Abstract
Early-life experience has long-term consequences on behavior and stress responsivity of the adult. We previously proposed that early-life experience results in stable epigenetic programming of glucocorticoid receptor gene expression in the hippocampus. The aim of this study was to examine the global effect of early-life experience on the hippocampal transcriptome and the development of stress-mediated behaviors in the offspring and whether such effects were reversible in adulthood. Adult offspring were centrally infused with saline vehicle, the histone deacetylase inhibitor trichostatin A (TSA), or the essential amino acid l-methionine. The animals were assessed in an unfamiliar open-field arena, and the hippocampal transcriptome of each animal was evaluated by microarray analysis. Here we report that TSA and methionine treatment reversed the effect of maternal care on open-field behavior. We identified >900 genes stably regulated by maternal care. A fraction of these differences in gene expression is reversible by either the histone deacetylase inhibitor TSA or the methyl donor l-methionine. These results suggest that early-life experience has a stable and broad effect on the hippocampal transcriptome and anxiety-mediated behavior, which is potentially reversible in adulthood.
Keywords: hypothalamic–pituitary–adrenal stress response, l-methionine, maternal behavior, microarray, trichostatin A
In primates and rodents, as in nonmammalian species, there are maternal effects on defensive responses in the adult offspring (1–3). In the rat, these effects are mediated by variations in maternal care such that maternal behavior stably alters the development of behavioral and endocrine responses to stress in the offspring through tissue-specific effects on gene expression (4, 5). Thus, the adult offspring of mothers that show increased pup licking/grooming and arched-back nursing (i.e., high LG-ABN mothers) over the first week of postnatal life exhibit reduced fearfulness and more modest hypothalamic–pituitary–adrenal (HPA) responses to stress.
Such maternal effects in the rat target neural systems that tonically inhibit corticotrophin-releasing factor (CRF) synthesis and release in the hypothalamus and amygdala, which serves to activate central noradrenaline in response to stress. Increased noradrenaline, in turn, regulates HPA activity through dynamic regulation of hypothalamic CRF and behavioral responses to stress. Glucocorticoids initiate tonic negative feedback inhibition over CRF synthesis and release and thus dampen HPA responses to stress (6). Glucocorticoid negative feedback is, in part, mediated by glucocorticoid binding to glucocorticoid receptors (GR) in a number of brain regions, including the hippocampus. As adults, the offspring of high LG-ABN mothers show increased hippocampal GR expression and enhanced glucocorticoid feedback sensitivity by comparison to adult animals reared by low LG-ABN mothers (4, 5). Predictably, adult offspring of high LG-ABN mothers show decreased hypothalamic CRF expression and more modest HPA responses to stress (4). Eliminating the difference in hippocampal GR levels abolishes the effects of early-life experience on HPA responses to stress in adulthood (7), suggesting that the difference in hippocampal GR expression serves as a mechanism for the effect of early-life experience on the development of individual differences in HPA responses to stress (3).
In the rat, increased fearfulness in response to stress is also associated with decreased hippocampal neurogenesis and synaptic density (8, 9). In addition to alterations in hippocampal GR expression, enhanced maternal LG-ABN behavior over the first week of life is associated with increased hippocampal neuronal survival, synaptogenesis, and improved cognitive performance under stressful conditions (4, 10, 11). These findings suggest a rather extensive influence of maternal care on hippocampal gene expression. Importantly, cross-fostering studies provide evidence of a direct relationship between maternal care and measures of hippocampal gene expression, behavioral responses to stress, and hippocampal development. Thus, the biological offspring of low LG-ABN mothers reared by high LG-ABN dams resemble the normal offspring of high LG-ABN mothers (and vice versa) (5). These findings suggest that variations in maternal behavior can directly program rudimentary defensive responses to stress and serve as a mechanism for the nongenomic transmission of individual differences in stress reactivity across generations (3, 5, 12).
Previous studies suggest that maternal programming of individual differences in gene expression and stress responses in the rat involves modifications of epigenetic mechanisms, including DNA methylation and histone modification of a nerve growth factor-inducible protein A (NGFI-A) transcription factor binding site on a brain-specific GR promoter (13). Increased maternal LG-ABN behavior during the first week of life causes DNA demethylation, increased histone acetylation and NGFI-A binding, and increased hippocampal GR expression (14). Accordingly, the NGFI-A binding site on the hippocampal GR promoter is methylated and hypoacetylated in offspring of low LG-ABN mothers and demethylated and hyperacetylated in offspring of high LG-ABN mothers. However, central infusion of the histone deacetylase inhibitor trichostatin A (TSA) eliminated the maternal effect on histone acetylation, DNA methylation, hippocampal GR expression, and HPA responses to stress in the adult offspring of low LG-ABN dams (13). In contrast, central infusion of the adult offspring of high LG-ABN mothers with the essential amino acid l-methionine, a precursor to S-adenosyl-methionine that serves as the donor of methyl groups, resulted in increased methylation of the NGFI-A binding site on the hippocampal GR promoter, decreased GR expression, and increased HPA responses to stress (15).
These findings refer to only a single genomic target, and it is unclear whether the effects of maternal care are limited to a single gene or whether they result in a global long-term reprogramming of the adult hippocampal transcriptome. Moreover, it remained unclear whether the expression of a maternally regulated gene would be affected by the same pharmacological manipulations that reverse the effects of maternal care on GR expression. To address these questions, we used microarray analysis to examine the effects of maternal care on global gene expression within the hippocampal transcriptome of the adult offspring.
Results
The Maternal Effect on Open-Field Behavior and Its Reversal by TSA and Methionine.
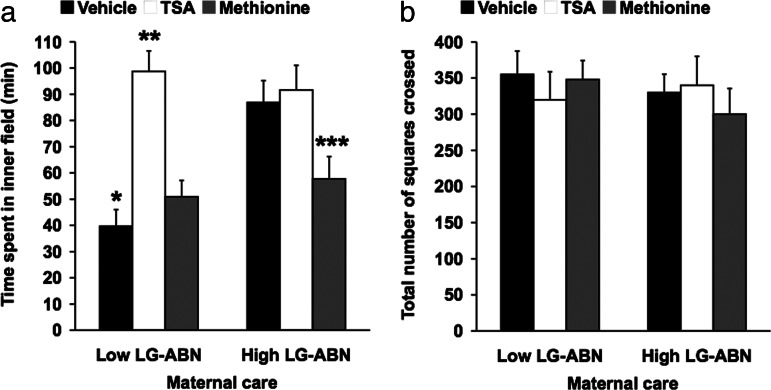
Temporal lobe-kindled rats express lower levels of hippocampal GR and are behaviorally more anxious in an open-field arena (16), suggesting that hippocampal gene expression may play a role in the development of anxiety-mediated behavior. In the rat, intracerebroventricular infusion of TSA or methionine can eliminate the maternal effect on hippocampal GR expression and endocrine and behavioral responses to stress (13). Here we examined the effects of TSA and methionine on anxiety-induced behavior in the open-field. To define the offspring of high and low LG-ABN mothers, we observed the maternal behavior in a cohort of 32 dams and devised the group mean and standard deviation for LG-ABN behavior over the first 10 days of life (17). High LG-ABN mothers were defined as females whose frequency scores for LG-ABN behavior were >1 SD above the mean. Low LG-ABN mothers were defined as females whose frequency scores for LG-ABN behavior were >1 SD below the mean. The postnatal day 90 (adult) male offspring of these high and low LG-ABN mothers were infused with saline vehicle, TSA (100 ng/ml), or methionine (100 μg/ml) once a day for 7 consecutive days, and 7 days after the final infusion the animals were tested in an unfamiliar open-field arena (Fig. 1). Previous studies have shown that the behavioral effects of the TSA and methionine infusion are sustained for at least 2 weeks after the final infusion (our unpublished data). Offspring of low LG-ABN mothers spent less time exploring the unfamiliar inner field than did offspring of high LG-ABN mothers (∗∗, P < 0.001). However, TSA treatment reversed this effect, and adult offspring of low LG-ABN mothers treated with TSA displayed behavior similar to that of offspring of high LG-ABN mothers (∗∗, P < 0.001). Conversely, methionine reversed the behavior of adult offspring of high LG-ABN mothers. Methionine-treated adult offspring of high LG-ABN mothers exhibited the behavior of offspring of low LG-ABN mothers (∗∗, P < 0.001). Group differences as a function of maternal care were observed in the length of time the animals spent exploring in the inner field, the critical measure of fear behavior (Fig. 1a), although no differences were observed in overall activity (Fig. 1b). ANOVA revealed a main effect of group (F = 13.86, P < 0.001) and treatment (F = 10.44, P < 0.01) and a group × treatment interaction effect (F = 12.96, P < 0.001). These findings suggest that anxiety-induced behavioral responses programmed early in life are potentially reversible in adult life through manipulations known to alter DNA methylation.
Fig. 1.
TSA and methionine eliminate the maternal effect on behavioral responses to novelty stress. Shown is open-field behavior of TSA-treated (100 ng/ml), methionine-treated (100 μg/ml), and vehicle-treated adult offspring of high and low LG-ABN mothers during a 10-min testing session (n = 10 animals per group). (a) Mean ± SEM time spent exploring in the inner arena. ∗, P < 0.001, vehicle-treated adult offspring of low LG-ABN mothers vs. vehicle-treated offspring of high LG-ABN dams; ∗∗, P < 0.001, TSA-treated adult offspring of low LG-ABN mothers vs. vehicle- and methionine-treated offspring of low LG-ABN dams; ∗∗∗, P < 0.001, methionine-treated adult offspring of high LG-ABN mothers vs. vehicle- and TSA-treated offspring of high LG-ABN dams. (b) Mean ± SEM activity in the arena (n = 10 animals per group).
TSA and Methionine Effects on Differential Gene Expression in the Hippocampus.
These long-term effects on behavior are predicted to result from long-term stable reprogramming of gene expression profiles. To determine the influence on the hippocampal transcriptome, the vehicle and TSA-treated offspring of low LG-ABN mothers and the vehicle- and methionine-treated offspring of high LG-ABN dams were killed, and Affymetrix microarrays were used to monitor changes in hippocampal expression of 31,099 unique mRNA transcripts. The four different treatment groups were compared with their respective control groups: (i) vehicle-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams, (ii) TSA-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams, and (iii) methionine-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of high LG-ABN dams. Transcripts with similar profiles within the same treatment group (i.e., same direction of expression) were averaged, and only transcripts with mean expression levels significantly (Student’s t test, P < 0.05) altered ≥1.5-fold between at least one group were included in subsequent analysis. In total, 935 different transcripts were present at significantly (P < 0.05) different levels among the three groups, representing 3% of the total number of transcripts analyzed, whereas 97% of the transcripts remained unaltered. Similarity in transcript expression levels among the four treatment groups was visualized by hierarchical clustering. Only those genes with a known biological function were included in the analysis. Accordingly, genes with unknown functions or classified as ESTs were excluded, leaving 100 well characterized genes for two-way hierarchal clustering (Fig. 5, which is published as supporting information on the PNAS web site). The clustering analysis shows that 16 transcripts (45.71% of the 35 maternal-responsive genes) were altered by maternal care alone, 31 transcripts (54.39% of the 57 TSA-responsive genes) were altered by TSA treatment alone, and 22 transcripts (53.66% of the 41 methionine-responsive genes) were altered by methionine treatment alone. Thus, approximately half of those genes affected within each treatment group were uniquely responsive to either maternal care, TSA, or methionine treatment. The analysis also reveals that 12 transcripts were altered by both maternal care and TSA treatment, 5 transcripts were altered by both maternal care and methionine treatment, and 12 transcripts were altered by both TSA and methionine treatment, whereas only 2 transcripts were altered by maternal care, TSA, and methionine treatment. Together these results illustrate that the effects of maternal care, TSA, and methionine treatment on the hippocampal transcriptome are gene-specific.
TSA and Methionine Effects on Gene Activation and Repression.
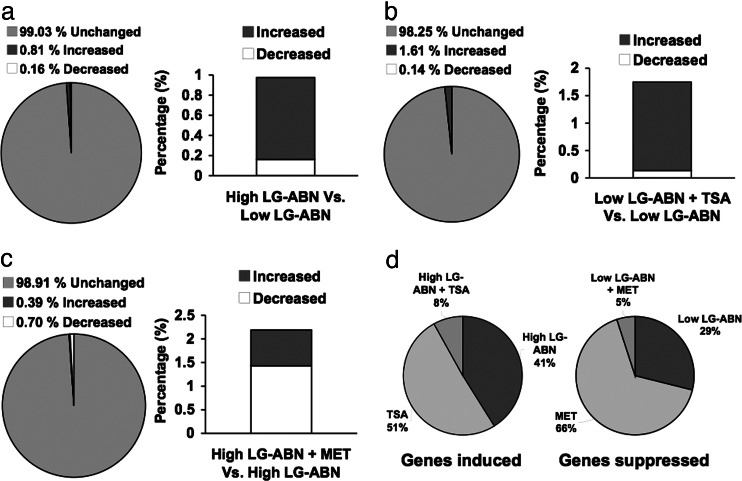
To examine the influence of maternal care, TSA treatment, and methionine on the direction of gene expression, the four different treatment groups were again compared with their respective control groups as described above. We first determined the effects of maternal care. A total of 253 transcripts (0.81%) were up-regulated and 50 transcripts (0.16%) were down-regulated in the offspring of high LG-ABN mothers in comparison to offspring of low LG-ABN dams, representing 0.97% (n = 303) of the total number of transcripts analyzed, whereas 99.03% (n = 30796) of the transcripts remained unaltered (Fig. 2a). Thus, a few hundred genes showed differences in expression in the adult offspring of high and low LG-ABN mothers. Comparison of the number of transcripts altered by TSA treatment of offspring of low LG-ABN mothers revealed that 501 transcripts (1.61%) were up-regulated and 42 transcripts (0.14%) were down-regulated by TSA, representing 1.75% (n = 543) of the total number of transcripts analyzed, whereas 98.25% (n = 30556) of the transcripts remained unaltered (Fig. 2b). Comparison of the number of transcripts altered by methionine treatment of offspring of high LG-ABN mothers revealed that 120 transcripts (0.39%) were up-regulated and 217 transcripts (0.70%) were down-regulated by methionine treatment, representing 1.08% (n = 337) of the total number of transcripts analyzed, whereas 98.92% (n = 30762) of the transcripts remained unaltered (Fig. 2c). The number of transcripts up-regulated by TSA treatment of offspring of low LG-ABN mothers (TSA-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams) was significantly (t = 42.29, P < 0.001) greater (≈2-fold) than the number of transcripts that were up-regulated by maternal care (vehicle-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams) (Fig. 2d Left). Thus, TSA has a broader impact on gene expression than maternal care early in life, which is expected because TSA is a global inhibitor of histone deacetylases. As anticipated from a manipulation that could increase DNA methylation, which is known to result in gene silencing, the number of transcripts down-regulated in methionine-treated offspring of high LG-ABN mothers (methionine-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of high LG-ABN dams) was significantly (t = 35.94, P < 0.01) greater in comparison to the number of transcripts down-regulated by low maternal care (vehicle-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of high LG-ABN dams) (Fig. 2d Right).
Fig. 2.
Direction of gene expression in hippocampal tissue from TSA-treated (100 ng/ml), methionine-treated (100 μg/ml), and vehicle-treated adult offspring of high and low LG-ABN mothers (n = 3 animals per group). (a) Percentage of mRNA transcripts increased, decreased, or unchanged by high LG-ABN. (b) Percentage of mRNA transcripts increased, decreased, or unchanged by TSA treatment. (c) Percentage of mRNA transcripts increased, decreased, or unchanged by methionine treatment. (d) Percentage of mRNA transcripts increased by high LG-ABN, TSA treatment, or both (Left) and the percentage of mRNA transcripts decreased by low LG-ABN, methionine treatment, or both (Right).
These data suggest first that maternal care during early life programs the expression of hundreds of genes in the adult offspring. Second, because the differences in gene expression are maintained well after the stimulus has gone, an epigenetic reprogramming of these genes might take place in response to maternal care to maintain their differential expression into adulthood. Third, the fact that the expression of a number of genes that were normally programmed by maternal care were induced in low LG-ABN offspring by TSA or repressed in the offspring of high LG-ABN offspring by methionine suggests that the effect of early-life experience on the hippocampal transcriptome is potentially reversible in adulthood. Fourth, our analysis reveals the general direction of these changes in gene expression. Whereas high LG-ABN mainly affects stimulation of gene expression in comparison with low LG-ABN and, as expected, TSA treatment results predominantly in activation of gene expression, methionine treatment primarily silences gene expression as expected from the general silencing effect of DNA hypermethylation.
Cellular Functions of Genes Affected by Maternal Care.
The maternal care-, TSA-, and methionine-regulated RNA transcripts are involved in several different classes of cellular function. A distinct class of regulated transcripts are derived from genes encoding protein products involved in general cellular metabolism and energy production, which include several glycolytic enzymes, ATPases, ATP synthases, and enzymes involved in lipid metabolism and mitochondrial components. A second class of molecules is comprised of factors involved in signal transduction pathways and includes membrane-bound receptors, intracellular messenger molecules, kinases, phosphatases, and transcription factors. A third main class of regulated transcripts consists of predominantly ribosomal proteins, but also nucleolar proteins, endoplasmic reticulum-localized proteins, and lysosomal membrane glycoproteins that are involved in protein synthesis, turnover, and folding as well as intracellular trafficking of proteins. A fourth functional class consists of transcripts with diverse functions in the control of neuronal development, including extracellular matrix proteins and other structural molecules that define the architecture of the synaptic connections in the brain (cytoskeletal proteins).
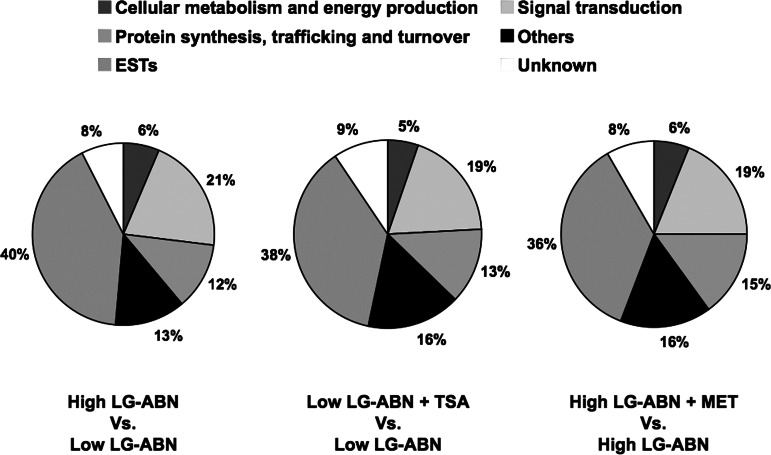
The most striking observation is that maternal care-, TSA-, and methionine-regulated RNA transcripts are all involved in similar classes of cellular function (Fig. 3). However, when examined across all of the categories, the distribution of genes up-regulated in the offspring of TSA-treated low LG-ABN mothers is significantly different in comparison to the distribution of genes up-regulated in the offspring of vehicle-treated high LG-ABN dams (χ2 = 12.425, P = 0.035). Thus, although both TSA treatment and high maternal LG-ABN behavior affected genes from similar categories, TSA treatment induced expression of a collection of unique transcripts that were different from those that were induced in the offspring of high LG-ABN mothers. Furthermore, the distribution of genes down-regulated in the offspring of methionine-treated high LG-ABN mothers is significantly different in comparison to the distribution of genes down-regulated in the offspring of vehicle-treated low LG-ABN dams (χ2 = 6.513, P = 0.026). Thus, although both methionine treatment and low maternal LG-ABN behavior affected genes from similar categories, methionine treatment suppressed expression of a collection of unique transcripts that were different from those that were suppressed in the offspring of low LG-ABN dams.
Fig. 3.
Distribution of maternal care-, TSA-, and methionine-responsive genes over different functional classes.
To examine the effects of maternal care, TSA, and methionine treatment on RNA transcript levels within the different functional categories, the three treatment groups were compared with their respective control groups, as previously described. Comparison of the transcripts altered in hippocampal tissue from offspring of vehicle-treated high LG-ABN mothers (vehicle-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams) revealed that 19 transcripts (6%) were involved in cellular metabolism and energy production, 63 transcripts (21%) were involved in signal transduction, and 36 transcripts (12%) were involved in protein synthesis, trafficking, and turnover (Fig. 3 Left). Comparison of the transcripts altered in hippocampal tissue from offspring of TSA-treated low LG-ABN mothers (TSA-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams) revealed that 28 transcripts (5%) were involved in cellular metabolism and energy production, 103 transcripts (19%) were involved in signal transduction, and 71 transcripts (13%) were involved in protein synthesis, trafficking, and turnover (Fig. 3 Center). Comparison of the transcripts altered in hippocampal tissue from offspring of methionine-treated high LG-ABN mothers (methionine-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of high LG-ABN dams) revealed that 21 transcripts (6%) were involved in cellular metabolism and energy production, 63 transcripts (19%) were involved in signal transduction, and 51 transcripts (15%) were involved in protein synthesis, trafficking, and turnover (Fig. 3 Right). To date only 90% of the estimated 2.8-Gb genome of the rat has been sequenced (www.hgsc.bcm.tmc.edu/projects/rat). Therefore, it is difficult to determine with great accuracy the normal pattern of gene distribution from the entire rat genome. However, the pattern of the functional distribution of genes found to be affected by our treatment is substantially different from the functional distribution of the 31,099 genes probe sets on the microarray. From the 31,099 unique probe sets on the rat 230.2 Affymetrix GeneChip, 21% encode protein products involved in general cellular metabolism and energy production, 12% encode protein products involved in signal transduction, 6% encode proteins involved in protein synthesis, trafficking, and turnover, 24% encode protein products involved in cellular scaffolding, 3% encode protein products with unknown functions, and 34% are ESTs. This discrepancy, between the distribution of genes with specific cellular functions in the genome and the distribution of genes of specific cellular functions affected by our treatment, suggests that the different treatments do have a specific effect on genes with distinct functions.
Together, these changes in mRNA expression demonstrate a clear effect of maternal care, TSA, and methionine treatment on the adult hippocampal transcriptome.
Effects of TSA and Methionine on ATRX, Reelin, and Vof-16 Gene Expression.
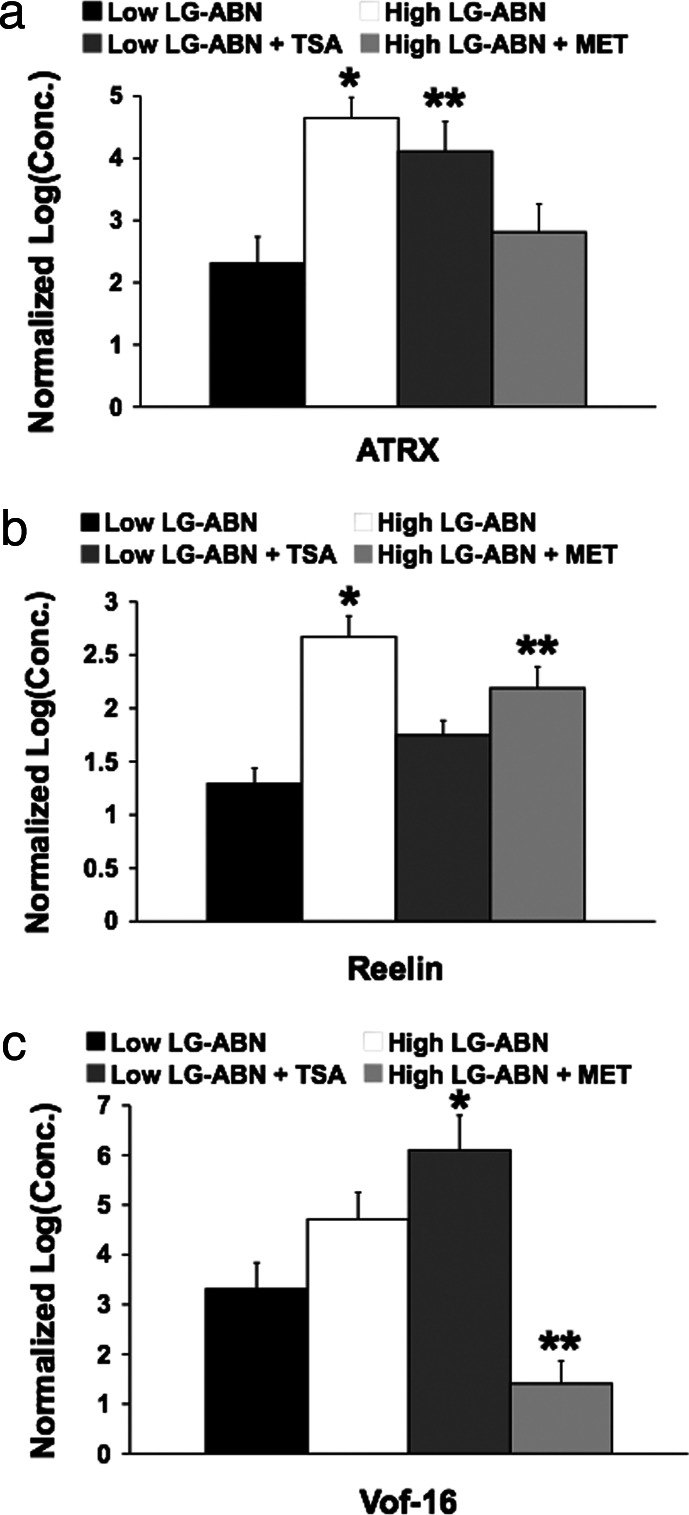
To validate the microarray data, three randomly chosen genes (shown in Fig. 5) expressed within the different functional categories were subject to quantitative real-time PCR analysis. Predesigned primers were used for amplifying the transcripts analyzed by the rat 230.2 GeneChip (Fig. 4). The results presented show that, for these three randomly chosen genes, the expression patterns obtained by using quantitative real-time PCR are similar to those obtained by using the microarray chips (compare the expression patterns in Fig. 4 with those shown for these genes in Fig. 5). For ATRX expression, ANOVA revealed a main effect of group (F = 8.83, P < 0.05) and treatment (F = 7.29, P < 0.01) and a main group × treatment interaction effect (F = 6.74, P < 0.05). Post hoc analysis revealed that the level of ATRX expression was significantly (∗, P < 0.001) greater in the vehicle-treated offspring of high LG-ABN mothers than in the vehicle-treated offspring of low LG-ABN dams and methionine-treated offspring of high LG-ABN mothers (Fig. 4a). The level of ATRX expression was also significantly (∗∗, P < 0.01) greater in the TSA-treated offspring of low LG-ABN mothers in comparison to the vehicle-treated offspring of low LG-ABN dams. No significant differences were found in the levels of ATRX expression between the vehicle-treated offspring of high LG-ABN mothers and TSA-treated offspring of low LG-ABN dams, or the vehicle-treated offspring of low LG-ABN mothers and methionine-treated offspring of high LG-ABN dams. For Reelin expression, ANOVA revealed main effects of group (F = 8.76, P < 0.05) and treatment (F = 7.02, P < 0.01) and a main group × treatment interaction effect (F = 5.73, P < 0.05). Post hoc analysis revealed that the level of Reelin expression was significantly (∗, P < 0.05) greater in the vehicle-treated offspring of high LG-ABN mothers in comparison to any other group. The level of Reelin expression was also significantly (∗∗, P < 0.01) greater in the methionine-treated offspring of high LG-ABN mothers in comparison to the vehicle-treated offspring of low LG-ABN mothers (Fig. 4b). No significant differences in the levels of Reelin expression were found between the vehicle- and TSA-treated offspring of low LG-ABN dams, or the TSA-treated offspring of low LG-ABN dams and methionine-treated offspring of high LG-ABN mothers. For Vof-16 expression, ANOVA revealed main effects of group (F = 9.03, P < 0.05) and treatment (F = 6.72, P < 0.01) and a main group × treatment interaction effect (F = 4.82, P < 0.05). Post hoc analysis revealed that the level of Vof-16 expression was significantly (∗, P < 0.01) greater in the TSA-treated offspring of low LG-ABN mothers in comparison to vehicle-treated offspring of low LG-ABN dams. The level of Vof-16 expression was significantly (∗∗, P < 0.001) lower in the methionine-treated offspring of high LG-ABN mothers in comparison to any other group (Fig. 4c). No significant differences in the level of Vof-16 expression were found between the vehicle-treated offspring of high and low LG-ABN dams, or the vehicle-treated offspring of high LG-ABN mothers and TSA-treated offspring of low LG-ABN dams.
Fig. 4.
Methionine and TSA treatment eliminates the maternal effect on genes expressed within different functional classes. Shown is quantitative real-time PCR analysis of hippocampal gene expression in TSA-treated (100 ng/ml), methionine-treated (100 μg/ml), and vehicle-treated adult offspring of high and low LG-ABN mothers (n = 3 animals per group). (a) Mean ± SEM ATRX expression. ∗, P < 0.001, vehicle-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams and methionine-treated offspring of high LG-ABN mothers; ∗∗, P < 0.01, TSA-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams. (b) Mean ± SEM Reelin expression. ∗, P < 0.05, vehicle-treated offspring of high LG-ABN mothers vs. all other groups; ∗∗, P < 0.01, methionine-treated offspring of high LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN mothers. (c) Mean ± SEM Vof-16 expression. ∗, P < 0.01, TSA-treated offspring of low LG-ABN mothers vs. vehicle-treated offspring of low LG-ABN dams; ∗∗, P < 0.001, methionine-treated offspring of high LG-ABN mothers vs. all other groups.
In summary, high licking and grooming increased hippocampal ATRX and Reelin expression with no significant effect on Vof-16 expression, and TSA treatment increased hippocampal ATRX and Vof-16 gene expression with no significant effect on Reelin expression, whereas low LG-ABN and methionine treatment decreased expression of all three genes. The results suggest that the genes are indeed regulated by one or more treatment, underscoring the validity of the microarray data.
Discussion
We have shown that gene expression is significantly altered in the hippocampus of adult rats as a function of maternal care early in life. These differences in gene expression may, at least in part, form the molecular basis for the effect of early-life experience on the development HPA responses to stress in the offspring that are endured throughout life. The majority of mRNA transcripts that showed group differences in expression were for those that encoded signal transduction proteins involved in the pathways that regulate brain formation and function. For example, our studies reveal a naturally occurring difference in reelin (probe set ID 1373957_at) expression within the adult offspring as a function of early-life experience. Reelin–integrin receptor interactions mediate activity-regulated cytoskeletal (Arc) protein synthesis in synaptoneurosomes (18). Therefore, Reelin may regulate developmental processes such as synaptogenesis and axon pruning as well as synaptic plasticity through life. Interestingly, we previously showed that the adult offspring of high LG-ABN mothers have increased hippocampal synaptic density and enhanced performance in paradigms that test hippocampal-dependent learning and memory (8, 11, 19). Furthermore, mRNA transcripts that differed in expression as a function of early-life experience encoded proteins associated with mitochondrial-mediated neuronal injury. For example, ionotrophic NMDA2C (probe set ID 1368306_at) and metabotrophic 5 (probe set ID 1369355_at) glutamate receptors were down-regulated in adult offspring of low LG-ABN mothers. Together with our previous findings of maternal care effects on neuron survival in the offspring (10), these data suggest that hippocampal neurons in the offspring of low LG-ABN mothers are more vulnerable to loss through apoptosis.
TSA and methionine treatment altered the expression of genes that were previously shown to be involved in human neurodegeneration and dementia. For example, enhanced expression levels of ischemia-related factor vof-16 (probe set ID 1377778_at) is associated with neuronal damage and impairment of learning and memory performance (20); hippocampal ATRX (probe set ID 1385006_at) dysregulation is associated with numerous forms of X-linked mental retardation (21); overexpression of S100 calcium-binding protein A4 (probe set ID 1367846_at) is observed in several neurodegenerative disorders (22); and reduced dihydropyrimidinase-related protein 2 (probe set ID 1380728_at) is associated with age-related dementia. Therefore, the rat model of natural variations in maternal care could be potentially useful in the study of gene–environment–therapeutic interactions of genomic regions known to be involved in human disease.
The mechanism responsible for the long-term programming of these genes by maternal care remains to be determined by future experiments. Nevertheless, based on our previous studies with the GR gene, we propose that epigenetic reprogramming in response to maternal care is involved in the observed effects on gene expression (13). A signaling pathway triggered by maternal LG-ABN behavior induces changes in chromatin structure and DNA modification, which then maintains this differential expression profile. The proposal that histone acetylation is involved is supported by the fact that some of the genes affected by maternal care are also induced by the histone deacetylase inhibitor TSA. The fact that the methyl donor l-methionine inhibits some of the genes induced by high maternal LG-ABN behavior supports the involvement of either DNA or histone methylation. Interestingly, although the effects of maternal care, TSA, and methionine involve a large number of genes, the process is exquisitely specific. The vast majority of the genome is not affected, suggesting that these treatments, despite their global nature, do not result in a general collapse of gene expression programming. Although the basis for this specificity remains unknown, this observation has important implications on the future therapeutic utility of these and similar treatments.
Genomic programs could be modified by genetic alterations, which are transmitted in the germ line. Genetic polymorphisms have attracted significant attention as a possible mechanism underlying interindividual behavioral differences and pathologies (23). Our data illustrate a new mechanism by which widespread and stable lifelong interindividual variation in gene expression in the brain might emerge. This mechanism does not require germ-line transmission and could be elicited by natural variations in maternal behavior early in life. The main difference between genetic and epigenetic variation is the potential for reversal with the appropriate manipulation, as illustrated here. Our data demonstrate the profound effects that early-life environment might have on the functioning of the genome and its lifelong consequences for behavior into adulthood.
Materials and Methods
Animals and Maternal Behavior.
The animals used in all studies were derived from Long–Evans hooded rats born in our colony from animals originally obtained from Charles River Canada (St. Catherine’s, Québec). All procedures were performed according to guidelines developed by the Canadian Council on Animal Care and protocol approved by the McGill University Animal Care Committee. Maternal behavior was scored as described in ref. 17. For further methodological details, see Supporting Methods, which is published as supporting information on the PNAS web site.
Intracerebroventricular Infusions.
A stainless steel guide cannula (22-gauge; 8-mm length; Plastic One) was placed in the left lateral ventricle (1.5 mm posterior to bregma, 2.0 mm lateral to midline, and 3.0 mm ventral to the brain surface). After a 7-day recovery period, animals received a single infusion of 2 μl of TSA (100 ng/ml in saline), l-methionine (100 μg/ml in saline), or saline vehicle alone through the infusion cannula every day for 7 consecutive days.
Open-Field Test.
The naive animal was removed from the home cage and placed directly into one corner of the open field (120 cm × 120 cm) (9). The floor was divided into a grid of 8 × 8 squares. Movement of the animal in the arena during the 10-min testing session was recorded. After 10 min, the animal was removed and returned to the home cage, and the open-field arena was cleaned to prevent olfactory cues from affecting the behavior of subsequently tested rats. An observer blind to the experimental conditions coded the videotapes by using a DOS-based program. Exploration was defined as the time spent in the inner 6 × 6 squares, whereas overall activity was defined as the number of squares crossed during the testing session.
Microarray Analysis.
Microarray experiments were performed at the McGill University and Génome Québec Innovation Centre by using the Affymetrix Rat Genome 230.2 GeneChip according to standard Affymetrix protocols (GeneChip Analysis Technical Manual, Rev. 5). Twelve independent microarray experiments were performed by using total hippocampal RNA preparations from 12 different animals (n = 3 animals per treatment). Bioconductor software (24) processed raw data for background adjustment, quantile normalization, and summarization (25). Genes were considered differentially expressed if they showed (i) average log2(treated/control) ratio between experimental groups > 0.585 and (ii) reported a t test P value <0.05. Hierarchical clustering was done by using genesis software (IBMT-TUG, Graz, Austria) (26).
Quantitative Real-Time PCR Analysis.
Samples were analyzed in triplicate by using total hippocampal RNA preparations from 12 different animals (n = 3 animals per treatment). Predesigned primers were used for amplifying vof-16 (UniGene accession no. Rn.38750), ATRX (UniGene accession no. Rn.107838), and Reelin (UniGene accession no. Rn.98353) RNA transcripts as rendered on the rat 230.2 chip (RT Real-Time Gene Expression Assay Kits, version 2.0, SuperArray Bioscience). Primers for β-actin (UniGene accession no. Rn.94978) were used for normalization. PCR mixtures (20 μl) were loaded into LightCycler capillaries (Roche Molecular Biochemicals) as described in Supporting Methods.
Supplementary Material
Acknowledgments
These studies were supported by grants from the Canadian Institutes for Health Research (CIHR; to M.J.M. and M.S.) and from the National Cancer Institute of Canada (to M.S.). M.J.M. is supported by a CIHR Senior Scientist Award, and the project was supported by a Distinguished Investigator Award from the National Alliance for Research on Schizophrenia and Affective Disorders (to M.J.M.).
Abbreviations
- GR
glucocorticoid receptor
- TSA
trichostatin A
- LG-ABN
licking/grooming and arched-back nursing
- HPA
hypothalamic–pituitary–adrenal.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Agrawal A. A. Science. 2001;294:321–326. doi: 10.1126/science.1060701. [DOI] [PubMed] [Google Scholar]
- 2.Higley J. D., Hasert M. F., Suomi S. J., Linnoila M. Proc. Natl. Acad. Sci. USA. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meaney M. J. Annu. Rev. Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 4.Liu D., Diorio J., Tannenbaum B., Caldji C., Francis D., Freedman A., Sharma S., Pearson D., Plotsky P. M., Meaney M. J. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 5.Francis D., Diorio J., Liu D., Meaney M. J. Science. 1999;286:1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 6.De Kloet E. R., Vreugdenhil E., Oitzl M. S., Joels M. Endocr. Rev. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- 7.Meaney M. J., Aitken D. H., Viau V., Sharma S., Sarrieau A. Neuroendocrinology. 1989;50:597–604. doi: 10.1159/000125287. [DOI] [PubMed] [Google Scholar]
- 8.Bredy T. W., Grant R. J., Champagne D. L., Meaney M. J. Eur. J. Neurosci. 2003;18:2903–2909. doi: 10.1111/j.1460-9568.2003.02965.x. [DOI] [PubMed] [Google Scholar]
- 9.Caldji C., Tannenbaum B., Sharma S., Francis D., Plotsky P. M., Meaney M. J. Proc. Natl. Acad. Sci. USA. 1998;95:5335–5340. doi: 10.1073/pnas.95.9.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver I. C., Grant R. J., Meaney M. J. J. Neurochem. 2002;82:998–1002. doi: 10.1046/j.1471-4159.2002.01054.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu D., Diorio J., Day J. C., Francis D. D., Meaney M. J. Nat. Neurosci. 2000;3:799–806. doi: 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- 12.Fleming A. S., O’Day D. H., Kraemer G. W. Neurosci. Biobehav. Rev. 1999;23:673–685. doi: 10.1016/s0149-7634(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 13.Weaver I. C., Cervoni N., Champagne F. A., D’Alessio A. C., Sharma S., Seckl J. R., Dymov S., Szyf M., Meaney M. J. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 14.McCormick J. A., Lyons V., Jacobson M. D., Noble J., Diorio J., Nyirenda M., Weaver S., Ester W., Yau J. L., Meaney M. J., et al. Mol. Endocrinol. 2000;14:506–517. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- 15.Weaver I. C., Champagne F. A., Brown S. E., Dymov S., Sharma S., Meaney M. J., Szyf M. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalynchuk L. E., Meaney M. J. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2003;27:1225–1234. doi: 10.1016/j.pnpbp.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Champagne F. A., Francis D. D., Mar A., Meaney M. J. Physiol. Behav. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 18.Dong E., Caruncho H., Liu W. S., Smalheiser N. R., Grayson D. R., Costa E., Guidotti A. Proc. Natl. Acad. Sci. USA. 2003;100:5479–5484. doi: 10.1073/pnas.1031602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredy T. W., Humpartzoomian R. A., Cain D. P., Meaney M. J. Neuroscience. 2003;118:571–576. doi: 10.1016/s0306-4522(02)00918-1. [DOI] [PubMed] [Google Scholar]
- 20.Tohda M., Watanabe H. Biol. Pharm. Bull. 2004;27:1228–1235. doi: 10.1248/bpb.27.1228. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons R. J., Higgs D. R. Am. J. Med. Genet. 2000;97:204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Heizmann C. W., Fritz G., Schafer B. W. Front. Biosci. 2002;7:d1356–d1368. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- 23.Moret C. IDrugs. 2004;7:558–562. [PubMed] [Google Scholar]
- 24.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Sturn A., Quackenbush J., Trajanoski Z. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.