Abstract
Mutations in RRP6 result in the accumulation of aberrant polyadenylated transcripts from small nucleolar RNA genes. We exploited this observation to search for novel noncoding RNA genes in the yeast genome. When RNA from rrp6Δ yeast is compared with wild-type on whole-genome microarrays, numerous intergenic loci exhibit an increased mutant/wild type signal ratio. Among these loci, we found one encoding a new C/D box small nucleolar RNA, as well as a surprising number that gave rise to heterogeneous Trf4p-polyadenylated RNAs with lengths of ≈250–500 nt. This class of RNAs is not easily detected in wild-type cells and appears associated with promoters. Fine mapping of several such transcripts shows they originate near known promoter elements but do not usually extend far enough to act as mRNAs, and may regulate the transcription of downstream mRNAs. Rather than being uninformative transcriptional “noise,” we hypothesize that these transcripts reflect important features of RNA polymerase activity at the promoter. This activity is normally undetectable in wild-type cells because the transcripts are somehow distinguished from true mRNAs and are degraded in an Rrp6p-dependent fashion in the nucleus.
Keywords: compound promoters, cryptic RNAs, genome annotation, RNA decay
The ability to sequence complete genomes has demanded comprehensive approaches to gene discovery and genome annotation, often using computational predictions. Eventually, experimental data must be used to assign gene function to the genome. Because transcription is the primary biosynthetic event required for gene function, identifying transcribed regions is the foundation of genome annotation.
Numerous methods allow the genome-scale annotation of transcribed regions. For example, cDNA cloning strategies or serial analysis of gene expression (SAGE) capture evidence of transcription not dependent on prior information, such as ORF prediction (1). However, transcripts present in low abundance may be missed unless cloning biases are controlled and exhaustive numbers of clones are studied.
More recently, “whole genome” or “genomic tiling” microarrays have been used to identify transcribed regions (2, 3). Such arrays have the advantage of not relying on the current genome annotation and are not limited by “clone coverage” problems of EST or serial analysis of gene expression approaches. Because of this, they have been used to map RNAs from the human (2, 4) and other genomes (5–7). One limitation to this approach is its sensitivity, which depends on how well an expressed RNA can be captured by the microarray. This property is likely governed by combined effects of the abundance of the transcript in the sample, its labeling efficiency, and the ability of probes on the array to capture the labeled target.
Based on our observations of global changes in RNA levels for introns in dbr1Δ yeast and for small nucleolar RNA (snoRNA)-containing introns in yeast using the rrp6Δ mutation (8, 9), we reasoned that locating genes or gene elements whose processing or decay requires specific factors (e.g., the debranching enzyme for introns, or Rrp6p for snoRNAs) could be accelerated by combining whole-genome microarrays with mutants in RNA processing pathways. Array elements representing genomic regions that produce transcripts whose abundance or labeling efficiency depend on proper RNA processing would be expected to capture different amounts of target in mutant vs. wild type comparisons, thus revealing their location. To test this idea, we searched for new small RNAs using strains lacking Rrp6p, a component of the nuclear exosome responsible for 3’ end processing of small stable transcripts such as small nuclear RNAs (snRNAs), snoRNAs, and rRNAs (10). Mutations in RRP6 lead to the accumulation of snRNAs, snoRNAs, and rRNAs with 3’ extensions and poly(A) tails (11–13), thus increasing their labeling efficiency in the mutant sample.
In this article, we compare RNA from wild type cells with RNA from a strain lacking RRP6 using microarrays containing the complete genome. Recently, an approach using arrays that carried probes only for previously discovered RNA transcripts found that many intergenic RNAs represented by serial analysis of gene expression transcripts are increased in the rrp6Δ mutant (13). Because we used arrays covering the entire genome, we discovered numerous regions not previously known to be transcribed. In one such region, we uncovered a new C/D box snoRNA, as we had hoped. The other class of RNA was unexpected and consists of transcripts that appear to originate from promoter regions of standard protein-encoding genes. In wild-type cells, these transcripts are decayed through a Trf4p-Rrp6p pathway (13–16). We propose that these transcripts are produced at mRNA promoters across the genome as a consequence of RNA polymerase II activity that normally does not lead to functional mRNA.
Results
Expression of RNAs from Many Genomic Loci Increases upon Loss of RRP6.
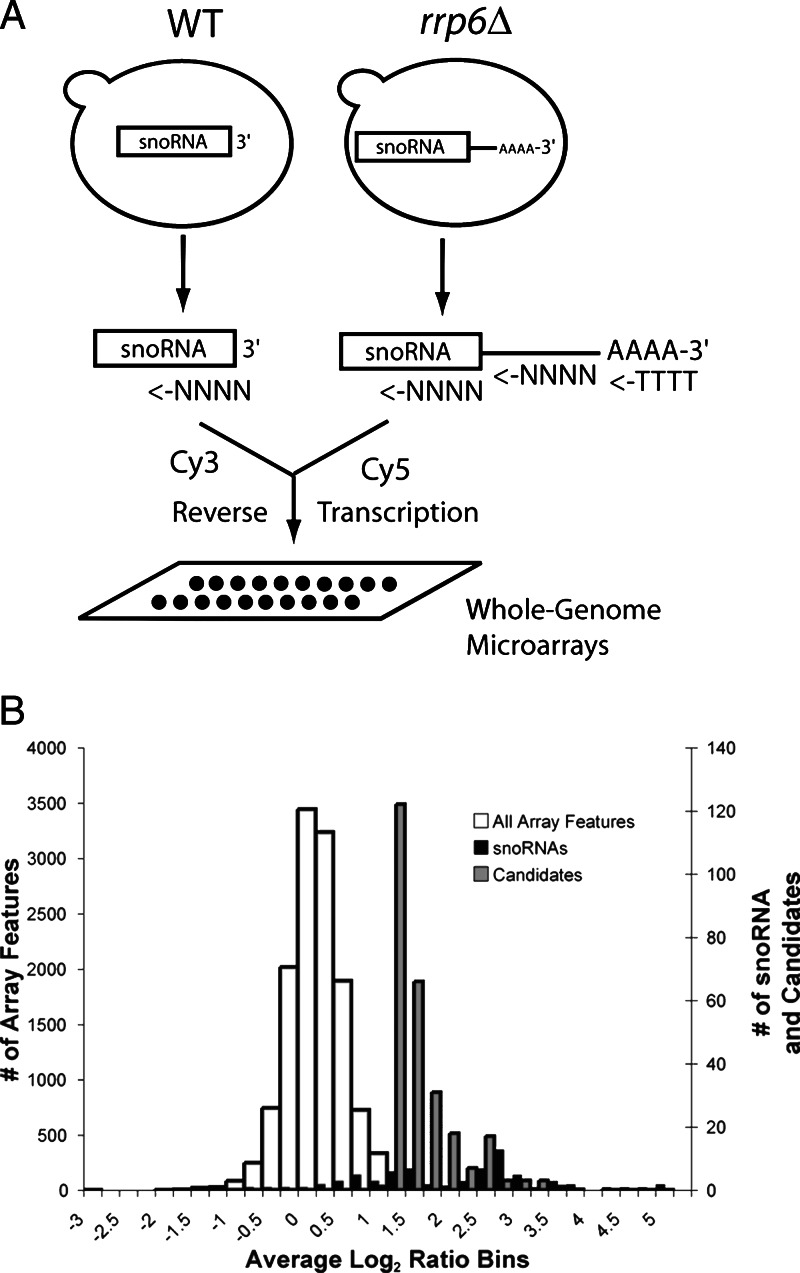
Our initial goal was to use the observations of Van Hoof et al. that noncoding RNAs accumulate as polyadenylated species (11) and a whole-genome microarray (17) to identify new stable RNAs across the entire yeast genome (Fig. 1A). Because reverse transcriptions were primed with a mixture of oligo(dT) and random hexamers, array signals should increase for transcripts whose poly(A) content might change upon loss of Rrp6p, even if the overall level of RNA does not, due to more efficient Cy-dye labeling of poly(A)+ RNAs in the presence of oligo(dT). After normalization, we find that 97% (12,754) of the total 13,137 array features have log2 ratios within two standard deviations (SD = 0.5) of the mean (mean = +0.03) (Fig. 1B, white bars). However, 2.6% (350) of the loci exhibit log2 ratios more than 2 SDs above the mean in the rrp6Δ mutant (Table 1, which is published as supporting information on the PNAS web site). This group is dominated (51/350) by known snoRNAs (Fig. 1B, black bars; 20 known snoRNAs are below this cutoff) or regions 3’ of known snoRNAs (28/350; Table 2, which is published as supporting information on the PNAS web site). Spliceosomal snRNAs U4 and U6 were also among those detected. Although polyadenylation of rRNAs occurs in the rrp6Δ mutant (12), we observed no increase in signal for rRNA features, possibly because rRNA is saturated. These results are consistent with reports indicating that snoRNAs and snRNAs accumulate as polyadenylated products in the rrp6Δ mutant (10, 11) and demonstrate that small stable RNA loci in the genome can be easily distinguished by using whole-genome microarrays and an rrp6Δ mutant strain. To begin identifying new small RNAs, we studied several of the ≈160 unannotated intergenic loci in this group (Fig. 1B, gray bars).
Fig. 1.
Genomic loci affected by loss of RRP6. (A) Scheme used to identify RNAs enriched or hyperadenylated in the rrp6Δ mutant. (B) Histogram of microarray results. Shown is the distribution of the average log2 ratio signal for the following: all array features, white bars (mean = 0.03, SD = 0.5); snoRNA-containing features, black bars; unannotated intergenic features above 1.04 (>2 SD), gray bars. Note that the axis for all of the array features (primary y axis) differs from that for the snoRNA and intergenic features (secondary y axis).
A New C/D Box snoRNA Is the Guide for 18S rRNA Methylation at Position A436.
Intergenic region iYKL006c-a gave a 6-fold increase in signal in the rrp6Δ mutant. Northern blots probed with this region uncovered a small stable RNA ≈106 nt in length (Fig. 2A, lane 1). This RNA appears slightly longer in the rrp6Δ strain (lane 2), consistent with the proposed role for Rrp6p in trimming the last three nucleotides (10, 11). In addition, a slower migrating heterogeneous RNA accumulates (Fig. 2A, lane 2). This RNA binds oligo(dT) indicating that it is polyadenylated (data not shown), as are other snoRNAs in strains lacking RRP6 (see below) (11). We mapped this RNA, called snR87, to the Crick strand of chromosome XI between positions 430850 and 430670, a highly conserved region in related yeasts (Fig. 7, which is published as supporting information on the PNAS web site). Using an rnt1Δ strain, we find that the 5’ end of this snoRNA is generated in part by Rnt1p processing, as for other snoRNAs (Fig. 7) (18, 19).
Fig. 2.
C/D box snoRNA snR87 guides methylation of 18S rRNA A436. (A) Northern blot of transcripts in iYKL006C-a. Total RNA from wild-type (lane1) and rrp6Δ (lane 2) strains was fractioned on a 6% acrylamide gel and transferred. The membrane was probed with a dsDNA probe spanning the region. Mature snR87 (rectangle) and precursors or aberrant products are indicated at the right. (B) Test for methylation of 18S rRNA near nucleotide A436. Total RNA from wild type (lanes 1–3) and snr87Δ (lanes 4–6) were reverse-transcribed under limiting dNTP levels and run on a 6% sequencing gel. The stops generated corresponding to known sites of methylation A420 and A436 are indicated at the right. (C) Predicted alignment of snR87 with 18S rRNA near A436. The C/D box signature motifs are indicated.
The sequence of snR87 contains C/D box snoRNA motifs and complementarity to 18S rRNA, characteristics of methylation guide RNAs for rRNA (20) (Fig. 2C). The rRNA complementary region suggested that snR87 might guide methylation of A436, an rRNA methylation site without a known C/D box snoRNA. To test this, we compared methylation at A436 in wild type with a strain deleted for snR87 (Fig. 2B). In wild type, reverse transcriptase stops occur at positions just downstream of the known methylation sites at A436 and A420 (lanes 4–6). In contrast, no stop consistent with methylation is seen at A436 in the snR87Δ strain, although the stop at A420 is readily apparent (lanes 1–3). This demonstrates the requirement for snR87 in 2’-O-methylation of 18S rRNA position A436 and validates this genomic approach for finding noncoding RNAs by testing the prediction of van Hoof (11) on a genomic scale.
Many Genomic Regions Encode Heterogeneous RNAs That Appear Unstable in the Presence of Rrp6p and Trf4p.
Probing of other regions uncovers another class of noncoding RNA distinct from snoRNAs, snRNAs, and rRNAs (Fig. 3A, lane 2). Accumulation of these RNAs, which range in size from 250 to 800 nt, is caused by loss of RRP6 because no RNA was seen in the wild-type strain. Furthermore, each of these RNAs is enriched in oligo(dT) selected RNA (lane 3), indicating that they are polyadenylated in the rrp6Δ mutant. Similar findings (13) were recently reported for regions of the yeast genome previously shown to be transcribed by using serial analysis of gene expression (1).
Fig. 3.
RNAs accumulate in the absence of RRP6 or TRF4. Northern blots detect transcripts from genomic loci. (A) rrp6Δ. Five micrograms of total RNA from wild-type (lane 1) and rrp6Δ (lane 2) strains, 300 ng of poly(A)+ RNA from rrp6Δ strain (lane 3), and 5 μg of poly(A)− RNA from rrp6Δ strain (lane 4) were loaded onto a 1% agarose-formaldehyde gel and probed with the indicated regions. (B) trf4Δ. Five micrograms of total RNA from wild-type (lane 1) and trf4Δ (lane 2) strains, 300 ng of poly(A)+ RNA from trf4Δ strain (lane 3), and 5 μg of poly(A)− RNA from trf4Δ strain (lane 4) was used. Annotations for the region are indicated on the left, and the probed region is indicated by the black bar. Dubious ORFs are shaded in gray. The name of the candidate region and size of the heterogeneous RNAs are indicated on the right.
Recently, a decay complex now referred to as TRAMP was described (13–16). Trf4p is a component of this complex that acts by adding 10–40 adenines onto the 3’ end of RNAs before their decay by the exosome (13–16). To test whether loss of Trf4p could cause accumulation of the RNAs we identified, we blotted RNA from a trf4Δ strain and probed with several genomic regions. RNA from each of these regions is stable in the trf4Δ mutant (Fig. 3B, lane 2) and is not enriched in poly(A)+ fractions (lane 4), in contrast to the rrp6Δ mutant. These results are consistent with a model in which Trf4p marks this class of RNAs for nuclear decay through addition of a polyA tail followed by Rrp6p-mediated degradation (see ref. 13).
The SER3 Promoter-Associated SRG1 RNA Accumulates in the rrp6Δ Mutant.
SRG1 RNA is a noncoding promoter-associated RNA whose expression regulates the adjacent SER3 gene (21). We asked whether the SRG1 RNA accumulates in the rrp6Δ mutant. Although the SER3 promoter region encoding SRG1 RNA did not exhibit a change in signal log2 ratio, the SER3 ORF 3’ of SRG1 increased 4-fold in the rrp6Δ mutant, comparable to the 3’ regions of the snoRNA genes (Table 2). Transcription of SRG1 spans the SER3 promoter and extends into the 5’ end of the SER3 coding region (21). We saw significant accumulation of SRG1 RNA in the rrp6Δ strain compared with wild type on a Northern blot (Fig. 4A). Although SRG1 RNA is reported to be polyadenylated (21), we did not observe efficient enrichment in the poly(A)+ fraction from wild type using our method (Fig. 4A), although there is a small increase in the amount of polyadenylated SRG1 the rrp6Δ mutant. In contrast, snR128 extended transcripts are well selected on oligo(dT) (Fig. 4A, compare lane 2 with lane 5 for each RNA). For both RNAs the loss of Trf4p leads to more modest accumulation of RNA that is not enriched in the poly(A)+ fraction (Fig. 4A). We conclude that SRG1 RNA decay occurs via the exosome.
Fig. 4.
SRG1 is a promoter associated RNA whose decay requires Rrp6p. (A) Northern blot to detect the levels of poly(A)+ and poly(A)− SRG1 RNA (Upper): 3 μg of total RNA from wild type, trf4Δ, and rrp6Δ (lanes 1, 4, and 7), 100 ng of poly(A)+ RNA from wild type, trf4Δ, and rrp6Δ (lanes 2, 5, and 8), and 3 μg of poly(A)− RNA from wild type, trf4Δ, and rrp6Δ (lanes 3, 6, and 9). The membrane was probed with an SRG1-specific oligo. (Lower) A duplicate membrane probed with an oligo complementary to snR128. (B) Northern blot analysis of SER3 in different yeast mutants. Three micrograms of total RNA from wild-type (lane 1), trf4Δ (lane 2), rrp6Δ (lane 3), heat-shifted spt5-194 (lane 4), heat-shifted spt5-194, rrp6Δ (lane 5), and snf2Δ (lane 6) yeast were fractioned on a 1% agarose formaldehyde gel. The membrane was probed with an SRG1+SER3 oligo complementary to a region contained in both SRG1 RNA and SER3 mRNA.
Mutations in components of the chromatin remodeling machinery lead to SER3 induction and a partial loss of SRG1 RNA accumulation (22), as well as an increase in aberrant transcripts that appear to originate at promoter-like sequences within coding regions (23). An increase in this latter class of transcripts also occurs upon loss of elongation factors (24). To test whether rrp6Δ-mediated increase in SRG1 RNA perturbs SER3 expression we measured levels of SRG1 and SER3 mRNA by Northern blot (Fig. 4B) and primer extension (see Fig. 8, which is published as supporting information on the PNAS web site). As shown previously (22), loss of Snf2p function leads to dramatic accumulation of SER3 mRNA and an apparent reduction in SRG1 RNA (compare lane 1 and lane 6). Loss of Rrp6p (lane 3) or Trf4p (lane 4) increases the level of SRG1 (see also Fig. 4A); however, SER3 mRNA appears unchanged. Loss of Spt5p leads to SER3 mRNA induction, without a large increase in SRG1 (lane 4). In the spt5-194, rrp6Δ double mutant, both RNAs accumulate (lane 5) (see also Fig. 8). These results argue that both correct chromatin remodeling (22) and active transcription elongation are required for SRG1-mediated repression of SER3, and that both of these requirements suggest that it is the act of transcription that mediates repression (21, 22, 24, 25). This conclusion suggests that locating other members of this class of RNAs may reveal new instances of this type of promoter regulation.
Heterogeneous RNAs Span the Promoters of LEU4 and IMD2.
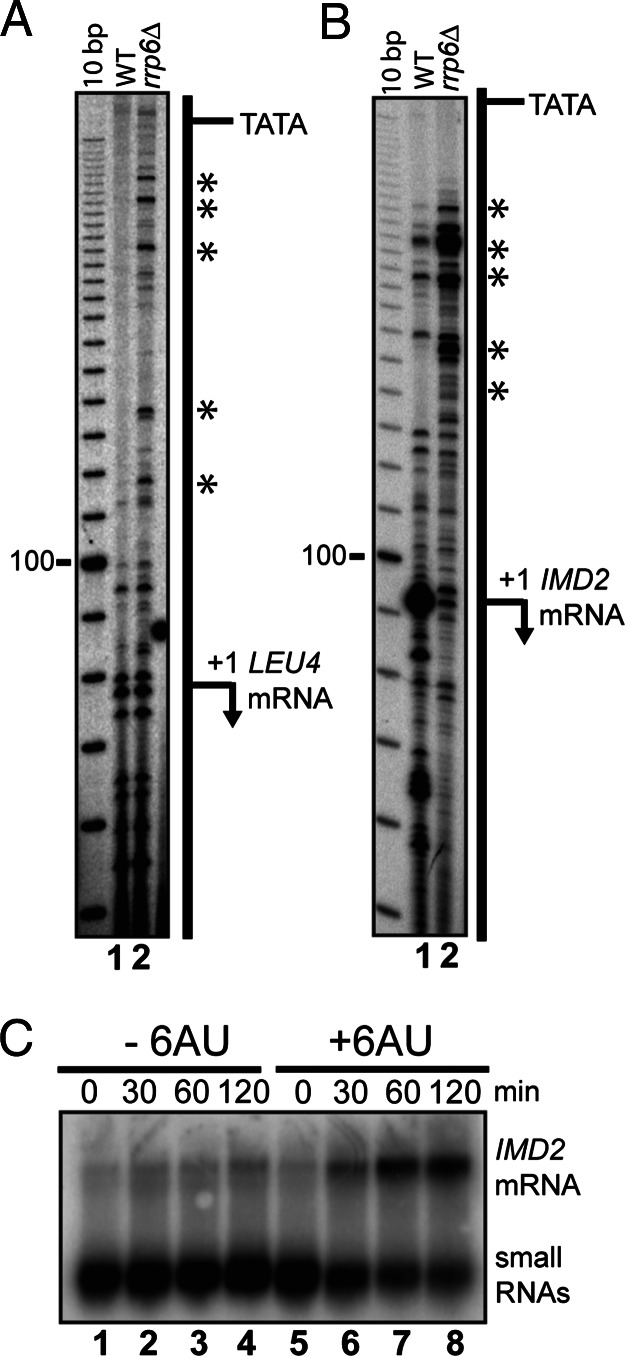
To explore the possibility that additional members of this class of RNAs span promoters of other genes, we decided to map precisely the 5’ ends of two RNAs that accumulate in the rrp6Δ mutant. Heterogeneous small RNAs arising from sequences containing the promoters for the LEU4 and IMD2 genes accumulate in the absence of rrp6Δ (Table 1 and Fig. 3A). We detect transcripts in wild type whose 5’ ends correspond to the reported initiation sites for LEU4 and IMD2 (Fig. 5 A and B, lanes 1). Transcripts that initiate upstream of the normal LEU4 and IMD2 mRNAs are readily detectable in the rrp6Δ mutant (Fig. 5 A and B, lanes 2). Several IMD2 region RNAs observed to accumulate in the rrp6Δ mutant are detectable in wild-type cells, suggesting that they are synthesized in the presence of Rrp6p. In addition, it appears that the level of RNA representing the normal IMD2 mRNA start site is reduced. We conclude that the loss of RRP6 causes stable accumulation of transcripts that initiate within the promoters of the LEU4 and IMD2 genes, and span the promoters in the same direction as the normal mRNA transcript.
Fig. 5.
RNAs map to promoters of IMD2 and LEU4. Primer extensions to map the 5’ end of RNA associated with the promoters of LEU4 (A) and IMD2 (B) from wild-type (lane 1) and rrp6Δ (lane 2) strains. The positions of upstream starts uncovered in the absence of RRP6 are indicated by an asterisk. The positions of TATA boxes tested in refs. 29 and 30 are indicated. Primer sequences are described in Supporting Methods. (C) Northern blots on 5 μg of total RNA from rrp6Δ strain grown in SCD-Ura media at times 0, 30, 60, and 120 min after treatment with 6-azauracil (lanes 5–8) or untreated (lanes 1–4).
IMD2 mRNA is induced by low levels of GTP in wild-type cells, a condition that can be created by addition of 6-azauracil to yeast cultures (26). To determine whether the RNAs that overlap the IMD2 promoter are regulated in the same fashion as the IMD2 mRNA, we treated rrp6Δ mutant cells with 6-azauracil and analyzed the RNA by Northern blotting (Fig. 5C). The IMD2 mRNA is dramatically induced under these conditions (Fig. 5C, lanes 5–8), consistent with previous observations (26). The levels of the small RNAs from the IMD2 promoter are high in the rrp6Δ mutant as shown previously (Figs. 3 and 5) and seem to decrease somewhat over the course of the experiment (Fig. 5C). This effect may be due to the general inhibition of transcription caused by 6-azauracil (27) or could reflect down-regulation of the IMD2 promoter-associated RNA. Rrp6p is clearly not required for the induction of IMD2 expression, and it seems unlikely that the IMD2 promoter-associated RNAs are transacting repressors of IMD2. We conclude that the transcription of the IMD2 promoter-associated RNAs is regulated differently from the IMD2 mRNA, despite apparently arising from closely overlapping regions of the chromosome. This arrangement is at least superficially similar to SRG1 and SER3, suggesting that regulation of the IMD2 small RNA by guanosine levels could lead to IMD2 induction.
Heterogeneous Unstable RNAs Most Frequently Arise from Standard mRNA Promoters Genome-Wide.
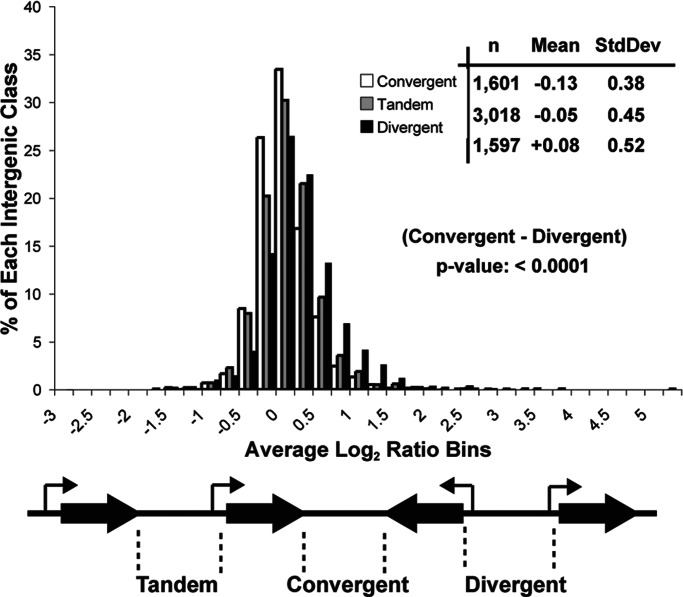
Given the overlap of the small RNAs with the promoters for LEU4 and IMD2 mRNAs, we asked whether a high intergenic signal in the rrp6Δ mutant is generally associated with promoter-containing intergenic regions as compared with other intergenic regions. Reasoning that yeast promoter elements are likely to be more common upstream of genes, we sorted intergenic regions into one of three classes (convergent, n = 1,601; tandem, n = 3,018; or divergent, n = 1,597), depending on the orientation of the two annotated genes on either side of the intergenic region (Fig. 6 and Table 3, which is published as supporting information on the PNAS web site). We expected promoters to occur more often in divergent regions, next most often in regions between tandem genes, and least often in regions between convergent genes. As a group, intergenic regions situated between divergently transcribed genes show the most increased transcription in the rrp6Δ mutant (Fig. 6). Intergenic regions between tandemly transcribed genes are moderately affected, and those situated between convergently transcribed genes are the least affected (Fig. 6). To test the null hypothesis that the difference between these distributions could be due to chance, we applied an unpaired t test between the convergent and divergent distributions. The likelihood (p value) that these distributions are the same is <0.0001. These results suggest that in the rrp6Δ mutant, promoter-associated RNAs distinct from true mRNAs are stabilized. In some regions not expected to contain mRNA promoters, a similar process appears to happen (e.g., iYCRCΔ6; Fig. 3), suggesting that regions with sequence features that resemble promoters may be functional but not normally detectable in wild-type cells due to the action of Rrp6p and Trf4p (see ref. 13).
Fig. 6.
Promoter-containing intergenic regions accumulate the most RNA in the absence of RRP6. Shown is a histogram representing the distribution of the average signal log2 ratio for all intergenic features as indicated below. The results of a two-tailed unpaired t test comparing convergent–divergent distributions are shown in the Inset.
Discussion
We have analyzed regions of the yeast genome whose expression is altered in the rrp6Δ mutant, as detected by microarrays representing the complete genome (Fig. 1). As expected (28), many of the most affected genomic regions encode known snoRNAs (Fig. 1 and Table 2). In the unannotated regions, we found one new C/D box snoRNA (Figs. 2 and 7), as well as an unanticipated class (Fig. 3) of promoter-associated RNAs (Figs. 4–6 and 8) whose accumulation is prevented by components of the Trf4p/Rrp6p decay pathway (Fig. 3) (13–16).
A New Class of RNA Transcripts with an Uncertain Relationship to Native Gene Function.
Our findings (Fig. 3) support the existence of a new class of heterogeneous transcripts ranging in size from 250–800 bp in length. These RNAs are difficult to detect convincingly in wild-type cells (Fig. 3 A and B); however, deletion of RRP6 results in the strong accumulation of this class of RNAs as polyadenylated species (Fig. 3A) (see ref. 13). Because previous work used arrays that carried probes only for previously discovered RNA transcripts (13), the genomic extent of such transcripts could not be determined. Because we have used arrays covering the entire genome, we have discovered many previously unannotated RNAs (Fig. 3 and Table 1). We propose that this class of unstable transcripts are produced at normal mRNA promoters as well and in many cases can link their association with the mRNA promoter regions of standard genes across the genome (Figs. 5–7). These RNAs arise as a consequence of RNA polymerase II activity that normally does not lead to, and may in fact repress, the synthesis of functional mRNA (Figs. 4 and 5) (21–25). This observation may explain in part why microarray data combined with SVM analysis recognized an rrp6Δ phenotype consistent with transcriptional defects, rather than decay (8).
Relationships Between SRG1 and Promoter-Associated RNAs.
Transcription of SRG1 is required to repress production of the SER3 mRNA by a transcriptional interference mechanism (21, 22). We find that the IMD2 and LEU4 promoters also have promoter-associated transcripts (Fig. 5). However, unlike the SRG1-SER3 system, mutations in the TATA boxes upstream of the promoter-associated RNAs do not result in induction of IMD2 or LEU4 mRNAs (29, 30). This suggests either that repression of IMD2 and LEU4 does not require intergenic transcription or that the intergenic transcripts and their linked mRNAs (LEU4 and IMD2) are both dependent on the same core promoter elements. Despite arising from within the promoter, the IMD2 promoter-associated RNAs do not respond to the same transcription regulatory mechanisms as the IMD2 mRNA. For example, the IMD2 promoter-associated RNA is robustly transcribed under conditions in which transcription of IMD2 mRNA is repressed (Fig. 5C). Unlike IMD2 mRNA, the IMD2 promoter-associated RNA appears to be repressed in response to 6AU (Fig. 5C) [although inhibition of overall transcription is a consequence of lowering intracellular NTP levels (27), we do not yet know whether this RNA is more inhibited than the average RNA by 6AU]. In contrast, the IMD2 mRNA is readily induced in wild-type cells (31), and this does not require Rrp6p (Fig. 5C). Given the common origin but independent regulation of these two classes of transcripts, it is possible that this region contains two promoters in tandem, similar to SRG1-SER3. We suggest that the production of unstable RNAs is a property of compound promoters, and that in some cases they indicate the presence of an auxiliary promoter that helps preserve a repressed or activated transcriptional state independent of the production of mRNA. In some cases the repressed state must be maintained by active transcription because loss of both chromatin remodeling factors and elongation factors can lead to inappropriate induction (Fig. 5) (21–25).
Materials and Methods
Additional details are available in Supporting Methods, which is published as supporting information on the PNAS web site.
Yeast Strains.
The following yeast strains were ordered from Open Biosystems. Wild-type strain BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) was compared with strains YSC1021-551682 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, rrp6::kanMX) and YSC1021-554220 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, trf4::kanMX). The strain containing the snr87 deletion, YWD452, is described in ref. 32. Strain DY884 is described in ref. 33 and is deleted for IMD1, IMD3, and IMD4. Strain CD4KO was made through a cross between strain DY884 and YSC1021-551682 and is deleted for RRP6, IMD1, IMD3, and IMD4. Strain FY300 carries the spt5-194 allele (MATa, his4-912Δ, lys2-128Δ, leu2Δ1, ura3-52, spt5-194). Strain GHY1344 carries the spt5-194 and rrp6Δ alleles (MATa, lys2-128Δ, leu2Δ, ura3Δ, spt5-194, rrp6::KanMX). Before RNA isolation, strains carrying the spt5-194 allele were heat-shifted to 39°C for 30 min. Stain GHY15 contains the SNF2 mutant allele (MATa, his4-194Δ, his3Δ200, lys2-128Δ, ura3-52, trp1Δ63, snf2Δ1::HIS3).
Oligonucleotides.
Oligonucleotides were ordered from Sigma-Genosys in pairs designed to amplify the part of each candidate locus except 50–100 bp from the ends of the flanking ORFs. This approach was used where possible to avoid hybridization to the 5’ or 3’ UTRs of mRNAs coming from known ORFs. Sequences are available in Supporting Methods.
RNA Isolation.
RNA was isolated by using a hot phenol treatment as described in ref. 9 and treated with RQ1 DNase (Promega). Oligo(dT) affinity chromatography was done by using Qiagen Oligotex kits.
Whole-Genome Microarray Analysis.
Total RNA (20 μg) was reverse-transcribed in the presence of Cy3- or Cy5-dUTP by using a mixture of oligo(dT) and random hexamers as described in ref. 9. The labeled cDNAs were hybridized to whole-genome PCR microarrays (gift of J. DeRisi, University of California, San Francisco) overnight at 62°C. Arrays were washed and scanned by using a GenePix 4000A scanner (Axon Instruments). Data were normalized to the average of all features on the array. The log2 ratios are averages from two reverse-labeled experiments. Intergenic assignments as convergent, divergent, or tandem were made by using the October 2003 annotation of the yeast genome and a program written by Leslie Grate (Table 3). Array data were deposited in the Gene Expression Omnibus under accession numbers GSE3813 and GSM86408.
Northern Blots.
Agarose-formaldehyde (1.5%, 2.2 M) gels were blotted onto nylon (Hybond-N, Amersham Pharmacia) in 10× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7). RNA was UV-crosslinked to the nylon by using the Stratagene Stratalinker. Acrylamide (6%), 7 M urea gels were electrophoretically transferred onto nylon (Hybond-N, Amersham Pharmacia) in 1× TBE (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3) at 400 mA for 4 h and UV-crosslinked. After transfer, membranes were stained with methylene blue to verify loading and transfer. Membranes were hybridized for 12 h at 42°C in 500 mM phosphate buffer, 7% (wt/vol) SDS, 1 mM EDTA (pH 8.0), and 1% (wt/vol) BSA. About 106 cpm of probe was added to each hybridization mix. After incubation, the membranes were washed twice at room temp in 2× SSC and once in 0.1× SSC/0.1% SDS at 65°C, exposed to storage phosphor screens, and scanned with a phosphorimager for analysis.
Radioactive probes were generated by denaturing 100 ng of gel-purified PCR product in the presence of 5 pmols of primer as indicated at 60°C for 10 min. After cooling on ice for 5 min, extensions were carried out in 50 mM Tris·Cl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.05 mg/ml BSA, 5 units of exonuclease-free Klenow, 50 μCi (1 Ci = 37 GBq) of [α-32P]dATP, 80 μM dCTP, 80 μM dGTP, 80 μM dTTP, and 0.5 μM dATP at 37°C for 15 min. After extension, unincorporated nucleotides were removed by passing the probes over a G-50 Sephadex resin.
Mapping Ribose Methylation by Primer Extension.
Primer extension using reverse transcriptase under limiting dNTPs to test for ribose methylation in 18S rRNA was done as described in ref. 20. The position of the modification stop was determined by measuring the distance from the 5’ end of the priming oligo using a radioactive 10-bp marker.
5’-End Mapping of Promoter-Associated RNAs IMD2 and LEU4.
Ten micrograms of total RNA from the indicated strains was annealed with 0.3 pmols of32P-kinased oligo at 60°C for 10 min and then snap-cooled on ice. After reverse transcription [30 min at 42°C in 50 mM Tris·Cl (pH 8.0)/60 mM NaCl/5 mM MgCl2/10 mM DTT/0.4 mM dNTPs/50 units of SuperScript II reverse transcriptase (Invitrogen)], the reactions were extracted, ethanol-precipitated, and resolved on a 6% polyacrylamide, 7 M urea gel.
Induction of IMD2 by 6-Azauracil Treatment.
Yeast strain YSC1021-551682 was grown to saturation in SCD-Ura, diluted to an OD600 of 0.8, and induced with 6-azauracil (75 μg/ml) at 26°C for the times indicated in Fig. 4. Total RNA (5 μg) from each time point was loaded onto an agarose-formaldehyde gel, blotted, and probed with DNA generated from amplification of chromosome 8 between positions 553716 and 554377 and Klenow extension by using 5’-gtggagttattaattgcactg as the priming oligo.
Supplementary Material
Acknowledgments
We thank Joe DeRisi for providing microarrays, Tyson Clark and Karen Artiles for hybridizations in the early stages of this work, Leslie Grate for computational help, Wayne Decatur for strain YWD452, Todd Lowe for sharing his knowledge of snoRNAs, Grant Hartzog and John Tamkun for insights into transcription, and Marv Wickens for encouraging words. This work was supported by National Institutes of Health Grant GM040478.
Abbreviations
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
Footnotes
References
- 1.Velculescu V. E., Zhang L., Zhou W., Vogelstein J., Basrai M. A., Bassett D. E., Jr., Hieter P., Vogelstein B., Kinzler K. W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P., Cawley S. E., Drenkow J., Bekiranov S., Strausberg R. L., Fodor S. P., Gingeras T. R. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 3.Schadt E. E., Edwards S. W., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K. W., Russell A., Li G., et al. Genome Biol. 2004;5:R73. doi: 10.1186/gb-2004-5-10-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Wang X., Xia M., Stolc V., Su N., Peng Z., Li S., Wang J., Wang X., Deng X. W. Genome Biol. 2005;6:R52. doi: 10.1186/gb-2005-6-6-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stolc V., Samanta M. P., Tongprasit W., Sethi H., Liang S., Nelson D. C., Hegeman A., Nelson C., Rancour D., Bednarek S., et al. Proc. Natl. Acad. Sci. USA. 2005;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M. F., Rifkin S. A., Hua S., Herreman T., Tongprasit W., Barbano P. E., et al. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 8.Burckin T., Nagel R., Mandel-Gutfreund Y., Shiue L., Clark T. A., Chong J. L., Chang T. H., Squazzo S., Hartzog G., Ares M., Jr. Nat. Struct. Mol. Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 9.Clark T. A., Sugnet C. W., Ares M., Jr. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 10.Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Hoof A., Lennertz P., Parker R. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuai L., Fang F., Butler J. S., Sherman F. Proc. Natl. Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., Libri D., Jacquier A. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 14.Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer V. R., Horak C. E., Scafe C. S., Botstein D., Snyder M., Brown P. O. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 18.Chanfreau G., Legrain P., Jacquier A. J. Mol. Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 19.Ghazal G., Ge D., Gervais-Bird J., Gagnon J., Abou Elela S. Mol. Cell. Biol. 2005;25:2981–2994. doi: 10.1128/MCB.25.8.2981-2994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe T. M., Eddy S. R. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 21.Martens J. A., Laprade L., Winston F. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 22.Martens J. A., Wu P. Y., Winston F. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martens J. A., Winston F. Genes Dev. 2002;16:2231–2236. doi: 10.1101/gad.1009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan C. D., Laprade L., Winston F. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt S., Prestel M., Paro R. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw R. J., Reines D. Mol. Cell. Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exinger F., Lacroute F. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 28.van Hoof A., Frischmeyer P. A., Dietz H. C., Parker R. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y., Kohlhaw G. B. J. Biol. Chem. 1995;270:5270–5275. doi: 10.1074/jbc.270.10.5270. [DOI] [PubMed] [Google Scholar]
- 30.Escobar-Henriques M., Daignan-Fornier B., Collart M. A. Mol. Cell. Biol. 2003;23:6267–6278. doi: 10.1128/MCB.23.17.6267-6278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw R. J., Wilson J. L., Smith K. T., Reines D. J. Biol. Chem. 2001;276:32905–32916. doi: 10.1074/jbc.M105075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schattner P., Decatur W. A., Davis C. A., Ares M., Jr., Fournier M. J., Lowe T. M. Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyle J. W., Shaw R. J., Reines D. J. Biol. Chem. 2003;278:28470–28478. doi: 10.1074/jbc.M303736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.