Abstract
Although leukocytes of the innate immune system, including eosinophils, contain within their granules preformed stores of cytokines available for selective and rapid release, little is known about the mechanisms governing the mobilization and secretion of these cytokines. Here we show that a cytokine receptor, the IL-4 receptor α chain, mediates eotaxin-stimulated mobilization of preformed IL-4 from eosinophil granules into secretory vesicles. Eosinophils contain substantial intracellular quantities of several granule- and vesicle-associated cytokine receptors, including IL-4, IL-6, and IL-13 receptors as well as CCR3. Both IL-4 and IL-4 receptor α chain colocalized in eosinophil granules; and after eotaxin-stimulation, IL-4 receptor α chain, bearing bound IL-4, was mobilized into secretory vesicles. These findings indicate that intracellular cytokine receptors within secretory vesicles transport their cognate cytokines requisite for the secretion of cytokines preformed in innate immune leukocytes.
Keywords: intracellular cytokine receptor, intracellular cytokine trafficking, piecemeal degranulation, vesicular transport
Eosinophils, cells of the innate immune system, have varied effector and immunomodulatory functions in health and in the pathogeneses of asthma, allergies, and other diseases (1–4). Many activities of eosinophils are mediated by preformed proteins stored within cytoplasmic-specific granules. These eosinophil granules contain four quantitatively dominant cationic proteins, including major basic protein (MBP), whose extracellular release is associated with classical effector and proinflammatory roles of eosinophils (1). Eosinophil-specific granules, notably, are also sites of storage of many, if not all, of the several dozen cytokines that human eosinophils quite distinctly contain in preformed intracellular pools (5). These cytokines include IL-4 (6), IL-6 (7), IFN-γ (8), and the chemokines eotaxin (CCL11) and RANTES (regulated on activation, normal T cell expressed and secreted, CCL5) (9–11). With this armamentarium of diverse, preformed cytokines, eosinophils have the capacity to exert immunomodulatory actions in innate immunity by drawing upon preformed granule stores of cytokine proteins for their rapid release, thus bypassing the need for de novo synthesis requiring transcription and/or mRNA translation (4, 5). Rapid secretion of preformed cytokines by eosinophils can be important for innate immune responses. For example, murine eosinophils rapidly released IL-4, which initiated a type 2 response to parasite antigens (12). Eosinophils, even in mice deficient in B and T cells, as well as basophils, were the major IL-4-producing cells recruited to the lungs of Nippostrongylus brasiliensis-infected mice (13–15). Innate immune cells, including IL-4-expressing eosinophils, initiated polarized type 2 responses independent of adaptive immunity (13–15). Mediator release is highly agonist-specific [i.e., eotaxin elicits release of IL-4 but not IL-12 or IFN-γ (refs. 16 and 17 and data not shown)], raising the issue of how parsimony and selectivity in stimulus-induced mediator release are achieved. In contrast to eosinophil degranulation based on an exocytotic process by which whole granules fuse with the plasma membrane to extrude their entire contents, there is increasing recognition that eosinophils selectively release granule-derived components. Ultrastructural observations of tissue eosinophils in many inflammatory and allergic disorders have documented that eosinophil proteins may be differentially mobilized from within granules (18–20). This granule secretion process, termed piecemeal degranulation (PMD), relies on vesicular transport of preformed proteins from within granules to the plasma membrane for their extracellular release. We have identified a structural basis for eosinophil PMD with the delineation that, in response to eotaxin stimulation, IL-4 is mobilized into a vesiculotubular network within granules and then transported through the cytoplasm in both small round vesicles and larger curved and elongated tubular vesicles (21, 22). This secretion draws on preformed stores of IL-4 because neither actinomycin D nor cycloheximide, which were used to block gene transcription or protein translation, respectively, diminished eotaxin-elicited IL-4 release, and this secretion is selective because IL-12, also stored within eosinophil granules, was not secreted (17). Notably, within secretory vesicles, IL-4 was observed to be predominantly membrane-bound (22). Thus, we hypothesized that a membrane-bound mechanism of transport mediates the selectivity of preformed cytokine secretion.
In a little-appreciated correlation, eosinophils contain a multitude of preformed cytokines and chemokines and at the same time express receptors for most, if not all, of these cytokines and chemokines. Eosinophils can respond to IL-4, and both IL-4 receptor α chain (IL-4Rα) and the receptor common gamma (γc) chain have been detected at low levels on eosinophil plasma membranes, suggesting expression of at least the type I IL-4 receptor (23, 24). The expression of multiple receptors for cytokines secreted by eosinophils might simply reflect a capacity for autocrine signaling. Not recognized to date, however, are potential additional distinct roles for cytokine receptor components in contributing to the cytokine ligand-specific transport and secretion of preformed cytokines. Indeed, in all cells, intracellular transport processes governing cytokine secretion remain poorly understood.
Here, we demonstrate receptor protein-mediated mechanisms that mediate stimulus-induced mobilization and trafficking of IL-4 from granules into secretory vesicles. We show that in addition to nominal surface expression, all components of functional IL-4 receptor complexes, as well as the IL-6 receptor α chain and the chemokine receptor, CCR3, are predominantly expressed intracellularly within eosinophils. IL-4Rα and CCR3 proteins are principally resident within eosinophil granules. Furthermore, we demonstrate that, upon eosinophil stimulation, IL-4Rα, the IL-4-binding chain of the IL-4 receptor, is localized to the membranous tubulovesicular network within granules, is mobilized out of granules in concert with IL-4, and is bound to IL-4 within secretory vesicular compartments. This work provides delineation of the vesicular and membrane-bound, receptor-mediated mechanisms for the trafficking, transport, and secretion of a preformed eosinophil granule-derived cytokine, identifies previously unrecognized intracellular pools of cytokine receptors within human eosinophils, and reveals a unique mechanism contributing to the specificity of cytokine secretion.
Results
IL-4 Receptor Components Are Preformed Within Eosinophils, and IL-4Rα Is Predominantly Granule-Associated.
We previously demonstrated that eotaxin, signaling through CCR3, induced the selective release of IL-4 from preformed stores within eosinophils (17). Using α-IL-4 immunonanogold EM, we localized IL-4 to budding vesicles and vesicles trafficking through the cytoplasm. Intriguingly, in contrast to MBP staining within the lumen of eosinophil secretory vesicles (22), IL-4 labeling within both budding and free vesicles was preferentially associated with vesicle membranes (see Fig. 5, which is published as supporting information on the PNAS web site). Because IL-4 lacks a membrane-spanning or insertion region, IL-4’s association with vesicle and granule membranes suggested the presence of an IL-4-docking molecule. Thus, we hypothesized that a membrane-associated IL-4-docking protein orchestrates the selective sequestering and packaging of granule-derived IL-4 into secretory vesicles destined for release.
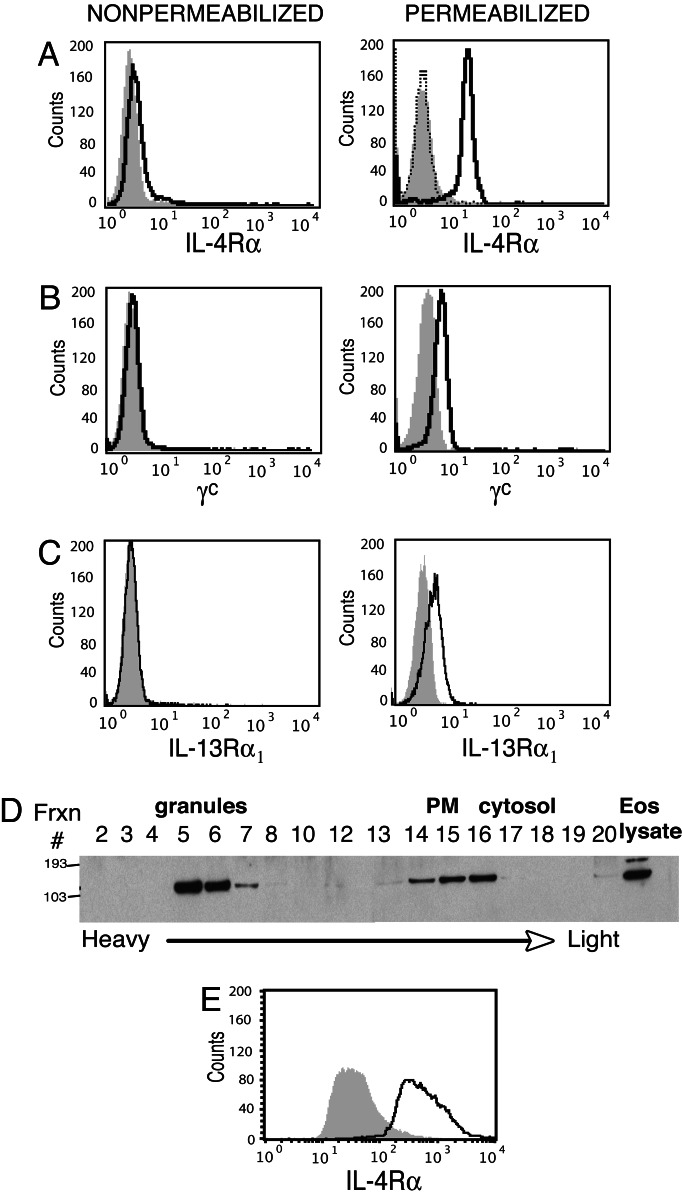
As noted, receptors specific for many eosinophil-derived mediators are expressed on human eosinophils, including the type I IL-4 receptor. IL-4Rα is the IL-4-binding component of all three types of functional IL-4 receptor complexes currently described, forming a heterodimer with γc or IL-13Rα1 chains to form type I or II receptors, respectively, or binding both accessory chains to form the type III receptor (25). To investigate the potential for IL-4 receptor components to function in the selective mobilization of its ligand, we used flow cytometry in parallel with immunoblotting of subcellular fractions to determine whether the expression of IL-4 receptor components was restricted to the cell surface. In agreement with a previous study reporting low IL-4Rα cell surface expression on eosinophils (23), freshly isolated, nonpermeabilized eosinophils expressed undetectable-to-low levels of IL-4Rα (Fig. 1A). However, eosinophils permeabilized with 0.1% saponin exhibited robust intracellular IL-4Rα staining, the specificity of which was ascertained by complete neutralization through preincubation of the anti-IL-4Rα Ab with its specific blocking peptide immunogen (Fig. 1A). Likewise, substantial intracellular IL-4Rα was demonstrable with Abs specific for peptides of the amino or carboxyl termini of IL-4Rα and with eosinophils from normal, atopic, and hypereosinophilic donors (data not shown). In addition, although eosinophil plasma membrane expressions of γc and IL-13Rα1 were low-to-undetectable, after eosinophil permeabilization with 0.1% saponin, substantial intracellular pools of γc and IL-13Rα1 were routinely detected (Fig. 1 B and C).
Fig. 1.
Human eosinophils contain preformed stores of IL-4Rα, γc, and IL-13Rα1 chains. (A–C) By flow cytometry, eosinophil expression of IL-4Rα (A), γc (B), or IL-13Rα1 (C) before (Left) or after (Right) saponin permeabilization. (D) Eosinophil subcellular fractions were probed with anti-IL-4Rα Ab. PM, plasma membrane. (E) Expression of IL-4Rα by saponin permeabilized eosinophil granules measured by flow cytometry. Shaded histograms, control Ab; solid lines, Ab; dotted line in A, anti-IL-4Rα Ab neutralized by prior absorption with its immunogen peptide.
To determine the localization of the previously unrecognized intracellular pool of IL-4Rα, we analyzed IL-4Rα expression within subcellular fractions by immunoblotting. As expected, IL-4Rα was detected in low-density fractions containing plasma membranes (Fig. 1D). Notably, the greatest concentrations of IL-4Rα protein consistently localized to granule-containing fractions (Fig. 1D). Isolated granules from these fractions were further analyzed by flow cytometry and, after saponin permeabilization, they uniformly contained IL-4Rα (Fig. 1E). The identity and purity of isolated granules were confirmed by light microscopy and EM of isolated fractions and by MBP and CD63 staining by flow cytometry (see Fig. 6, which is published as supporting information on the PNAS web site). These findings provide one of the first documentations of substantial intracellular, preformed stores of cytokine receptors within human eosinophils and localize the IL-4-binding IL-4Rα chain to specific granules, sites at which IL-4 sorting and mobilization likely occur.
Eotaxin Induces Mobilization of Il-4Rα from Granules to Intragranular and Cytoplasmic Vesiculotubular Structures in Parallel with IL-4.
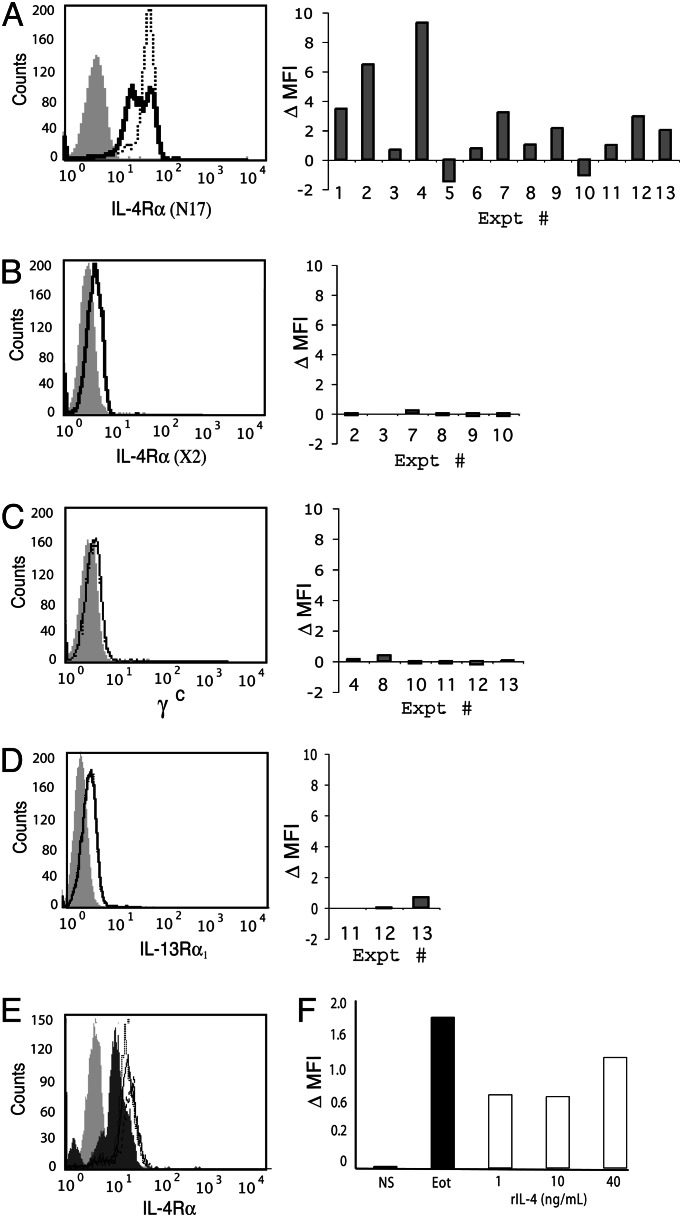
To investigate a functional relationship between granule-stored IL-4Rα and IL-4 secretion, intracellular IL-4 receptor expression was analyzed after eotaxin stimulation. Quantitative analyses of intracellular IL-4Rα by flow cytometry of 0.1% saponin-permeabilized cells revealed, in eosinophils from 11 of 13 donors, increased intracellular IL-4Rα expression after 30 min of eotaxin stimulation (Fig. 2A), a time concordant with increasing eotaxin-elicited extracellular IL-4 secretion (17). The increase in IL-4Rα expression was often detectable as early as 10 min after eotaxin stimulation (data not shown). Notably, we found that eotaxin stimulation acted preferentially to enhance detection of the IL-4Rα chain because γc exhibited contrasting patterns of intracellular expression when compared with IL-4Rα after eotaxin stimulation (compare C and A in Fig. 2). Nonparametric analysis simultaneously comparing data sets generated for IL-4Rα, γc, and IL-13Rα1 expression reveals a statistically significant difference between IL-4Rα and γc (P < 0.05). In contrast, differences between IL-4Rα and IL-13Rα1 expression (Fig. 2D) do not reach statistical significance, perhaps reflecting mobilization of the latter in response to eotaxin stimulation in transport of preformed IL-13, a concept discussed in greater detail below.
Fig. 2.
IL-4Rα, but not γc, is mobilized with eotaxin stimulation. Eosinophils were incubated for 30 min in the presence (dotted line) or absence (solid line) of eotaxin (A–D); pretreated with actinomycin D (solid line), cycloheximide (dashed line), or medium alone (dotted line) before eotaxin stimulation (E); or stimulated with increasing concentrations of rhIL-4 (F). Intracellular cytokine receptors were detected within saponin-permeablized cells with Abs recognizing an amino-terminal peptide distinct from the IL-4-binding site of IL-4Rα (N17) (A, E, and F), the IL-4-binding epitope of IL-4Rα that is inaccessible when IL-4 is bound (clone X2) (B), γc (C), or IL-13Rα1 (D). In A–D, histograms representing individual experiments are shown (Left), and data from all experiments are shown in the bar graphs (Right). Data in bar graphs represent the difference in geometric mean fluorescence intensity (MFI) between non- and eotaxin-stimulated cells after subtraction of MFI values obtained from specific isotype controls. Numbers along x axis identify individual experiments for comparison among panels. Lightly shaded peaks, control Ab; darkly shaded peak in E, no pretreatment and no stimulation. In F, data are calculated by the following equation: (geometric MFI-relevant Ab − geometric MFI isotype control)/(geometric MFI isotype control).
The rapidity with which intracellular IL-4Rα expression increased (within 10–30 min) argues for a direct effect of eotaxin signaling rather than a secondary consequence mediated by autocrine-acting extracellular IL-4 secreted in response to eotaxin (17). We ascertained this finding by investigating the potential of IL-4 to increase intracellular IL-4Rα expression within this time frame by stimulating eosinophils with either recombinant human (rh)IL-4 or eotaxin in parallel for 30 min before detecting intracellular levels of IL-4Rα by flow cytometry. Even at concentrations as high as 40 ng/ml (≈3 nM), exogenous rhIL-4 was unable to fully elicit increases in intracellular IL-4Rα receptor expression induced by eotaxin stimulation (Fig. 2F).
Pretreatment of eosinophils with actinomycin D or cycloheximide before eotaxin stimulation did not inhibit increases in IL-4Rα detection, indicating that the increases in intracellular IL-4Rα were not attributable to enhanced gene transcription or translation of new or nascent mRNA transcripts, respectively (Fig. 2E). Rather, increases in IL-4Rα likely reflected an eotaxin-dependent mobilization of preformed intracellular IL-4Rα from a relatively saponin-impermeable organelle (i.e., specific granules) to more effectively permeabilized cytosolic vesicular compartments. Supporting this interpretation, we were unable to visualize specific intragranular proteins (i.e., MBP) within intact eosinophils differentially permeabilized with various concentrations of saponin, whereas extragranular vesicular sources were readily detectable (data not shown).
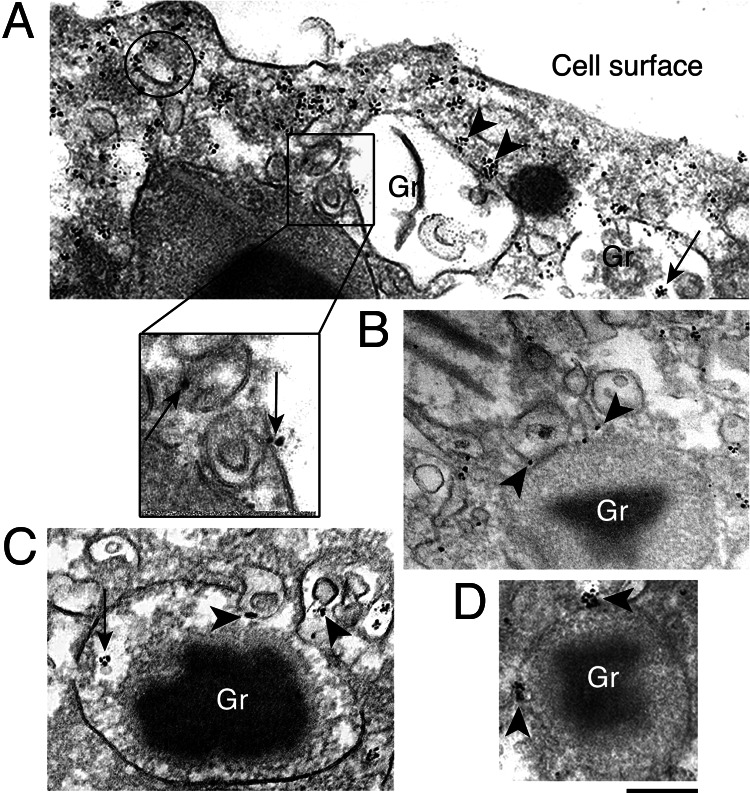
Immunogold EM analyses confirmed the increases in extragranular IL-4Rα expression after eotaxin stimulation and localized the mobilized IL-4Rα intracellular pool. Within granules demonstrating “core rearrangements” and other morphological patterns of PMD, IL-4Rα localization often appeared in clusters (Fig. 3A and C, arrows). At the granule periphery, IL-4Rα localization was demonstrable on outer granule membranes (Fig. 3 A and D, arrowheads) and frequently at sites where vesicles appeared to be budding or interacting (Fig. 3A, boxed area; Fig. 3 B and C, arrowheads). Of note, membranes of these vesicles were clearly labeled for IL-4Rα (Fig. 3A, boxed area; Fig. 3B, arrowheads) and moreover, showed the same morphology (Fig. 3A, boxed area; Fig. 3 B and C, arrowheads) as granule-attached, IL-4-positive vesicles (Fig. 5A, arrow). A striking association of IL-4Rα with cytoplasmic secretory vesicles was observed, often forming tight clusters at vesicle membranes (Fig. 3A, circle). In addition, clusters of IL-4Rα staining were prominent just beneath the plasma membrane (Fig. 3A). Thus, immunogold EM of eotaxin-stimulated cells supported the translocation of IL-4Rα from granules to secretory vesicles and revealed similar patterns of expression of IL-4 and IL-4Rα, with both molecules highly enriched on secretory vesicle membranes, further supporting the hypothesis that IL-4Rα functions in the selective transport of IL-4 by serving to anchor its ligand to membranes of transport vesicles.
Fig. 3.
IL-4Rα distribution in eotaxin-stimulated (100 ng/ml, 1 h) eosinophils by immunogold EM. (A) Clusters of IL-4Rα are indicated within granules (arrow), on granule outer membranes (arrowheads), on the surface of vesicles (circle), and underneath the plasma membrane. The boxed area shows membrane-bound, IL-4Rα-labeled vesiculotubular structures at the surface of an emptying granule (arrows). (B and C) Vesicles with IL-4Rα on their membranes (arrowheads) are seen attached to and apparently budding from specific granules. In C and D, IL-4Rα clusters are indicated within granules (C, arrow) and at the granule surface (D, arrowheads). Labeling was absent with control rabbit IgG. Gr, granule. [Scale bars: A, 750 nm; A Inset, 200 nm; B and C, 300 nm; and D, 400 nm.]
Mobilized IL-4Rα Binds IL-4 Within Vesicular Compartments.
To evaluate a direct interaction between the two proteins within vesicles, IL-4Rα mobilization was monitored in parallel with two epitope-distinct anti-IL-4Rα Abs. One Ab recognized an epitope within the amino terminus and distinct from the IL-4-binding domain (N17). The other Ab recognized the IL-4-binding site that is inaccessible when IL-4 is bound (the X2 clone) (26). Within 30 min of eotaxin stimulation, the amino-terminal-specific Ab uniformly detected increases in intracellular IL-4Rα expression. In contrast, the X2 clone, which competes with IL-4 for its binding site, failed to detect increases in IL-4Rα (P = 0.02) (compare A and B in Fig. 2), likely because IL-4 was bound to IL-4Rα.
Eosinophils Contain Preformed Intracellular Stores of IL-6 Receptor and CCR3.
Given the multitude of cytokines and chemokines preformed in eosinophils, receptor-mediated transport of cytokines might be a mechanism not unique for IL-4 secretion. Thus, we determined whether other receptor chains specific for eosinophil-derived cytokines were present preformed within eosinophils. IL-13Rα1 expression has been detected on human eosinophils (27), and as noted above, we identified an intracellular pool of IL-13Rα1 (Fig. 1C). Although this chain forms a heterodimer with IL-4Rα to form the type II IL-4-receptor complex, it also serves as the ligand-binding chain of a functional receptor for IL-13, a cytokine stored preformed within human eosinophils and released after activation (28, 29).
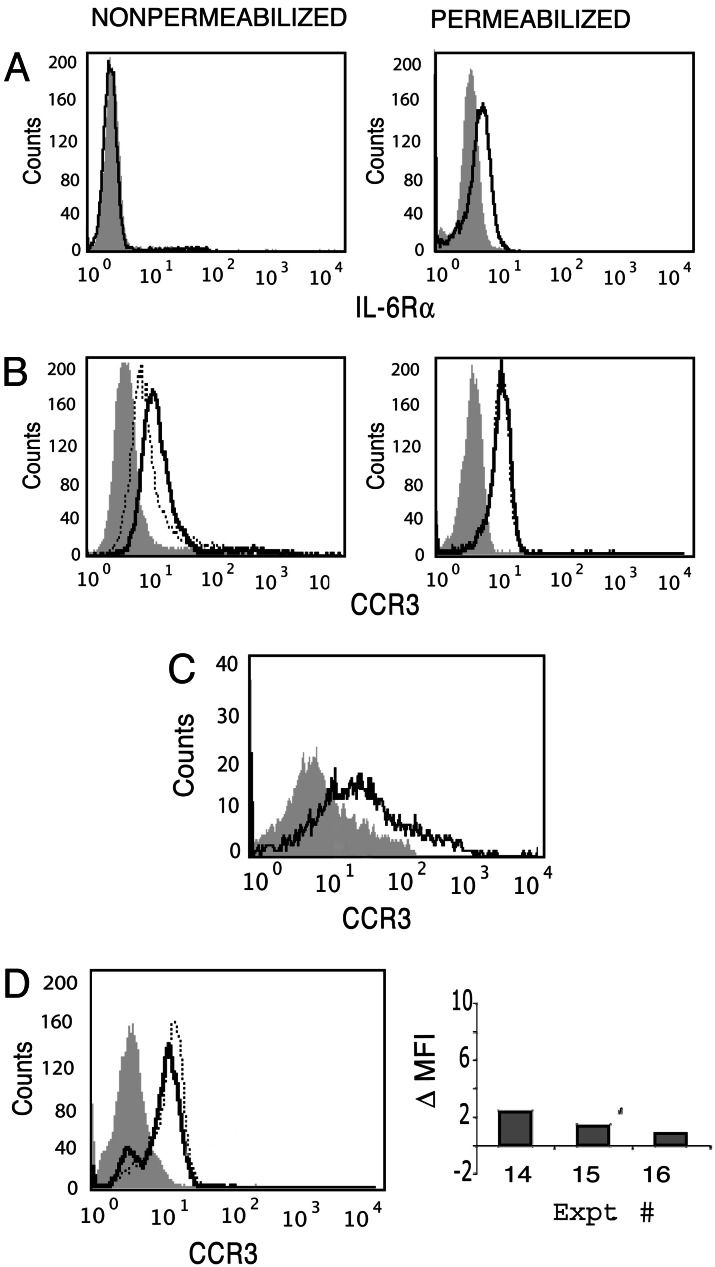
IL-6 has also been identified within eosinophil-specific granules and is secreted upon stimulation with IFN-γ (7). To date, no reports have addressed the existence of IL-6 receptor expression on human eosinophils. Although we found very low plasma membrane surface expression of the IL-6Rα chain on freshly isolated eosinophils, as observed for IL-4Rα, after permeabilization with 0.1% saponin, substantial intracellular anti-IL-6Rα chain staining was demonstrable within eosinophils (Fig. 4A).
Fig. 4.
Intracellular stores of IL-6Rα and CCR3 within human eosinophils. (A and B) Eosinophil expression, measured by flow cytometry, of IL-6Rα (A) and CCR3 (B) before (Left) and after (Right) saponin permeabilization. In B, the dotted line represents surface CCR3 blocked with nonlabeled anti-CCR3 before intracellular detection with labeled anti-CCR3. (C) Isolated granules were permeabilized and analyzed for CCR3 expression. (D) Intracellular CCR3 was detected in cells cultured for 30 min in the presence (dotted line) or absence (solid line) of IFN-γ. A histogram of one representative experiment and the change in MFI observed in three independent experiments (calculated as in Fig. 2 A–D) are shown. Shaded peaks represent control Ab.
Functional receptors for IL-4, IL-13, and IL-6 are all type I membrane-spanning glycoproteins sharing several features, including a need for homo- or heterodimerization for effective signaling. To determine whether intracellular storage and specifically granule localization of other distinct receptor subtypes occur, intra- and extracellular levels of the G protein-coupled receptor CCR3 were compared. Eosinophils respond rapidly to eotaxin and RANTES that bind their common, surface-expressed CCR3 receptor. In addition, both eotaxin and RANTES are stored preformed within eosinophil-specific granules (9–11). Similar to that observed for IL-6Rα, substantial CCR3 was detected within freshly isolated, saponin-permeabilized eosinophils (Fig. 4B). Blocking of plasma membrane-expressed CCR3 failed to diminish anti-CCR3 labeling, demonstrating that the enhanced CCR3 staining seen with permeabilized eosinophils was due to an intracellular pool of CCR3 (Fig. 4B). Analogous to our findings with intracellular IL-4Rα, specific granules, isolated by subcellular fractionation and 0.1% saponin-permeabilized, uniformly contained CCR3 detectable by flow cytometry (Fig. 4C). Further, upon stimulation with IFN-γ, a known stimulator of RANTES release from eosinophils (30), intracellular levels of CCR3 were found to increase (Fig. 4D). Additional studies focusing on eosinophil secretion of RANTES are needed to determine whether CCR3 traffics with its ligand in a manner akin to that observed for IL-4Rα and IL-4.
Discussion
Immune responses depend on the elaboration of cytokines and chemokines by cells of the adaptive and innate immune systems. Adaptive immune lymphocytes must, over time, be induced to clonally expand and differentiate before they can synthesize and secrete their cytokines. Some cells, such as natural killer T cells, contain preformed cytokine mRNA transcripts that expedite their capacity to secrete cytokines, although these would still require time during de novo cytokine synthesis (31). Leukocytes associated with innate immune responses, including eosinophils (5), mast cells (32), basophils (33, 34), and neutrophils (35), are also recognized as potential sources of cytokines. In eosinophils, there is extensive evidence that diverse cytokines are resident in preformed stores in sites that include eosinophil granules (5). Likewise, in mast cells and basophils, there is evidence, albeit not as extensive to date, that preformed cytokines are present in these cells (32, 34) and may be localized to granules (32).
In both cells of the adaptive and innate immune systems, the mechanisms underlying the intracellular transport and secretion of cytokines are ill understood. In cells of the innate immune system, a process of vesicular transport-mediated PMD has been recognized that may provide for the speedy secretion of granule-derived proteins into the local microenvironment; but this finding raises the question of how selectivity is achieved in eosinophil mediator release. PMD, based on in vivo ultrastructural observations, is most cogent for eosinophils (18–21) but also has been recognized for mast cells and basophils (36). To delineate mechanisms whereby preformed cytokines might be secreted by cells of the innate immune system, we focused on the mechanisms governing eosinophil secretion of preformed IL-4, a cytokine prominent in type 2 immune responses, that we have shown to be rapidly releasable through vesicular transport from human eosinophils (17).
Intriguingly, receptors specific for eosinophil-derived products, including IL-4, have been identified on eosinophils. Our observations of IL-4 localizing to membranes of secretory vesicles (22), suggesting participation of a docking molecule, led us to analyze IL-4 receptor expression during eotaxin-induced PMD of IL-4. Surprisingly, in addition to low-level plasma membrane expression, we detected substantial intracellular stores of each component of functional type I and II IL-4 receptors. The majority of intracellular IL-4Rα protein colocalized with granules in nonstimulated eosinophils. Thus, IL-4Rα expression coincided with the location of granule-stored IL-4 in sites at which potential cytokine sequestration and sorting mechanisms likely occur. Eotaxin stimulation induced IL-4Rα association with the intragranular tubulomembranous network arising in mobilized granules, a network we have implicated in the sequestration and sorting of granule products before secretion (21, 22).
Quantitative analyses of intracellular IL-4Rα expression revealed a salient increase evident within 10–30 min of eotaxin stimulation. We found no evidence of new gene transcription or mRNA translation of IL-4Rα in eotaxin-stimulated eosinophils, suggesting that the increase in expression resulted from an increased availability of the IL-4Rα chain to detecting antibodies, i.e., mobilization from a relatively saponin-impermeable compartment to a more readily permeabilized one. Consistent with this interpretation, the majority of IL-4Rα within nonstimulated cells colocalized with granules, which are relatively insensitive to saponin permeabilization (data not shown). Upon stimulation, mobilized IL-4Rα was strikingly associated with more saponin-sensitive vesicular compartments. Thus, there was a temporal coincidence of IL-4Rα and IL-4 mobilization from granules into the vesicles. These observations, combined with a clear association of IL-4 with secretory vesicle membranes, suggested that eotaxin-mobilized IL-4Rα functions as a transporter for IL-4 through the secretory pathway. In full confirmation, an anti-IL-4Rα mAb, whose binding is blocked if IL-4 is bound to IL-4Rα, did not detect increases in intracellular IL-4Rα, indicating that IL-4 was bound to IL-4Rα. In contrast to IL-4Rα, intracellular γc chain expression did not uniformly increase with eotaxin stimulation, demonstrating that the two chains are independently mobilized, with eotaxin selectively eliciting the translocation of the IL-4-binding IL-4Rα in the absence of the remainder of a functional receptor complex. Omission of the γc accessory chain would presumably allow IL-4 binding and transport without initiation of a signaling cascade.
In other cell types, both IL-4Rα and IL-13Rα2 have been recognized to be substantially intracellular, apparently without ligand-induced signaling (25, 37, 38). Although CCR3 is known to undergo internalization within eosinophils (39), neither the intracellular content nor localization of CCR3 within eosinophils had been ascertained. Of pertinence, however, is that immunogold EM has recently documented substantial intracellular CCR3 localization to mast cell granules (40), fully analogous to what we are recognizing in eosinophils. In neutrophils, IL-10 receptors have been localized to specific granules (41). Mast cells are sources of CCR3-binding chemokines (i.e., eotaxin and RANTES) (32), and neutrophils can express IL-10 (42). Thus, it is possible that the intracellular localization of cytokine receptors to granules of cells of the innate immune system, including mast cells and neutrophils, may have roles in the secretion of granule-derived cytokines from both these cells comparable with what we have recognized in eosinophils.
We have identified substantial, previously unrecognized, intracellular pools of cytokine receptors, including IL-4Rα, and suggest a previously unrecognized function for these chains in the secretory trafficking of cytokines, specifically of IL-4, in eotaxin-stimulated cells. Taken together, our data have led us to generate a model in which eotaxin signaling mobilizes granular stores of both IL-4 and IL-4Rα, with the latter interacting with a complex network of membranous domains within mobilized granules, potentially sequestering its cognate ligand and guiding its loading into secretory vesicles. IL-4 within granule-derived secretory vesicles remains largely bound to vesicular membranes through interaction with IL-4Rα while the vesicles traffic to the plasma membrane. Our identification of intracellular stores of several additional ligand-binding receptor chains specific for eosinophil-derived cytokines and chemokines suggests the potential for multiple preformed cytokines and chemokines to be selectively transported by their respective receptors. Thus, cytokine receptor chain involvement in cytokine secretion may provide a crucial component of the regulatory mechanisms governing specificity of rapid, stimulus-induced release of preformed immunomodulatory cytokines from human eosinophils, as well as other innate immune cells containing granule stores of preformed cytokines and chemokines.
Methods
Eosinophil Isolation and Stimulation.
As approved by the Committee on Clinical Investigation (Beth Israel Deaconess Medical Center), eosinophils from healthy donors were purified by negative selection (>99% pure) as described in ref. 17. Eosinophils (106 cells per ml) were stimulated with 100 ng/ml rh eotaxin (R & D Systems), 500 units/ml rhIFN-γ (BioSource International, Camarillo, CA), or medium alone (RPMI medium 1640/0.1% ovalbumin) at 37°C for the indicated time periods. In some experiments, cells were pretreated for 30 min at 37°C in the presence or absence of 10 μM actinomycin D (Sigma) or 10 μM cycloheximide (Sigma) before eotaxin stimulation. Cell viability after stimulation was >95%.
Ab Reagents.
Anti-human IL-4 (clone 3010.211) and irrelevant isotype control (clone 11711.11) mAbs (R & D Systems) were used for immunonanogold EM (2 μg/ml). Secondary Ab was goat anti-mouse Fab-conjugated to 1.4 nm of gold (1:100) (Nanogold; Nanoprobes, Stony Brook, NY). Reagents for EM and flow cytometry included rabbit Abs generated against peptides mapping to the amino (N17) and carboxyl (C20) termini of human IL-4Rα (Santa Cruz Biotechnology) used in parallel with control rabbit IgG (R & D Systems) at concentrations of 2 μg per 106 cells (flow cytometry) or at 1 μg/ml (EM). Secondary Ab for EM was 1.4 nm gold-conjugated goat anti-rabbit Fab fragments (1:100) (Nanoprobes). A competitive inhibitor of IL-4 binding (26) (clone X2; eBioscience, San Diego) and its isotype control (clone MOPC 21; BD Pharmingen) were used at concentrations from 2 to 4 μg per 106 cells. Anti-human γc (clone TUGh4; BD Pharmingen), anti-human IL-13Rα1 (clone 116730; R & D Systems), anti-human IL-6Rα (clone H300; Santa Cruz Biotechnology), and irrelevant isotype controls (clone A95–1, BD Pharmingen; clone 20116.11, R & D Systems; and rabbit IgG, R & D Systems, respectively) were used at 2–4 μg per 106 cells. Isolated granules were stained with mouse anti-human MBP mAb (clone AHE-2) and irrelevant isotype control (clone S1-68.1; BD Pharmingen), anti-human CCR3 (clone 61828) and irrelevant isotype control (clone 5447; R & D Systems), and anti-human CD63 (clone H5C6; BD Pharmingen) and irrelevant isotype control at final 2 μg/ml concentrations.
Immunogold EM.
Agar pellets containing intact eosinophils were processed (43), and preembedding immunogold EM was performed on frozen 10-μm sections. Steps, as described in ref. 44, were modified as follows: sections were incubated in PBS-BSA (0.02 M PBS/1% BSA) containing 0.1% gelatin for 20 min, followed by PBS-BSA plus 10% normal goat serum and incubated with primary Ab for 1 h. After blocking with PBS-BSA plus normal goat serum for 30 min, sections were incubated with secondary Ab for 1 h, washed in PBS-BSA, postfixed in 1% glutaraldehyde for 10 min, and incubated with HQ Silver enhancement solution (Nanoprobes) for 10 min. Sections were immersed in 5% Na2S2O3 for 5 min, postfixed in 1% OsO4 for 10 min, stained in 2% uranyl acetate for 5 min, and embedded as described in ref. 44. For controls, (i) primary Ab was replaced by an irrelevant Ab, and (ii) primary Ab was omitted. Thin sections were examined by EM (44).
Subcellular Fractionation and Immunoblotting.
Eosinophils, resuspended in disrupting buffer (30), were subjected to nitrogen cavitation at 800 psi for 10 min. Postnuclear supernatants recovered after centrifugation at 400 × g for 10 min were ultracentrifuged at 100,000 × g for 1 h at 4°C in linear OptiPrep gradients (Greiner Bio-One, Longwood, FL; 0–45% in disrupting buffer). Collected fractions (20 × 0.5 ml) were stored at −80°C. Granule-enriched fractions were identified by MBP staining (flow cytometry), eosinophil peroxidase reactivity (enzymatic assay), and morphology (EM); plasma membrane was detected with MHC I antibodies (immunoblotting); and cytosol-containing fractions were identified by measurement of lactate dehydrogenase (enzymatic assay).
For immunoblotting, 25 μl of each fraction (or whole-cell lysate) was mixed with reducing buffer, boiled for 5 min, and run on 4–12% Bis-Tris gels (Invitrogen) under denaturing conditions. Gels were transferred to poly(vinylidene difluoride) membranes (Pierce), blocked overnight with 5% milk, and probed with rabbit anti-IL-4Rα amino terminal Ab (N17, Santa Cruz Biotechnology) diluted 1:200 followed by anti-rabbit secondary Ab conjugated to HRP (Jackson ImmunoResearch). Membranes were developed by using West Pico chemiluminescence kits (Pierce).
Flow Cytometry of Whole Cells and Isolated Granules.
Live nonpermeabilized cells or isolated granules were incubated with primary and secondary Abs on ice in the absence of cell fixation. After staining, cells were stored in buffer containing 0.5% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA). For intracellular detection, cells or isolated granules were fixed for 5 min in 2% paraformaldehyde and permeabilized for 5 min on ice with 0.1% saponin before incubation with primary and secondary Abs in the presence of 0.1% saponin. After staining, samples were stored in saponin-free buffer (calcium- and magnesium-free Hanks’ balanced salt solution/0.5% BSA). Isotype control staining was included at each time point for each condition. Data were acquired by using a FACScan with cellquest acquisition and analysis software (Becton Dickinson).
Statistical Analyses.
Simultaneous comparison of change in geometric mean fluorescence intensity (MFI) detected with IL-4Rα (N17), γc, and IL-13Rα1 was performed by using a one-way ANOVA, nonparametric assay (Kruskal–Wallis) followed by Dunn’s post hoc test to determine significance. Competitive and noncompetitive Abs against IL-4Rα were compared by using a nonparametric test (Mann–Whitney U test).
Supplementary Material
Acknowledgments
We thank Tracey Sciuto, Rita Monahan-Earley, and Ellen Morgan for EM assistance. This work was supported by National Institutes of Health Grants AI20241, AI22571, AI51645, and HL70270. R.C.N.M. and S.A.C.P. were supported by the Conselho Nacional de Desenvolvimento Cientifico e Tecnológico of Brazil.
Abbreviations
- MBP
major basic protein
- RANTES
regulated on activation, normal T cell expressed and secreted
- PMD
piecemeal degranulation
- IL-4Rα
IL-4 receptor α chain
- γc
common gamma
- rh
recombinant human
- MFI
mean fluorescence intensity.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gleich G. J. J. Allergy Clin. Immunol. 2000;105:651–663. doi: 10.1067/mai.2000.105712. [DOI] [PubMed] [Google Scholar]
- 2.Humbles A. A., Lloyd C. M., McMillan S. J., Friend D. S., Xanthou G., McKenna E. E., Ghiran S., Gerard N. P., Yu C., Orkin S. H., Gerard C. Science. 2004;305:1776–1779. doi: 10.1126/science.1100283. [DOI] [PubMed] [Google Scholar]
- 3.Adamko D. J., Odemuyiwa S. O., Vethanayagam D., Moqbel R. Allergy. 2005;60:13–22. doi: 10.1111/j.1398-9995.2005.00676.x. [DOI] [PubMed] [Google Scholar]
- 4.Munitz A., Levi-Schaffer F. Allergy. 2004;59:268–275. doi: 10.1111/j.1398-9995.2003.00442.x. [DOI] [PubMed] [Google Scholar]
- 5.Lacy P., Moqbel R. Chem. Immunol. 2000;76:134–155. doi: 10.1159/000058782. [DOI] [PubMed] [Google Scholar]
- 6.Möller G. M., de Jong T. A., van der Kwast T. H., Overbeek S. E., Wierenga-Wolf A. F., Thepen T., Hoogsteden H. C. Am. J. Respir. Cell Mol. Biol. 1996;14:439–443. doi: 10.1165/ajrcmb.14.5.8624248. [DOI] [PubMed] [Google Scholar]
- 7.Lacy P., Levi-Schaffer F., Mahmudi-Azer S., Bablitz B., Hagen S. C., Velazquez J., Kay A. B., Moqbel R. Blood. 1998;91:2508–2516. [PubMed] [Google Scholar]
- 8.Woerly G., Roger N., Loiseau S., Dombrowicz D., Capron A., Capron M. J. Exp. Med. 1999;190:487–495. doi: 10.1084/jem.190.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying S., Meng Q., Taborda-Barata L., Corrigan C. J., Barkans J., Assoufi B., Moqbel R., Durham S. R., Kay A. B. Eur. J. Immunol. 1996;26:70–76. doi: 10.1002/eji.1830260111. [DOI] [PubMed] [Google Scholar]
- 10.Lim K. G., Wan H.-C., Bozza P. T., Resnick M. B., Wong D. T. W., Cruikshank W. W., Kornfeld H., Center D. M., Weller P. F. J. Immunol. 1996;156:2522–2527. [PubMed] [Google Scholar]
- 11.Nakajima T., Yamada H., Iikura M., Miyamasu M., Izumi S., Shida H., Ohta K., Imai T., Yoshie O., Mochizuki M., et al. FEBS Lett. 1998;434:226–230. doi: 10.1016/s0014-5793(98)00863-1. [DOI] [PubMed] [Google Scholar]
- 12.Sabin E. A., Kopf M. A., Pearce E. J. J. Exp. Med. 1996;184:1871–1878. doi: 10.1084/jem.184.5.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinkai K., Mohrs M., Locksley R. M. Nature. 2002;420:825–829. doi: 10.1038/nature01202. [DOI] [PubMed] [Google Scholar]
- 14.Voehringer D., Shinkai K., Locksley R. M. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 15.Min B., Prout M., Hu-Li J., Zhu J., Jankovic D., Morgan E. S., Urban J. F., Jr, Dvorak A. M., Finkelman F. D., LeGros G., Paul W. E. J. Exp. Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandeira-Melo C., Perez S. A., Melo R. C., Ghiran I., Weller P. F. J. Immunol. Methods. 2003;276:227–237. doi: 10.1016/s0022-1759(03)00076-0. [DOI] [PubMed] [Google Scholar]
- 17.Bandeira-Melo C., Sugiyama K., Woods L. J., Weller P. F. J. Immunol. 2001;166:4813–4817. doi: 10.4049/jimmunol.166.8.4813. [DOI] [PubMed] [Google Scholar]
- 18.Ahlstrom-Emanuelsson C. A., Greiff L., Andersson M., Persson C. G., Erjefalt J. S. Eur. Respir. J. 2004;24:750–757. doi: 10.1183/09031936.04.00133603. [DOI] [PubMed] [Google Scholar]
- 19.Erjefalt J. S., Greiff L., Andersson M., Adelroth E., Jeffery P. K., Persson C. G. Thorax. 2001;56:341–344. doi: 10.1136/thorax.56.5.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karawajczyk M., Seveus L., Garcia R., Bjornsson E., Peterson C. G., Roomans G. M., Venge P. Am. J. Respir. Cell Mol. Biol. 2000;23:521–529. doi: 10.1165/ajrcmb.23.4.4025. [DOI] [PubMed] [Google Scholar]
- 21.Melo R. C. N., Perez S. A. C., Spencer L. A., Dvorak A. M., Weller P. F. Traffic. 2005;6:866–879. doi: 10.1111/j.1600-0854.2005.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo R. C. N., Spencer L. A., Perez S. A. C., Ghiran I., Dvorak A. M., Weller P. F. Traffic. 2005;6:1047–1057. doi: 10.1111/j.1600-0854.2005.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois G. R., Schweizer R. C., Versluis C., Bruijnzeel-Koomen C. A. F. M., Bruijnzeel P. L. B. Am. J. Resp. Cell Mol. Biol. 1998;19:691–699. doi: 10.1165/ajrcmb.19.4.3208. [DOI] [PubMed] [Google Scholar]
- 24.Elovic A., Ohyama H., Sauty A., McBride J., Tsuji T., Nagai M., Weller P. F., Wong D. T. W. J. Immunol. 1998;160:6121–6127. [PubMed] [Google Scholar]
- 25.Hershey G. K. J. Allergy Clin. Immunol. 2003;111:677–690. doi: 10.1067/mai.2003.1333. [DOI] [PubMed] [Google Scholar]
- 26.Ramanathan L., Ingram R., Sullivan L., Greenberg R., Reim R., Trotta P. P., Le H. V. Biochemistry. 1993;32:3549–3556. doi: 10.1021/bi00065a005. [DOI] [PubMed] [Google Scholar]
- 27.Myrtek D., Knoll M., Matthiesen T., Krause S., Lohrmann J., Schillinger D., Idzko M., Virchow J. C., Friedrich K., Luttmann W. Immunology. 2004;112:597–604. doi: 10.1046/j.1365-2567.2004.01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woerly G., Lacy P., Younes A. B., Roger N., Loiseau S., Moqbel R., Capron M. J. Leukocyte Biol. 2002;72:769–779. [PubMed] [Google Scholar]
- 29.Schmid-Grendelmeier P., Altznauer F., Fischer B., Bizer C., Straumann A., Menz G., Blaser K., Wuthrich B., Simon H. U. J. Immunol. 2002;169:1021–1027. doi: 10.4049/jimmunol.169.2.1021. [DOI] [PubMed] [Google Scholar]
- 30.Lacy P., Mahmudi-Azer S., Bablitz B., Hagen S. C., Velazquez J. R., Man S. F. P., Moqbel R. Blood. 1999;94:23–32. [PubMed] [Google Scholar]
- 31.Stetson D. B., Mohrs M., Reinhardt R. L., Baron J. L., Wang Z. E., Gapin L., Kronenberg M., Locksley R. M. J. Exp. Med. 2003;198:1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galli S. J., Nakae S., Tsai M. Nat. Immunol. 2005;6:135–142. doi: 10.1038/ni1158. [DOI] [PubMed] [Google Scholar]
- 33.Gibbs B. F., Haas H., Falcone F. H., Albrecht C., Vollrath I. B., Noll T., Wolff H. H., Amon U. Eur. J. Immunol. 1996;26:2493–2498. doi: 10.1002/eji.1830261033. [DOI] [PubMed] [Google Scholar]
- 34.Mitre E., Taylor R. T., Kubofcik J., Nutman T. B. J. Immunol. 2004;172:2439–2445. doi: 10.4049/jimmunol.172.4.2439. [DOI] [PubMed] [Google Scholar]
- 35.Cassatella M. A. Adv. Immunol. 1999;73:369–509. doi: 10.1016/s0065-2776(08)60791-9. [DOI] [PubMed] [Google Scholar]
- 36.Dvorak A. M. Basophil and Mast Cell Degranulation and Recovery. New York: Plenum; 1991. [Google Scholar]
- 37.Friedrich K., Kammer W., Erhardt I., Brandlein S., Arnold S., Sebald W. Eur. J. Biochem. 1999;265:457–465. doi: 10.1046/j.1432-1327.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- 38.Daines M. O., Hershey G. K. J. Biol. Chem. 2002;277:10387–10393. doi: 10.1074/jbc.M108109200. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann N., Rothenberg M. E. J. Allergy Clin. Immunol. 2003;111:97–105. doi: 10.1067/mai.2003.3. [DOI] [PubMed] [Google Scholar]
- 40.Price K. S., Friend D. S., Mellor E. A., De Jesus N., Watts G. F., Boyce J. A. Am. J. Respir. Cell Mol. Biol. 2003;28:420–427. doi: 10.1165/rcmb.2002-0155OC. [DOI] [PubMed] [Google Scholar]
- 41.Elbim C., Reglier H., Fay M., Delarche C., Andrieu V., El Benna J., Gougerot-Pocidalo M. A. J. Immunol. 2001;166:5201–5207. doi: 10.4049/jimmunol.166.8.5201. [DOI] [PubMed] [Google Scholar]
- 42.Piskin G., Bos J. D., Teunissen M. B. Arch. Dermatol. Res. 2005;296:339–342. doi: 10.1007/s00403-004-0522-z. [DOI] [PubMed] [Google Scholar]
- 43.Dvorak A. M., Furitsu T., Letourneau L., Ishizaka T., Ackerman S. J. Am. J. Pathol. 1991;138:69–82. [PMC free article] [PubMed] [Google Scholar]
- 44.Feng D., Flaumenhaft R., Bandeira-Melo C., Weller P. F., Dvorak A. M. J. Histochem. Cytochem. 2001;49:293–304. doi: 10.1177/002215540104900303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.