Abstract
Type II collagen is the major cartilage matrix protein in the jawed vertebrate skeleton. Lampreys and hagfishes, by contrast, are thought to have noncollagenous cartilage. This difference in skeletal structure has led to the hypothesis that the vertebrate common ancestor had a noncollagenous skeleton, with type II collagen becoming the predominant cartilage matrix protein after the divergence of jawless fish from the jawed vertebrates ≈500 million years ago. Here we report that lampreys have two type II collagen (Col2α1) genes that are expressed during development of the cartilaginous skeleton. We also demonstrate that the adult lamprey skeleton is rich in Col2α1 protein. Furthermore, we have isolated a lamprey orthologue of Sox9, a direct transcriptional regulator of Col2α1 in jawed vertebrates, and show that it is coexpressed with both Col2α1 genes during skeletal development. These results reveal that the genetic pathway for chondrogenesis in lampreys and gnathostomes is conserved through the activation of cartilage matrix molecules and suggest that a collagenous skeleton evolved surprisingly early in vertebrate evolution.
Keywords: cartilage, development, gene duplication, gnathostome, phylogenetics
The earliest known vertebrates are jawless fishes that date to the Lower Cambrian (≈520 million years ago) (1). The remarkable preservation of these fossils has revealed the morphological patterns of the earliest vertebrate skeletons, however, little is known about the developmental processes or the molecular mechanisms underlying the evolutionary origin of the skeleton. In the cartilage of jawed vertebrates (gnathostomes), the major extracellular matrix molecule is type II collagen (Col2α1). By contrast, the living jawless fish (lampreys and hagfishes), which are sister groups to the gnathostomes, have been reported to lack collagen-based cartilage (2, 3). In lampreys and hagfishes, some skeletal elements are composed of unusual matrix proteins, called, respectively, lamprin and myxinin, which are distantly related to, but share some structural properties with, vertebrate elastin proteins (4, 5). This difference in skeletal structure has raised the hypothesis that the collagenous skeleton of vertebrates evolved relatively recently, within the gnathostome lineage (and is therefore a synapomorphy of this group). According to this hypothesis, the skeleton of the vertebrate common ancestor would have lacked collagen protein (3).
Collagens are estimated to have appeared in the late Proterozoic (≈800 million years ago) and then diversified into two major groups, the fibril-forming and nonfibril-forming collagens (6). The fibril-forming group, consisting of collagen types I, II, III, V, and XI, share a high degree of sequence and structural similarity and provide stiffness to a variety of tissues, whereas the nonfibril-forming group is structurally and functionally heterogeneous (6–8). The former then split into three phylogenetic clades, designated type A for collagens I, II, III, and Vα2, type B for collagens Vα1, Vα3, Vα4, Vα5, and XI, and type C for collagens XXIV and XXVII (6, 9). During chondrocyte differentiation in gnathostomes, expression of the Col2α1 gene leads to secretion of Col2α1 protein into the extracellular matrix, where it mineralizes. Expression of Col2α1 is regulated directly by Sox9, which binds to a chondrocyte-specific enhancer to activate Col2α1 transcription (10, 11). The Sox family of transcription factors consists of at least 10 subgroups, designated A–J, that are characterized by a specific 79-aa DNA-binding region, termed the high-mobility-group box (12). Sox9, a member of the SoxE subgroup, is required for Col2α1 expression and for chondrogenesis in jawed vertebrates (13, 14). Mutation of Sox9 in zebrafish disrupts the stacking of chondrocytes and the separation and shaping of individual cartilage elements and, in humans, causes campomelic dysplasia, which is characterized by slender, bowed long bones, scoliosis, and underdevelopment of the limb girdles and facial skeleton (15, 16).
Despite the different compositions of lamprey and gnathostome cartilage matrix, the genetic pathway that regulates early development of the cranial skeleton is well conserved (17–20). This conservation raises the question of how the same cascade of gene expression can lead to activation of different cartilage matrix gene targets in these two lineages. Although studies of lamprey cartilage matrix have identified noncollagenous proteins in particular skeletal elements (5, 21, 22), it is not clear whether these exist in place of, or in addition to, collagen. Indeed, comparative anatomical studies from the 19th century identified true hyaline cartilage in lampreys and noted striking structural similarities to gnathostome cartilage (23), suggesting that there may be hitherto undiscovered molecular similarities in lamprey and gnathostome cartilage matrix. Here we revisit the evolutionary origin of collagenous cartilage from a molecular developmental perspective, and we report that development of the lamprey skeleton involves type II collagen. Our experiments show that lampreys have two type II collagen genes, and that both are expressed in the developing skeleton. We also find Col2α1 protein in cranial and postcranial cartilages. We go on to show that lampreys have an orthologue of Sox9 that is coexpressed with both Col2α1 genes during development of the lamprey skeleton. Thus, we conclude that lampreys have collagen-based cartilage, and that the genetic pathway for chondrogenesis is conserved in lampreys and gnathostomes from earliest Sox9 expression through cartilage matrix gene activation. The results indicate that a collagenous skeleton evolved before the divergence of the lamprey and gnathostome lineages and suggest that collagen-based cartilage may be a unifying character of crown vertebrates.
Results
Lampreys Have Two Col2α1 Orthologues.
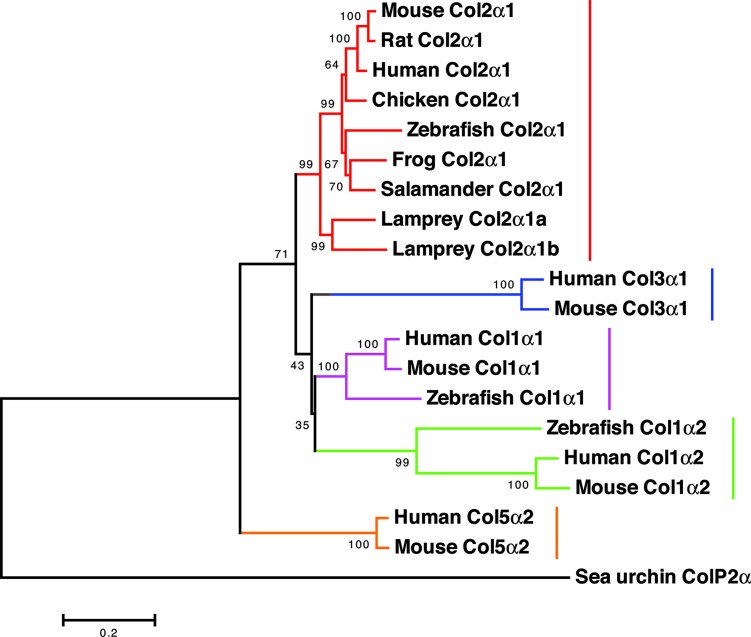
As a first step toward resolving whether collagen genes are involved in lamprey skeletal development, we searched for expressed orthologues of Col2α1 by using a degenerate PCR screen of a Petromyzon marinus embryonic cDNA library. We isolated two 1.74-kb clones with 75% nucleotide sequence identity to one another, and their deduced amino acid sequences were 80% identical to mouse Col2α1. Absence of gaps in the nucleotide alignments suggested that the two transcripts were not splice variants from a single gene. Molecular phylogenetic analyses with maximum parsimony (MP), minimum evolution (ME), maximum likelihood (ML), and Bayesian phylogenetics (BP) methods consistently placed the lamprey sequences in the vertebrate Col2α1 family and supported their position as a sister group to the gnathostome Col2α1 clade (Fig. 1; see also Fig. 6, which is provided as supporting information on the PNAS web site). These results indicate the presence of two Col2α1 genes in lamprey, which we designate Col2α1a and Col2α1b. These results represent a previously undescribed demonstration of true type II collagen orthologues outside of jawed vertebrates and reveal that an independent duplication of the ancestral Col2α1 gene occurred within the lamprey lineage.
Fig. 1.
Minimum evolution phylogeny for fibril A collagen proteins as obtained with JTT plus Γ distances (α = 0.906). Numbers indicate bootstrap scores for each node, based on 1,000 replicates, whereas branch lengths are proportional to expected replacements per site. This tree is rooted by sea urchin ColP2α. Equally and unequally weighted MP, ML, and BP also place the two lamprey sequences together at the base of the Col2α1 clade, with bootstrap scores or a posterior probability of 93%, 77%, 71%, and 87%, respectively. In contrast to this consistent support for a Col2α1 grouping, this ME tree is unique among the different phylogenies in its placement of the root along the Col5 clade. This discrepancy over the root is illustrated with the BP phylogeny and discussed further in Fig. 6.
Col2α1a and Col2α1b Are Expressed During Lamprey Chondrogenesis.
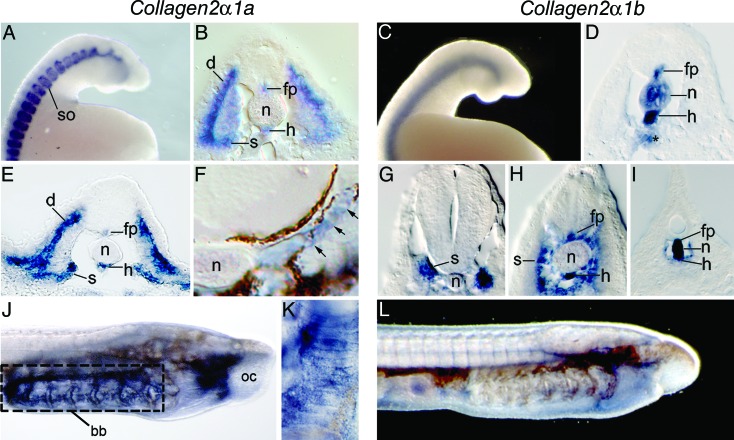
If the ancestral Col2α1 gene was involved in skeletogenesis in the common ancestor of lampreys and gnathostomes, then an evolutionary signature may be detectable during lamprey embryonic development. We therefore examined whether Col2α1a and Col2α1b are expressed in tissues that give rise to the lamprey skeleton. Using whole mount in situ hybridization, we found that Col2α1a is expressed during somitogenesis in a segmental pattern along the lamprey trunk (Fig. 2A). Histological sections showed that Col2α1a transcripts in the somites were localized ventromedially to the sclerotome, which gives rise to the axial skeleton, and dorsolaterally to the dermatome, which gives rise to the dermis (Fig. 2B). In the midline, lamprey Col2α1a expression was observed in the floor plate of the neural tube and in the hypochord, but not in the notochord (Fig. 2B). This pattern represents a subset of the zebrafish Col2α1 expression pattern, which occurs along the midline in three domains, the floor plate, notochord, and hypochord (24). The second lamprey orthologue, Col2α1b, was expressed in a pattern similar to Col2α1a along the anteroposterior axis and in the midline; however, Col2α1b transcripts also localized to the notochord and to endoderm immediately ventral to the hypochord (Fig. 2 C and D).
Fig. 2.
Col2α1a and Col2α1b expression during lamprey development. Whole mount in situ hybridization of lamprey embryos at stages 23 (A–D), 25 (E), 26 (G–I), and 30 (F and J–L). Anterior is to the right in (A, C, J, and L); All sections are transverse with dorsal to the top. (A and B) Col2α1a expression is evident in the somites and within the dermatome and sclerotome. Expression in the midline is restricted to floor plate and hypochord (B). (C and D) Col2α1b is expressed in the floor plate, notochord, hypochord, and dorsal endoderm (asterisk in D). (E) Col2α1a expression in the dermatome, sclerotome, floor plate, and hypochord. (F) Col2α1a expression in a prevertebral condensation (arrows). (G–I) Sections through the hindbrain (G), mid-trunk (H), and tail (I) show an anterior to posterior retraction of the Col2α1b domain in the notochord, hypochord, and floor plate. (J) Col2α1a is expressed throughout the branchial skeleton (boxed) and posterior to the oral cavity. (K) Col2α1a expression in a stack of chondrocytes in a branchial bar. (L) Col2α1b is not detected in the branchial skeleton. bb, branchial basket; d, dermatome; fp, floor plate; h, hypochord; n, notochord; oc, oral cavity; s, sclerotome; so, somite.
During differentiation of the lamprey skeleton, both Col2α1a and Col2α1b were expressed in chondrogenic cells. The axial skeleton of lampreys consists of a series of paired cartilaginous vertebrae or arcualia, which are serially repeated on either side of the notochord and have long been thought to be homologous to the neural arches of gnathostome vertebrae (25). Col2α1a expression persisted in sclerotomal cells (Fig. 2E) and could be seen in the prevertebral condensations by stage 30 (Fig. 2F). Col2α1b expression appeared in the sclerotome by stage 26, but expression in the midline was down-regulated anteriorly (Fig. 2 G–I). In the developing head skeleton, Col2α1a expression was detected throughout the cartilaginous branchial basket and posterior to the oral cavity (Fig. 2J). Transcripts were most abundant in the chondrocyte stacks that make up each gill bar (Fig. 2K). By contrast, Col2α1b expression could not be detected in the pharyngeal arches (Fig. 2L). During development of the median fin skeleton, Col2α1a and Col2α1b were expressed, respectively, in the posterior and anterior parts of the median finfold (data not shown). Thus, both lampreys and jawed vertebrates exhibit widespread expression of Col2α1 genes during skeletal development.
Adult Lamprey Cartilage Contains Col2α1 Protein.
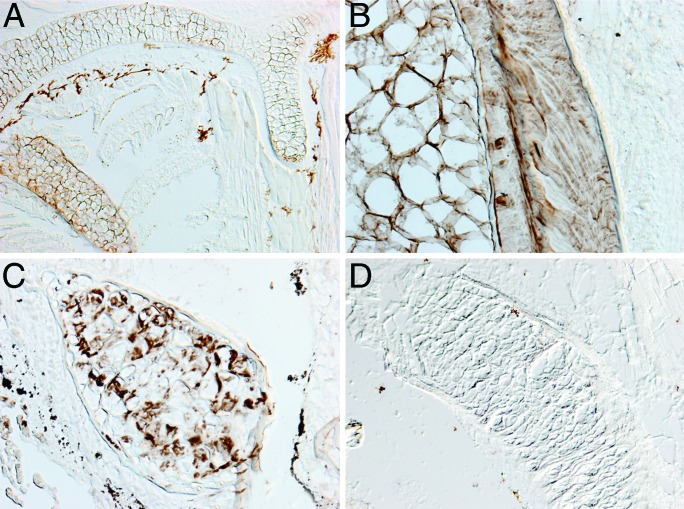
The discovery that two Col2α1 genes are transcribed during lamprey skeletal development raised the possibility that lampreys may possess a collagenous skeleton. As a direct test of whether lamprey differentiated cartilage contains type II collagen protein, we performed immunohistochemical analysis of an adult by using an antibody specific to Col2α1. We detected Col2α1 protein in the extracellular matrix of pharyngeal cartilages, notochord, notochordal sheath, and arcualia (Fig. 3), confirming that Col2α1 protein is abundant in the lamprey skeleton. These results demonstrate that a collagenous skeleton evolved before the divergence of the lamprey and gnathostome lineages and suggest that Col2α1 was involved in skeletal development at least as early as the common ancestor of crown vertebrates.
Fig. 3.
Col2α1 protein is abundant in adult lamprey cartilage. Immunohistochemical staining of lamprey cartilage with Col2α1 antibody. (A) Sagittal section through pharyngeal cartilage bars. (B) Transverse section through notochord. Note staining of matrix in notochord and notochordal sheath. (C) Transverse section through arcualia on ventrolateral side of notochord. (D) Control section through pharyngeal cartilage, after omission of the primary antibody.
Upstream of Col2α1: A Lamprey Orthologue of Sox9.
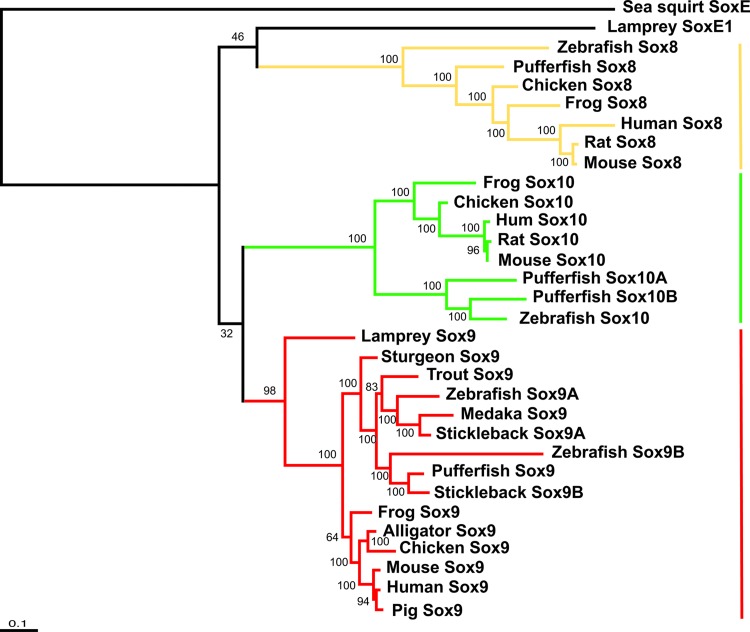
In gnathostomes, transcription of Col2α1 is regulated directly by Sox9, and these two genes are coexpressed during chondrogenesis (14, 15, 24, 26). Given the critical role of Sox9 in the development of cartilages derived from both neural crest and mesoderm, we next asked whether lampreys have a Sox9 orthologue. Using degenerate RT-PCR, we isolated a 1.3-kb clone whose inferred amino acid sequence most closely matched those of the gnathostome Sox9 proteins. For example, this inferred protein sequence was 96% identical to human Sox9 within the high-mobility-group box (Fig. 7, which is provided as supporting information on the PNAS web site). Furthermore, this amino acid sequence included the Sox9-specific signature motif (12, 27) that occurs immediately 3′ to the high-mobility-group domain. Molecular phylogenetic analyses consistently joined this lamprey protein to the base of the Sox9 clade, with BP providing a posterior probability of 98% for this assignment (Fig. 4). In concert with its overlapping gene expression pattern with that of Sox9 in jawed vertebrates (see below), these results collectively provide support for the designation of this clone as lamprey Sox9.
Fig. 4.
Extended majority-rule consensus tree for the BP analysis of the chordate SoxE proteins. Numbers indicate posterior probabilities for groups with >50% credibility and for those clades that are combinable with this first set. Branch lengths are proportional to the means of the posterior probability densities for their expected replacements per site. This tree is rooted by sea squirt SoxE. Equally and unequally weighted MP, ME, and ML also place the previously undescribed lamprey sequence at the base of the Sox9 clade, with bootstrap scores of 86%, 74%, 28%, and 75%, respectively. The surprisingly low bootstrap score for ME is related to our complete deletion of gapped positions in the pairwise distance calculations (Fig. 7). Despite this low score, the optimal ME phylogeny still supports as best a Sox9 assignment for the previously undescribed lamprey protein and is therefore consistent with the other phylogenetic results.
Sox9 Expression Colocalizes with Col2α1a and Col2α1b in the Developing Skeleton.
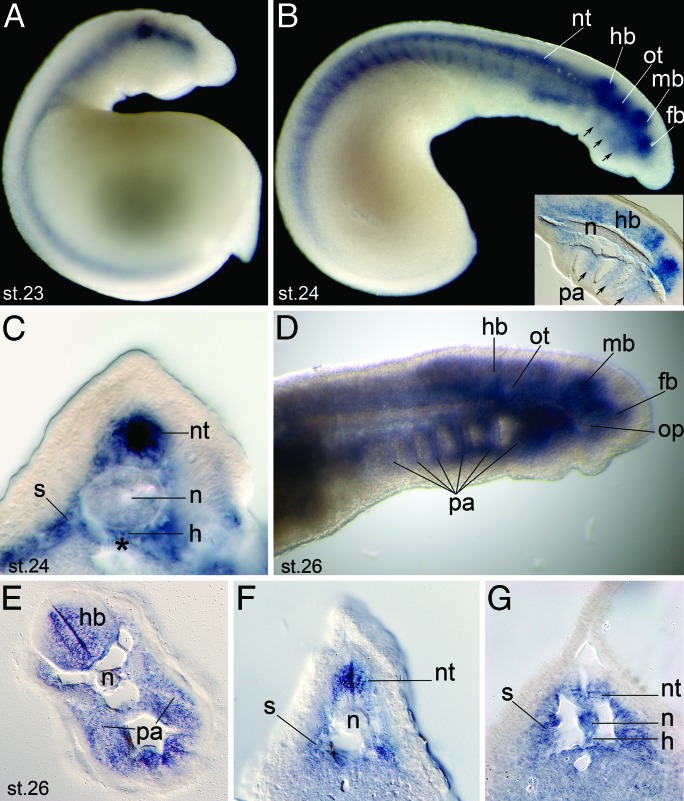
We next investigated whether lamprey Sox9 is expressed in a pattern consistent with a role in regulation of Col2α1. Lamprey Sox9 expression along the primary body axis resembled the lamprey Col2α1b pattern at stage 23 (compare Fig. 5A with Fig. 2C). Transcripts were localized to the ventral neural tube, notochord, and hypochord along the midline and the sclerotome and dorsal endoderm at stage 24 (Fig. 5C). Neural expression of Sox9 extended from the spinal cord to the forebrain (Fig. 5B). At stage 23, the pharyngeal arches were negative for Sox9; however, by stage 24, expression was detected in streams of neural crest cells extending from the hindbrain toward the arches (Fig. 5B). At stage 26, Sox9 was expressed throughout the developing branchial basket and in the otic and optic placodes (Fig. 5D). Like Col2α1b, Sox9 expression in the notochord later retracted from anterior to posterior, beginning at stage 25, but transcripts remained in sclerotome and neural tube (Fig. 5 E–G). Thus, lamprey Sox9 expression colocalizes with Col2α1 transcripts during chondrogenesis and closely follows the pattern described for gnathostomes (14, 26).
Fig. 5.
Sox9 expression during lamprey development. (A and B) Expression of lamprey Sox9 in embryos at stages 23 (A) and 24 (B). (B Inset) Sagittal section through hindbrain and pharyngeal arches are shown. Arrows indicate Sox9 expression in streams of neural crest cells invading pharyngeal arches. (C) Transverse section through trunk of embryo shown in B. Note Sox9 expression in the notochord, hypochord, neural tube, sclerotome, and dorsal endoderm (asterisk). (D) Sox9 expression in stage 26 lamprey head. (E–G) Sections through the hindbrain (E), mid-trunk (F), and tail (G) at stage 26 show an anterior to posterior retraction of the Sox9 domain in the notochord. Note the persistent expression in neural tube, pharyngeal arch mesenchyme, sclerotome, and hypochord. fb, forebrain; hb, hindbrain; h, hypochord; mb, midbrain; n, notochord; nt, neural tube; op, optic placode; ot, otic placode; pa, pharyngeal arches; s, sclerotome.
Discussion
The data presented here demonstrate that lampreys have two orthologues of Col2α1 that are expressed during development of the cartilagenous skeleton. Our discovery of type II collagen protein throughout the adult lamprey skeleton challenges the view that collagen-based cartilage is a gnathostome character and indicates that a collagenous skeleton evolved before the divergence of lampreys and gnathostomes. Expression of lamprey Sox9 in chondrogenic cells suggests that a common suite of genes is targeted during skeletal differentiation in jawed and jawless vertebrates. We conclude that a collagenous skeleton is a shared, derived feature of all vertebrates and not of jawed vertebrates only.
Molecular Evolution of Lamprey Collagens.
Our molecular phylogenetic analyses of fibril A collagens lead us to conclude that Col1 to Col5 duplicated before the speciation of lampreys and jawed vertebrates. Thus, one prediction of our phylogenetic results is that future studies will recover lamprey orthologues of Col1, Col3, and/or Col5, which should join at the bases of their respective gnathostome clades in the fibril A collagen tree (Fig. 1). An independent duplication of the Col2α1 gene then occurred in the lamprey lineage. The expression patterns of Col2α1a and Col2α1b suggest that subfunctionalization followed this duplication of the ancestral Col2α1 gene, with the ancestral expression pattern being partitioned between Col2α1a and Col2α1b. Force and colleagues (28, 29) proposed a mechanism by which duplicated genes are preserved during evolution by subfunctionalization, in which the combined expression patterns of both duplicated genes equal that of their single ancestral gene. Our finding that the expression domains of Col2α1a and Col2α1b in lampreys correspond to that of the single Col2α1 gene in jawed vertebrates is consistent with their duplication-degeneration-complementation model.
It is also noteworthy that collagen genes are physically linked to the Hox clusters in gnathostomes (6, 30). In lampreys, there are at least three Hox clusters (31–33). Our identification of two Col2α1 genes supports the suggestion that one of their three Hox clusters may have arisen by an independent duplication in the lamprey lineage (31–33). If type A fibril collagens are linked with Hox clusters in all chordates, then based on the presence of a single Hox cluster in amphioxus, we predict that (barring tandem duplications) cephalochordates may possess a single type A fibril collagen gene.
Evolution of Collagen-Based Cartilage.
Localization of type II collagen mRNA and protein in the lamprey skeleton reveals that a collagenous skeleton is not restricted to the gnathostome lineage, but instead is a character shared by the crown vertebrates. This result may provide a molecular explanation for Parker’s observation in 1883 that lampreys have hard hyaline cartilage (23). We suggest that the additional cartilage matrix molecules (e.g., lamprin and myxinin) of agnathans may represent derived character states that were added onto the more ancient collagenous skeleton. Expression of elastin-related molecules in a subset of lamprey cranial cartilages that also express Col2α1 may underlie the different structural and mechanical properties within the lamprey skeleton. In comparing lamprey Col2α1a and Col2α1b with zebrafish Col2α1, we detected only one major difference, the unexpected localization of lamprey Col2α1a to the dermatome. Given that vertebrate dermis is generally characterized by expression of type I collagen (8), this difference may represent a derived feature of lampreys.
Five independent molecular phylogenetic analyses joined lamprey Sox9 to the gnathostome Sox9 clade. Sox9 is expressed in strikingly similar patterns in embryonic lampreys and gnathostomes, including the zebrafish, which have two copies of the gene that collectively make up the generalized vertebrate expression domain (14). The coexpression of Sox9 with Col2α1 during skeletogenesis in both lineages raises the possibility that the regulatory relationship between these two genes had already been established in their common ancestor. In gnathostomes, Sox9 is a target of PTH-related protein (PTHrP), which regulates chondrocyte differentiation through a negative feedback loop with Indian hedgehog (34, 35). Interestingly, PTHrP expression has recently been detected in lamprey cartilage (36). Our discovery of conserved expression of Sox9 and Col2α1, taken together with the extensive conservation of upstream regulatory genes such as AP2, Dlx, Msx, Id, and PTHrP (17–20, 36), suggests that the genetic program for chondrogenesis, from the initial induction of chondrogenic mesenchyme to the synthesis of collagen matrix, was assembled surprisingly early in vertebrate evolution. Comparative analyses of fibrillar collagen and SoxE genes in hagfishes, amphioxus, ascidians, and hemichordates will further refine the evolutionary history of the collagenous skeleton.
Materials and Methods
Gene Cloning and Sequence Analyses.
Degenerate RT-PCR was performed to amplify fragments of lamprey Col2α1 and Sox9 orthologues from a P. marinus cDNA library (37). PCR products were cloned into pDrive vector (Qiagen, Valencia, CA) and sequenced in both directions. These sequences have been submitted to GenBank (accession numbers: DQ136023DQ136024–DQ136025). The inferred protein sequences for the previously undescribed lamprey cDNAs were initially assigned to the Col2α1 and Sox9 families on the basis of blast searches and conserved domains (12, 27, 38). These initial assignments were followed by estimates of their amino acid identities and phylogenetic relationships. Multiple sequence alignments for available fibrillar A collagens and SoxE proteins, including the previously undescribed lamprey sequences, were generated with clustal x and then refined according to their known tertiary structures (12, 27, 38). GenBank accession numbers for all sequences used in these phylogenetic analyses can be found in Table 1, which appears as supporting information on the PNAS web site. Phylogenetic analyses of these multiple protein alignments were conducted with MP, ME, ML, and BP methods (39). The MP analyses included both the equal and unequal weighting of amino acid replacements, with the latter relying on the “ProtPars” cost matrix. The former relied on branch-and-bound searches, whereas the latter was based on heuristic ones with tree-bisection-and-reconnection branch swapping and 1,000 starting trees that were generated from different random sequence additions. The pairwise distances in ME were corrected for multiple replacements with the JTT rate matrix and the gamma (Γ) distribution for site-to-site heterogeneity in rates. The ME analyses relied on heuristic searches with close-neighbor-interchanges starting from neighbor-joining trees. The ML and BP analyses also relied on Γ, but in combination with the improved WAG rate matrix available in their computer programs (but not in that for ME; see below). In ML and BP, the α parameter for Γ was estimated, whereas it was fixed to its ML estimates in ME. The ML analyses relied on a fast heuristic procedure that simultaneously searched for both optimal branch lengths and topologies. The BP analyses were based on three independent runs of 2 million generations and one additional confirmational run of 10 million generations apiece, with each run consisting of one cold and three heated chains (T = 0.2) and with samples taken from the former every 100 generations. The reliability of groups was evaluated in MP, ME, and ML with 1,000 bootstrap replicates apiece and, in BP, with posterior probabilities that were calculated after discarding the first 1,000 samples of each run as burnin. The MP, ME, ML, and BP analyses were conducted with paup*4.0b10 (40), mega3 (41), phyml2.4.4 (42), and mrbayes3.1.1 (43), respectively.
In Situ Hybridization.
Whole mount in situ hybridization was performed as described for chick embryos (44) with the following modifications: embryos were treated with proteinase K (10 μg/ml) for 15–30 min at room temperature, and 10% dimethylformamide was added to color reaction solution. For histological analysis, specimens were equilibrated in 15% sucrose then 30% sucrose in 20% gelatin, after which they were embedded in 20% gelatin for cryosectioning (10 μm).
Immunohistochemistry.
Lamprey specimens were fixed in 4% paraformaldehyde or 70% ethanol and processed for paraffin sectioning by using standard methods. Sections were cut at 6 μm, and antigen retrieval was performed by autoclaving slides in 0.01 M citrate buffer (pH 6). Antibody staining was performed by using the Vectastain ABC kit according to manufacturer’s instructions. Primary antibodies against human Col2α1 (Santa Cruz Biotechnology) were used at concentrations of 1:500–1:1,000.
Supplementary Material
Acknowledgments
We thank Renata Freitas (University of Florida) for assistance and advice; Jim Langeland (Kalamazoo College, Kalamazoo, MI) for generously sharing the cDNA library; Phyllis Luvalle (University of Florida) for sharing reagents; and Larry Page, Rob Robbins (Florida Museum of Natural History), and Gordon Weddle (Campbellsville University, Campbellsville, KY) for assistance with specimen collection.
Abbreviations
- BP
Bayesian phylogenetics
- ME
minimum evolution
- ML
maximum likelihood
- MP
maximum parsimony
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. A.S. is a guest editor invited by the Editorial Board.
References
- 1.Shu D.-G., Luo H.-L., Conway Morris S., Zhang X.-L., Hu S.-X., Chen L., Han J., Zhu M., Li Y., Chen L.-Z. Nature. 1999;402:42–46. [Google Scholar]
- 2.Wright G. M., Youson J. H. Am. J. Anat. 1983;167:59–70. doi: 10.1002/aja.1001670106. [DOI] [PubMed] [Google Scholar]
- 3.Wright G. M., Keeley F. W., Robson P. Cell Tissue Res. 2001;304:165–174. doi: 10.1007/s004410100374. [DOI] [PubMed] [Google Scholar]
- 4.Robson P., Wright G. M., Keeley F. W. Anat. Embryol. 2000;202:281–290. doi: 10.1007/s004290000113. [DOI] [PubMed] [Google Scholar]
- 5.Robson P., Wright G. M., Sitarz E., Maiti A., Rawat M., Youson J. H., Keeley F. W. J. Biol. Chem. 1993;268:1440–1447. [PubMed] [Google Scholar]
- 6.Morvan-Dubois G., Le Guellec D., Garrone R., Zylberberg L., Bonnaud L. J. Mol. Evol. 2003;57:501–514. doi: 10.1007/s00239-003-2502-x. [DOI] [PubMed] [Google Scholar]
- 7.Exposito J. Y., Cluzel C., Garrone R., Lethias C. Anat. Rec. 2002;268:302–316. doi: 10.1002/ar.10162. [DOI] [PubMed] [Google Scholar]
- 8.van der Rest M., Garrone R. FASEB J. 1991;5:2814–2823. [PubMed] [Google Scholar]
- 9.Boot-Handford R. P., Tuckwell D. S., Plumb D. A., Rock C. F., Poulsom R. J. Biol. Chem. 2003;278:31067–31077. doi: 10.1074/jbc.M212889200. [DOI] [PubMed] [Google Scholar]
- 10.Bell D. M., Leung K. K., Wheatley S. C., Ng L. J., Zhou S., Ling K. W., Sham M. H., Koopman P., Tam P. P., Cheah K. S. Nat. Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. Mol. Cell. Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowles J., Schepers G., Koopman P. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 13.Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 14.Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M., Postlethwait J. H. Development (Cambridge, U.K.) 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 15.Yan Y. L., Miller C. T., Nissen R. M., Singer A., Liu D., Kirn A., Draper B., Willoughby J., Morcos P. A., Amsterdam A., et al. Development (Cambridge, U.K.) 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- 16.Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E., et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 17.Neidert A. H., Virupannavar V., Hooker G. W., Langeland J. A. Proc. Natl. Acad. Sci. USA. 2001;98:1665–1670. doi: 10.1073/pnas.98.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohn M. J. Nature. 2002;416:386–387. doi: 10.1038/416386a. [DOI] [PubMed] [Google Scholar]
- 19.McCauley D. W., Bronner-Fraser M. Development (Cambridge, U.K.) 2003;130:2317–2327. doi: 10.1242/dev.00451. [DOI] [PubMed] [Google Scholar]
- 20.Shigetani Y., Sugahara F., Kawakami Y., Murakami Y., Hirano S., Kuratani S. Science. 2002;296:1316–1319. doi: 10.1126/science.1068310. [DOI] [PubMed] [Google Scholar]
- 21.McBurney K. M., Keeley F. W., Kibenge F. S., Wright G. M. Anat. Embryol. 1996;193:419–426. doi: 10.1007/BF00185873. [DOI] [PubMed] [Google Scholar]
- 22.McBurney K. M., Keeley F. W., Kibenge F. S., Wright G. M. Biotech. Histochem. 1996;71:44–53. doi: 10.3109/10520299609117130. [DOI] [PubMed] [Google Scholar]
- 23.Parker W. Philos. Trans. R. Soc. London B. 1883;174:411–457. [Google Scholar]
- 24.Yan Y. L., Hatta K., Riggleman B., Postlethwait J. H. Dev. Dyn. 1995;203:363–376. doi: 10.1002/aja.1002030308. [DOI] [PubMed] [Google Scholar]
- 25.Gadow H. F. The Evolution of the Vertebral Column: A Contribution to the Study of the Vertebrate Phylogeny. Cambridge: Cambridge Univ. Press; 1933. [Google Scholar]
- 26.Ng L. J., Wheatley S., Muscat G. E., Conway-Campbell J., Bowles J., Wright E., Bell D. M., Tam P. P., Cheah K. S., Koopman P. Dev. Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 27.Koopman P., Schepers G., Brenner S., Venkatesh B. Gene. 2004;328:177–186. doi: 10.1016/j.gene.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch M., O’Hely M., Walsh B., Force A. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey W. J., Kim J., Wagner G. P., Ruddle F. H. Mol. Biol. Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- 31.Irvine S. Q., Carr J. L., Bailey W. J., Kawasaki K., Shimizu N., Amemiya C. T., Ruddle F. H. J. Exp. Zoolog. B. Mol. Dev. Evol. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 32.Force A., Amores A., Postlethwait J. H. J. Exp. Zoolog. B. Mol. Dev. Evol. 2002;294:30–46. doi: 10.1002/jez.10091. [DOI] [PubMed] [Google Scholar]
- 33.Fried C., Prohaska S. J., Stadler P. F. J. Exp. Zoolog. B. Mol. Dev. Evol. 2003;299:18–25. doi: 10.1002/jez.b.37. [DOI] [PubMed] [Google Scholar]
- 34.Huang W., Chung U.-I., Kronenberg H. M., de Crombrugghe B. Proc. Natl. Acad. Sci. USA. 2001;98:160–165. doi: 10.1073/pnas.011393998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M., Tabin C. J. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 36.Trivett M. K., Potter I. C., Power G., Zhou H., Macmillan D. L., John Martin T., Danks J. A. Dev. Genes Evol. 2005:1–11. doi: 10.1007/s00427-005-0015-x. [DOI] [PubMed] [Google Scholar]
- 37.Tomsa J. M., Langeland J. A. Dev. Biol. 1999;207:26–37. doi: 10.1006/dbio.1998.9163. [DOI] [PubMed] [Google Scholar]
- 38.lkkila M., Melkoniemi M., Kvist L., Kuivaniemi H., Tromp G., Ala-Kokko L. Matrix Biol. 2001;20:357–366. doi: 10.1016/s0945-053x(01)00145-7. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 40.Swofford D. L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 41.Kumar S., Tamura K., Nei M. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 42.Guindon S., Gascuel O. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 43.Ronquist F., Huelsenbeck J. P. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 44.Nieto M. A., Patel K., Wilkinson D. G. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.