interfaces.
†The energy transfer efficiency of open interface.
‡The calculated distance of the open subunit interface.
§SA, streptavidin.
Several important conclusions can be drawn from the distance information. First, an open form of PCNA exists only in the ATP–RFC–PCNA complex. Previous biochemical studies have indicated that, whereas nucleotide-free RFC binds PCNA with moderate affinity (KD ∼ 20 nM), addition of ATP causes a strong increase in affinity, KD ∼ 1 nM) (17). In our current experiments, >80% of the PCNA is in a PCNA–RFC complex, even without ATP present. Therefore, our FRET data show that binding of PCNA to RFC does not suffice for ring opening, because, before ATP addition, PCNA stays closed in the presence of RFC. Rather, ATP is required for opening the PCNA interface by RFC. The formation of a ring-opened PCNA–RFC–ATP complex had previously been hypothesized (8) and here, through solution FRET studies, we provide direct evidence for such a ternary complex. The PCNA exists as an opened ring with a measured distance of 34 Å across the opened subunit interface, which represents an ≈21 Å gain, as compared with the interface distance found in a closed ring.
Second, the ring-opened PCNA–RFC–ATP intermediate complex is capable of loading and closing the PCNA ring onto DNA with a primer-template end. Upon addition of DNA blocked at both ends by a DNA flap and streptavidin bound to a biotin tag, a significant decrease of the open subunit distance from 34 to 17 Å was observed. The observed distance of 17 Å is close to that of the free PCNA ring in solution. Third, the loaded PCNA ring can slide off duplex DNA. A marked increase of averaged ET was observed when the open end of duplex DNA was blocked by binding of streptavidin to biotin tag (Table 1). This agrees with previous studies showing PCNA can freely slide on duplex DNA (18, 19).
From previous biochemical studies, it was proposed that ATP binding to yeast RFC powers the opening of a PCNA ring, whereas ATP hydrolysis is not required in this step. However, direct evidence has been lacking. In this study, we are able to address this intriguing question by using the nonhydrolyzable analog ATPγS in substitution for ATP and follow the PCNA subunit interface distance changes. The experiments were carried out in the same way as when ATP was used as cofactor. When labeled PCNA was mixed with yeast RFC in the absence of nucleotide, the AEDANS fluorescence stays largely unchanged. A subsequent addition of 1 mM ATPγS clearly decreased the AEDANS fluorescence (Fig. 3B), similar to the observation when 1 mM ATP was used (see above). We attributed the changes of AEDANS fluorescence to the opening of PCNA ring, hence the decrease in fluorescence ET. Next, DNA substrate was added to the above solution that contained the ATPγS–RFC–PCNA complex. If the PCNA ring can be closed after loading onto DNA, an increase of AEDANS fluorescence should be expected. However, no increase of AEDANS fluorescence was observed (Fig. 3B), in sharp contrast to observation with ATP. The slight decrease in AEDANS fluorescence was due to dilution and inner filter effects. Further addition of streptavidin brought no increase of the AEDANS fluorescence (Fig. 3B). The quantitative distance information derived from these studies demonstrates that ATPγS binding to RFC can power opening of the PCNA ring; however, ATPγS prohibits the closing of the PCNA ring onto DNA (Table 1).
Our FRET studies demonstrate that the binding of either ATP or ATPγS to RFC potentiates the opening of the PCNA interface and suggest that ring closure after DNA binding requires hydrolysis of the bound ATP, because closure is not effected in the presence of ATPγS. In addition, however, the details of opening may slightly differ between ATP and ATPγS, because the PCNA subunit interface opening detected in the presence of ATP is 34 Å compared with 27 Å when ATPγS was used. One important question regarding ATP in the clamp-loading process is the timing of ATP hydrolysis. In light of the different effects of ATP and ATPγS, we propose that ATP hydrolysis occurs after DNA binding and either before or concurrent with PCNA ring closing.
Kinetics of PCNA Loading by Stopped-Flow FRET.
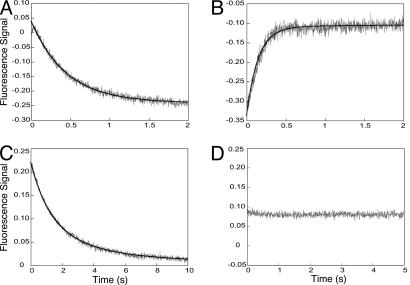
Steady-state FRET experiments provided us distance information regarding the PCNA subunit interface opening and closing. Next, the time dependence of the interactions between RFC, PCNA, and DNA in the clamp-loading process was investigated by using presteady-state kinetic approaches. When RFC and PCNA-WCad were mixed in the absence of ATP, no fluorescence signal change was observed, which was in good agreement with the steady-state FRET experiment (see above). Mixing the same protein solutions in the presence of 1 mM ATP showed a large time-dependent decrease in AEDANS fluorescence (Fig. 4A). The trace can be fitted with a single exponential equation to generate the observed rate constant of 2.23 ± 0.02 s−1. The rate is insensitive to RFC concentration, because lowering the [RFC] by 2-fold resulted in a similar fitted rate constant of 2.13 ± 0.05 s−1, suggesting a first-order kinetic nature of this process. This result agreed with the steady-state fluorescence experiment (see above) interpreted as the opening of the PCNA interface.
Fig. 4.
Stopped-flow fluorescence traces of mixing RFC versus PCNA-WCad in the presence of ATP (A), RFC-PCNA-WCad complex formed in the presence of ATP versus DNA bound with streptavidin (B), RFC versus PCNA-WCad in the presence of ATPγS (C), and RFC-PCNA-WCad complex formed in the presence of ATPγS versus DNA–streptavidin (D). The solid line represents the best fit of the trace to the exponential equations, as described in Results.
In the next step of clamp loading, the RFC–PCNA complex formed in the presence of 1 mM ATP, was mixed with streptavidin–DNA. An increase of the fluorescence signal was observed (Fig. 4B). The trace included two phases with observed rate constants of k1 = 7.2 ± 0.1 s−1 and k2 = 0.5 ± 0.1 s−1. Both rate constants were independent of the DNA and RFC–PCNA concentration (data not shown). The bimolecular association of the RFC–PCNA complex with DNA apparently is fast and was not observed on the time scale of our stopped-flow experiments. Our kinetic results suggest that the closing of the PCNA ring around DNA is not a single-step process.
The effect of ATPγS was also probed in the presteady-state kinetic experiments. Mixing RFC with PCNA-WCad in the presence of 1 mM ATPγS resulted in a fluorescence decrease (Fig. 4C), as observed when ATP was used as a cofactor. However, the ATPγS transient contains two phases compared with the single exponential with ATP. Furthermore, the fitted rate constants of the two phases (k1 = 0.88 ± 0.06 s−1, k2 = 0.28 ± 0.03 s−1) are significantly lower than the single rate with ATP (k = 2.23 s−1). We then mixed the ATPγS–RFC–PCNA complex with the streptavidin–DNA. No fluorescence signal changes were observed (Fig. 4D), in agreement with the steady-state fluorescence results, indicating that ATPγS is not effective in closing the clamp onto DNA.
Discussion
The clamp-loader proteins from every branch of life play an essential role in loading the circular clamp protein onto DNA with the concomitant expenditure of ATP. However, there exist intriguing differences in the details of how this task is fulfilled. In bacteriophage T4, ATP binding to the clamp-loader gp44/62 and subsequent ATP hydrolysis are required for opening of the cognate clamp gp45 (12). In contrast, the E. coli clamp-loader γ complex has been demonstrated to couple the binding of ATP to opening of β clamp, and no ATP hydrolysis is required (10, 11). The yeast clamp-loader RFC, as a valuable paradigm for the eukaryotic system, is of higher complexity compared with its prokaryotic counterparts, because binding of the required ATP molecules proceeds by a sequential process. Only two ATPs bind to high-affinity binding sites in free RFC. Upon binding of PCNA, a third ATP is bound, whereas a fourth essential ATP is bound upon engagement of the ATP–RFC–PCNA complex with DNA (8).
Precise structural information regarding a nucleotide-bound RFC–PCNA complex had been lacking until the report of the recent structure of the yeast RFC–PCNA complex crystallized in the presence of ATPγS (13). The pentameric RFC has ATPγS at each of its four ATPase sites and forms an inner chamber that presumably binds a duplex DNA and facilitates the DNA threading through the PCNA ring. This structure provides valuable information about the interactions between RFC and PCNA, as well as inferences about the RFC–DNA interaction. Surprisingly, the RFC-bound PCNA was found as a closed ring despite the binding of ATPγS at RFC ATPase sites and contrasts with the notion that binding of ATPγS to RFC powers the opening of the PCNA ring. Most likely, the presence of mutations in the “arginine fingers” of the ATP-binding sites of the RFC subunits prohibited PCNA ring opening, suggesting that the structure is more akin to an initial engagement complex between RFC and PCNA in the absence of ATP. Another possible explanation suggested by Bowman et al. (13) is that after initial ring opening, closure occurred without prior DNA binding or hydrolysis of bound ATPγS, thus resembling a late stage on the loading pathway. Interestingly, a very recent cryo-EM study of an archaeal ATPγS–RFC–PCNA–DNA complex did provide structural evidence for a ring-opened PCNA (20).
At present, our understanding of the eukaryotic clamp-loading process is still limited. In this study, we aimed to address the above discrepancy and some of the gaps in our knowledge by probing the dynamic conformational changes of PCNA in the clamp-loading process by using the FRET technique. By introducing a unique FRET pair across the PCNA subunit interface, we determined the subunit interface distances at different stages of clamp loading. For PCNA alone, the determined distance of 13 Å between two fluorophores (W185 and C107-AEDANS) is close to the distance (10 Å) derived from crystal structure of yeast PCNA. Thus PCNA alone exists as a closed-ring structure in solution, which was corroborated by hydrodynamic analysis (Supporting Text).
In the absence of nucleotide, we observed little fluorescence change upon mixing RFC and PCNA–AEDANS, which is in accord with the earlier binding studies showing that stable association between RFC and PCNA required either ATP or ATPγS (17). Inclusion of ATP resulted in a marked fluorescence decrease consistent with the opening of a PCNA subunit interface to a distance of 34 Å in the ATP–RFC–PCNA complex. Interestingly, whereas ATPγS binding to RFC is capable of inducing opening of PCNA, both the rates and extent of this process are affected, suggesting that ATPγS is only an approximate analog for effecting changes caused by ATP binding (Table 1). It should be noted that our calculation of the open subunit distance assumed that the remaining two PCNA interfaces stay closed without significant distance changes. A recent molecular dynamics simulation predicted possible conformational changes within a dimeric form of S. cerevisiae PCNA in the absence of constraints imposed by a closed ring structure. The simulation results revealed sizable conformational changes in the PCNA dimer structure. But at the subunit interface, the changes mainly involve small rotation of the two adjacent domains without breakage of the intermolecular β sheet (21). This molecular dynamics simulation supports our assumption that the distance across the unopened PCNA subunit does not undergo significant changes.
The open PCNA–RFC complex formed in the presence of ATP is functional in loading PCNA onto DNA. The blocking effect of streptavidin argues for the actual loading of PCNA onto DNA followed by PCNA sliding, instead of the mere dissociation of the RFC–PCNA complex. A markedly different scenario was found for the open PCNA–RFC complex formed in the presence of ATPγS, to which addition of the same DNA substrate had no effect on the PCNA interface distance. Two possibilities were considered. First, the ATPγS–RFC–PCNA complex is not able to interact with DNA. Second, although the interaction occurred, the PCNA interface remained open. We can rule out the first possibility based on a previous binding study showing that, in the presence of ATPγS, the RFC–PCNA complex strongly bound to DNA and the RFC was retained within the complex (17, 22). Thus, we propose the formation of an open PCNA–RFC(ATPγS)–DNA complex where the open PCNA trimer possibly encircles duplex DNA. Indeed, a similar archaeal complex structure was observed recently by EM (20).
The differing effects of ATP and ATPγS as cofactors suggest that ATP hydrolysis may be needed for clamp closing. The closing of PCNA ring onto duplex DNA could be either a “passive” process, driven by the electrostatic interactions between the positively charged PCNA inner ring and negatively charged DNA backbone, or an “active” process that requires the mechanic forces generated by the conformation change of RFC as the result of ATP hydrolysis. Our results favor the latter mechanism. Our present data do not address whether additional ATP hydrolysis is required for the ejection of clamp loader (as proposed for the E. coli clamp-loading system).
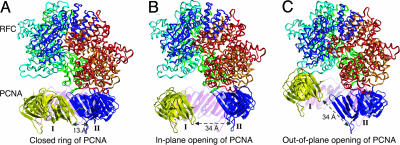
The present FRET studies allowed us to build an open PCNA–RFC complex structure based on our distance information and the coordinates of the RFC–PCNA complex determined by x-ray crystallography. We first tested an in-plane opening of the PCNA ring. The opening of the PCNA ring was achieved by in-plane rotating of the subunits I and II outwardly while keeping the intermolecular β sheet formed between subunits III and I/II intact (Fig. 5). Furthermore, the dihedral angles of backbone bonds in the interdomain loop were adjusted to give the final open PCNA structure. It should be noted that these structures do not necessarily represent an energy-minimized structure. The gap resulting from in-plane opening of PCNA ring is determined by measuring the closest distance between the two neighboring subunits. The gap distance determined in the in-plane open PCNA–RFC complex is ≈27 Å, which is within the range of the simulated distance of 20∼30 Å (21). Given a diameter of 20 Å for B form duplex DNA, we consider this gap distance sufficient to allow the passage of duplex DNA. We also explored the out-of-plane ring conformation of opened PCNA (Fig. 5). A previous study demonstrated that such an open ring conformation exists in the T4 clamp-loading pathway (23). We constructed this model by twisting the intersubunit β sheet formed between subunits and adjusting the dihedral angle of the interdomain loop to keep the distance between W185 and C107 close to 34 Å. Interestingly, with a right-handed opening of the PCNA ring, a gap of ≈5 Å can be generated. Our modeling is in accord with EM results on the archaeal RFC–PCNA–DNA complex showing an open PCNA structure with an estimated gap size of 5 Å (20). We also tested a left-handed ring opening of PCNA. Although the resulting gap size is larger, we consider the left-handed opening of the PCNA ring was less likely, because previous molecular simulations suggested this open ring conformation is energetically unfavorable (21).
Fig. 5.
Modeling of the opened PCNA–RFC structure. (A) The S. cerevisiae RFC–PCNA crystal structure. The RFC five subunits are rendered as ribbon and colored as red, gold, green, cyan, and blue, respectively. The PCNA exists as a closed trimer with three subunits colored as yellow (subunit I), blue (subunit II), and magenta (subunit III), respectively. (B) Modeled in-plane opening PCNA structure with 34-Å distance between Trp-185 and Cys-107-AEDANS. (C) Modeled out-of-plane opening PCNA structure with 34-Å distance between Trp-185 and Cys-107-AEDANS.
Our modeling studies, guided by the distance information obtained from FRET studies, showed an in-plane opening of PCNA generates a gap large enough to encircle duplex DNA. In contrast, the out-of-plane opening resulted in a gap too small to allow the passage of duplex DNA. Therefore, we consider that the in-plane opening of PCNA represents the active conformation for clamp loading. A recent EM image of archaeal RFC–PCNA–DNA complex captured an intriguing open PCNA with a spiral configuration, which encircled a duplex DNA. The distance of the gap is too small (5 Å), as noticed by Miyata et al. (20), for duplex DNA to pass through. Thus this PCNA conformation is unlikely to represent an open PCNA configuration suitable for encircling DNA. Here we propose that the archaeal PCNA configuration observed by Miyata et al. (20) represents an intermediate along the pathway for the closing of PCNA ring on DNA.
Our stopped-flow FRET studies also provided useful information regarding possible intermediate species in PCNA loading. Rapid-mixing RFC(ATP) with PCNA resulted in a single exponential fluorescence change, suggesting the opening of PCNA by RFC(ATP) most likely is limited by a single conformation change step. However, PCNA closing after mixing RFC(ATP)–PCNA with DNA suggested a more complex pathway where at least one clamp-loading intermediate exists. We propose that the intermediate may represent a partially closed PCNA ring, possibly adapting an out-of-plane conformation. Indeed, in bacteriophage T4, we have demonstrated a clamp-loading sequence that involves sequential in-plane opening of gp45 ring, an out-of-plane closing of the clamp onto DNA followed by a final in-plane closing of clamp (23).
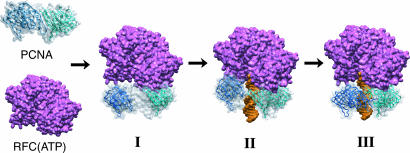
Based on the collective data, we propose a RFC-catalyzed PCNA loading process, as illustrated in Fig. 6. Once bound to RFC, an in-plane opening of PCNA ring occurs, resulting in a gap of 34 Å (Fig. 6I), which will allow the threading of DNA through an open PCNA ring and binding to the inner chamber of RFC complex. In a subsequent step an out-of-plane closing of PCNA occurs resulting in a spiral PCNA ring with a much reduced gap distance (Fig. 6II). The final step involves an in-plane closing of the PCNA ring around DNA (Fig. 6III). It should be noted that the intermediate complex, as depicted in Fig. 6III, may be transient, because the final in-plane closing of PCNA may be followed by the fast ejection of the RFC complex. Further experiments using more than one pair of FRET reporters at the PCNA interface should test the existence of an out-of-plane PCNA intermediate along the PCNA-loading pathway.
Fig. 6.
The proposed steps of RFC catalyzed PCNA loading. (I) In-plane opening PCNA complexed with RFC(ATP). (II) Out-of-plane closing of PCNA onto duplex DNA. (III) Final in-plane closing of PCNA onto DNA.
Materials and Methods
Purification of Wild-Type and Mutant S. cerevisiae PCNA.
The wild-type PCNA gene was cloned into a pET-22b vector (Invitrogen) and overexpressed in E. coli essentially as described before, except that reducing agents were omitted during the final purification step (Q Sepharose) (24). The yeast PCNA mutants were constructed by using the QuikChange protocol (Stratagene) and purified by using the same protocol as described for wild-type PCNA.
Labeling the PCNA Mutants with N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine (1,5-IAEDANS).
The mutant PCNA-WC and -C were dialyzed against 20 mM Tris (pH 7.4) with 0.2 M NaCl/10% glycerol. A 10-fold molar excess of 1,5-IAEDANS was added to the PCNA mutant protein solution at 150 μM monomer concentration. The labeling reaction was allowed to proceed for 4 h at room temperature. The excessive labeling reagent was removed by subjecting the reaction product to FPLC Superose 6 chromatography. The extent of cysteine labeling was determined by measuring the AEDANS dye concentration at 336 nm (ε = 5.43 mM−1·cm−1). The protein concentration was determined by using the Bradford assay with wild-type yeast PCNA as standard.
Steady-State Fluorescence Measurements.
Steady-state fluorescence spectroscopy was measured on an SLM-8000C spectrofluorometer (SLM Instruments, Urbana, IL). The buffer solution contained 30 mM Hepes (pH 7.5), 10 mM MgOAc, 100 mM NaCl, 0.05% ampholyte, and 1 mM DTT. The assay solution typically contains 250 nM AEDANS-labeled PCNA mutants and 250 nM RFC equilibrated at 25°C. To this solution, 1 mM ATP or ATPγS, 250 nM Bio62/34/36 DNA, and 250 nM streptavidin were sequentially added, and fluorescent spectra were taken after each addition with 290- and 340-nm excitation, respectively. The fluorescence intensity was corrected for dilution and inner filter effects by using the formula F = Fobs antilog(Aexc/2), where Aexc is the absorbance at the excitation wavelength (25). Equations for calculating ET and R are given in Supporting Text.
Stopped-Flow Fluorescence Spectroscopy.
Stopped-flow fluorescence experiments were performed on an Applied Photophysics (Surrey, U.K.) SX.18MV stopped-flow reaction analyzer in fluorescence mode at 25°C. The reaction mixture was excited at 290 nm, and the AEDANS fluorescence was detected by using a 450-nm cutoff filter. The final data trace was an average of at least five runs. The data traces were fitted to equations that describe either a single- or a double-exponential by using the software provided to obtain an observed rate constant (kobs).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants GM32431 and GM13306.
Abbreviations
- PCNA
proliferating cell nuclear antigen
- RFC
replication factor C
- AEDANS
5-[2(acetyl)aminoethyl]aminonaphthalene-1-sulfonate
- ET
energy transfer efficiency.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Garg P., Burgers P. M. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Kuriyan J., O'Donnell M. J. Mol. Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 3.Trakselis M. A., Benkovic S. J. Structure (Cambridge, U.K.) 2001;9:999–1004. doi: 10.1016/s0969-2126(01)00676-1. [DOI] [PubMed] [Google Scholar]
- 4.Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 5.Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Burgers P. M. J. Biol. Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 7.Cullmann G., Fien K., Kobayashi R., Stillman B. Mol. Cell. Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes X. V., Schmidt S. L., Burgers P. M. J. Biol. Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 9.Majka J., Burgers P. M. Prog. Nucleic Acid Res. Mol. Biol; 2004. pp. 227–260. [DOI] [PubMed] [Google Scholar]
- 10.Hingorani M. M., O'Donnell M. J. Biol. Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 11.Bertram J. G., Bloom L. B., Hingorani M. M., Beechem J. M., O'Donnell M., Goodman M. F. J. Biol. Chem. 2000;275:28413–28420. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- 12.Trakselis M. A., Berdis A. J., Benkovic S. J. J. Mol. Biol. 2003;326:435–451. doi: 10.1016/s0022-2836(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 13.Bowman G. D., O'Donnell M., Kuriyan J. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 14.van der Meer B.W., Coker G., III, Chen S.-Y. in Resonance Energy Transfer: Theory and Data. New York: VCH; 1994. [Google Scholar]
- 15.Alley S. C., Abel-Santos E., Benkovic S. J. Biochemistry. 2000;39:3076–3090. doi: 10.1021/bi992377r. [DOI] [PubMed] [Google Scholar]
- 16.Kaboord B. F., Benkovic S. J. Curr. Biol. 1995;5:149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 17.Gomes X. V., Burgers P. M. J. Biol. Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- 18.Burgers P. M., Yoder B. L. J. Biol. Chem. 1993;268:19923–19926. [PubMed] [Google Scholar]
- 19.Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyata T., Suzuki H., Oyama T., Mayanagi K., Ishino Y., Morikawa K. Proc. Natl. Acad. Sci. USA. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazmirski S. L., Zhao Y., Bowman G. D., O'Donnell M., Kuriyan J. Proc. Natl. Acad. Sci. USA. 2005;102:13801–13806. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsurimoto T., Stillman B. J. Biol. Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 23.Trakselis M. A., Alley S. C., Abel-Santos E., Benkovic S. J. Proc. Natl. Acad. Sci. USA. 2001;98:8368–8375. doi: 10.1073/pnas.111006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayyagari R., Impellizzeri K. J., Yoder B. L., Gary S. L., Burgers P. M. Mol. Cell. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakowicz J. R. Principles of Fluorescence Spectroscopy. New York: Plenum; 1983. p. 44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.