Abstract
The nitric oxide-mediated actions are mostly due to cyclic GMP (cGMP) formation, but cGMP-independent mechanisms, such as tyrosine nitration, have been suggested as potential signaling pathways modulating the NO-induced responses. However, the mechanisms that lead to tyrosine nitration in platelets are poorly studied, and the protein targets of nitration have not been identified in these cells. Therefore, we have used the model of platelet adhesion to fibrinogen-coated plates to investigate the cGMP-independent mechanisms of the NO-donor sodium nitroprusside (SNP) that leads to inhibition of platelet adhesion. SNP concentration-dependently inhibited platelet adhesion, as observed at 15-min and 60-min adhesion. Additionally, SNP markedly increased the cGMP levels, and the soluble guanylate inhibitor ODQ nearly abolished the SNP-mediated cGMP elevations in all experimental conditions used. Nevertheless, ODQ failed to affect the adhesion inhibition obtained with 1.0 mM SNP at 15 min. On the other hand, superoxide dismutase or peroxynitrite (ONOO−) scavenger epigallocatechin gallate significantly reversed the inhibition of platelet adhesion by SNP (1 mM, 15 min). Western blot analysis in SNP (1 mM, 15 min)-treated platelets showed a single tyrosine-nitrated protein with an apparent mass of ≈105 kDa. Nanospray LC-MS/MS identified the human α-actinin 1 cytoskeletal isoform (P12814) as the protein contained in the nitrated SDS gel band. Thus, tyrosine nitration of α-actinin, through ONOO− formation, may be a key modulatory mechanism to control platelet adhesion.
Keywords: protein nitration, sodium nitroprusside, peroxynitrite, human platelet, soluble guanylate cyclase
Blood platelets play a key role in maintaining the integrity of the vascular system through their ability to arrest bleeding and to promote repair of injured blood vessels (1). Platelet adhesion is the first step to begin the haemostatic process, and fibrinogen is responsible to mediate both platelet adhesion and aggregation through binding to the platelet membrane glycoprotein IIb-IIIa (integrin αIIbβ3) (2). In nonactivated platelets, the majority of GPIIb/IIIa is in a low-affinity state. However, they are capable of binding directly to immobilized fibrinogen and von Willebrand factor, because in this nonactivated state, the platelets attach to domains of fibrinogen distinct of those seen after platelet activation (3).
The nitric oxide (NO)-dependent inhibition of platelet adhesion to the subendothelium is essential in preventing excessive aggregation and thrombus formation. It is well accepted that NO-mediated actions are due to activation of the soluble guanylate cyclase leading to an increase of cyclic GMP (cGMP; ref. 4). The NO donor sodium nitroprusside (SNP) has been shown to spontaneously release NO for prolonged time periods (5, 6), leading to inhibition of platelet adhesion in vivo (7) and in vitro (8, 9). Although the reduction of platelet adhesion by SNP is accompanied by increased cGMP levels, cGMP-independent mechanisms have also been proposed to mediate the NO-induced antiplatelet activity (10, 11). The cGMP-independent mechanisms have not been clearly characterized, but some evidence suggests that tyrosine nitration may account for the inhibition of cellular function. Nitration of proteins is described as a residue-, protein- and tissue-selective process, which generally leads to inhibition of protein function (12). It should be noted that not all proteins are targets for nitration, nor are all tyrosine residues in target proteins nitrated (13).
Peroxynitrite (ONOO−), a reactive oxidant produced by the reaction of NO with superoxide anions (O2−), is capable of nitrating aromatic amino acids in proteins, particularly tyrosine, leading to inhibition of several enzymes (14–17). Although a large number of studies investigated the effects of NO donors on platelet aggregation, only few in vitro studies (9, 18) addressed the role of NO in the platelet adhesion. Therefore, in this study, we used the model of platelet adhesion to fibrinogen-coated wells (19, 20), which mimics the adhesion of platelets to vascular matrices at the site of vascular injury to investigate the cGMP-dependent and -independent mechanisms underlying the inhibitory effects of the NO donor SNP on human platelet adhesion. We have demonstrated here that the inhibitory effect of SNP on platelet adhesion is mediated by both cGMP-dependent and -independent mechanisms. The cGMP-independent mechanism occurs at a high concentration of SNP after a short period of incubation and involves the nitration of α-actinin cytoskeletal protein. It is known that platelets have important roles in pathologies like atherosclerosis, ischemia-reperfusion, sepsis, and diabetes, where the production of reactive nitrogen and oxygen species are abundant. Tyrosine nitration therefore may be a key modulatory mechanism to control platelet functions.
Results
Effect of SNP on Acid Phosphatase Activity and Platelet Viability.
Because the adhesion assay is based on acid phosphatase activity, a set of experiments was initially carried out to determine the direct effect of SNP on phosphatase activity. Nonactivated or thrombin (50 milliunits/ml)-activated platelets (1.2 × 108 platelets/ml; 50 μl per well) were added to uncoated plates and incubated with SNP (0.1 or 1 mM) for 15 or 60 min, after which phosphatase activity was measured. Addition of SNP (0.1 and 1 mM) for either 15 or 60 min did not modify this enzyme activity (data not shown; n = 3). In addition, the MTT reduction assay showed that neither the short (15 min) nor the prolonged (60 min) exposure of human platelets to SNP (0.1 and 1 mM) caused any toxic effect (data not shown).
Adhesion of Nonactivated and Thrombin-Activated Platelets to Fibrinogen-Coated Wells.
Significant platelet adhesion (6 × 106 platelets per well) was observed when nonactivated platelets were kept on plates for 15 min (10 ± 0.5%) or 60 min (30 ± 1%). The adhesion was significantly increased when platelets were activated with 50 milliunits/ml of thrombin (50 ± 5% and 55 ± 4% for 15 min and 60 min, respectively; P < 0.05). Nonactivated and thrombin-stimulated platelets attached to the plates neither at 15 nor at 60 min if left 24 h at room temperature before the adhesion assays (data not shown).
Inhibitory Effect of SNP on Platelet Adhesion.
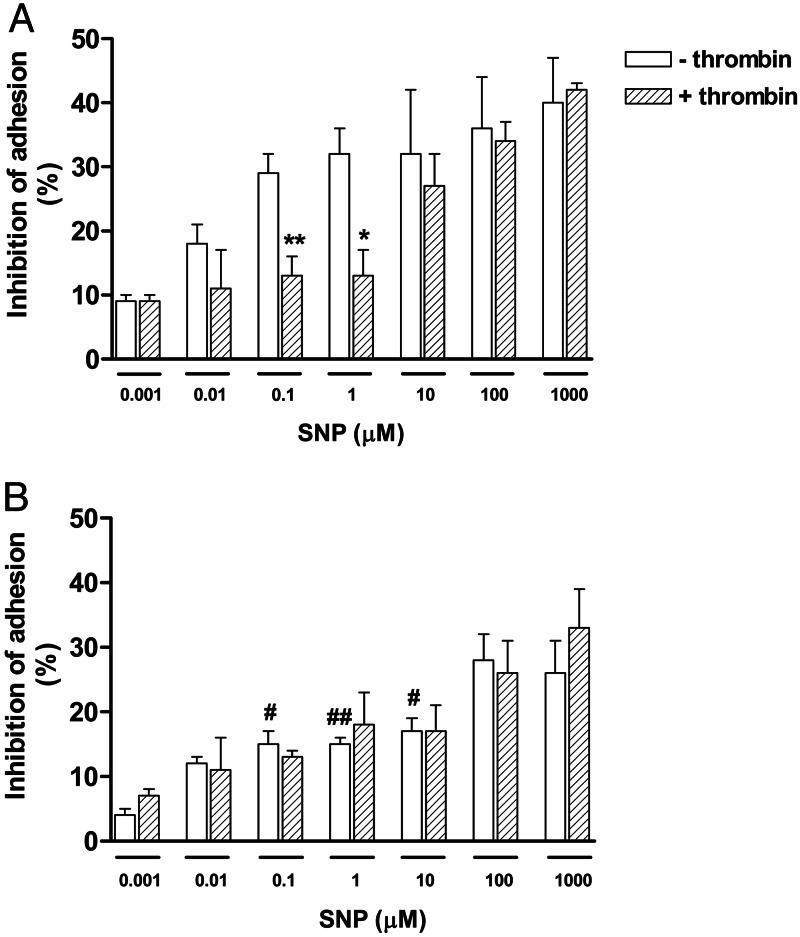
When platelets were allowed to adhere to fibrinogen for 15 min, the addition of SNP (0.001–1,000 μM) concentration-dependently inhibited both nonactivated and thrombin-stimulated platelet adhesion. The inhibition of nonactivated platelets was significantly higher than that of activated cells at 0.1 and 1 μM of SNP (Fig. 1A; P < 0.05).
Fig. 1.
Inhibitory effects of SNP on human platelet adhesion to fibrinogen. Platelets (6 × 106 platelets per well), in the absence or presence of thrombin (50 milliunits/ml), were incubated with SNP (0.001–1 mM) for 15 min (A) or 60 min (B) before platelet adhesion measurement. Results represent the mean ± SEM of three separate experiments, each performed in triplicate. ∗, P < 0.05 and ∗∗, P < 0.01, compared with the respective adhesion in the absence of thrombin. #, P < 0.05 and ##, P < 0.01, compared with the respective adhesion in 15 min.
The platelet adhesion at 60 min showed that SNP (0.001–1,000 μM) also concentration-dependently inhibited both nonactivated and thrombin-stimulated platelet adhesion to the same extent (Fig. 1B).
The inhibition of platelet adhesion by SNP was significantly higher at 15 min (Fig. 1A) compared with 60 min (Fig. 1B) in nonactivated cells, whereas in thrombin-activated cells, no differences were observed.
Effect of the Soluble Guanylate Cyclase Inhibitor ODQ.
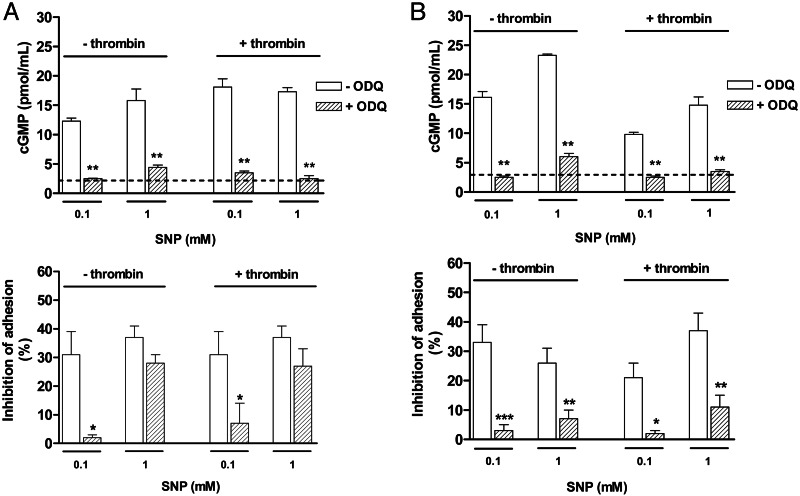
Fig. 2A shows that, at 15 min, the inhibition of platelet adhesion by 0.1 mM SNP was accompanied by marked increases in cGMP levels in nonactivated and thrombin-activated cells. Pretreatment of platelets with ODQ (10 μM) nearly abolished SNP (0.1 mM)-mediated cGMP elevation and inhibition of platelet adhesion. Likewise, the inhibition of platelet adhesion achieved with 1 mM SNP was accompanied by marked increases in cGMP levels. Pretreatment of platelets with ODQ prevented the elevation of cGMP levels but failed to significantly affect the inhibitory effect of SNP (1 mM) on cell adhesion (Fig. 2A).
Fig. 2.
Effect of the soluble guanylate cyclase inhibitor ODQ on cGMP levels (Upper) and inhibition of platelet adhesion (Lower) by SNP at 15 (A) and 60 (B) min. Nonactivated or thrombin (50 milliunits/ml)-activated platelets (1.2 × 108 platelets/ml) were incubated with SNP (0.1 and 1 mM) for 15 or 60 min in the presence or absence of ODQ (10 μM). Dashed lines represent basal levels of cGMP. The results are shown as the means ± SEM (n = 3). ∗, P < 0.05, ∗∗, P < 0.01, and ∗∗∗, P < 0.001, compared with the experiments in the absence of ODQ.
When platelets were allowed to adhere for 60 min (Fig. 2B), addition of SNP (0.1 and 1.0 mM) increased the cGMP levels in nonactivated and thrombin-activated cells to the same extent, as that seen at 15 min measurement. Pretreatment of platelets with ODQ restored the cGMP to basal levels, and markedly reduced the inhibitory effect of SNP on cell adhesion in both nonactivated and thrombin-activated cells (Fig. 2B).
Effect of Superoxide Dismutase (SOD).
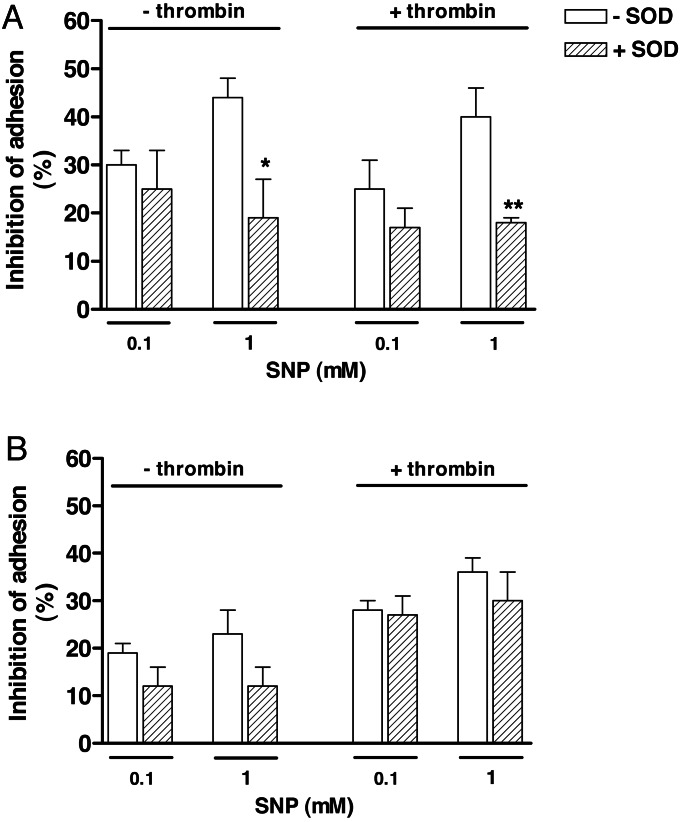
Pretreatment of platelets with SOD (100 units/ml) alone significantly inhibited the platelet adhesion at 15 min (17 ± 6% and 28 ± 4% for nonactivated and thrombin-activated platelets, respectively) and 60 min (10 ± 4% and 16 ± 5% for nonactivated and thrombin-activated platelets, respectively).
On the other hand, SOD (100 units/ml) failed to modify the inhibitory responses of 0.1 mM SNP at 15 min in nonactivated and thrombin-activated platelets, whereas at 1 mM SNP, the inhibitory effect was significantly reduced by SOD (Fig. 3A). At 60 min, SOD did not significantly affect the inhibition of platelet adhesion by SNP in any condition used (Fig. 3B).
Fig. 3.
Effect of SOD on inhibition of platelet adhesion by SNP at 15 (A) and 60 (B) min. Nonactivated or thrombin (50 milliunits/ml)-activated platelets (1.2 × 108 platelets/ml) were incubated with SNP (0.1 and 1 mM) for either 15 or 60 min in the presence of SOD (100 units/ml). Results represent the means ± SEM (n = 3). ∗, P < 0.05 and ∗∗, P < 0.01, compared with the adhesion in absence of SOD.
Effect of Epigallocatechin Gallate (ECG).
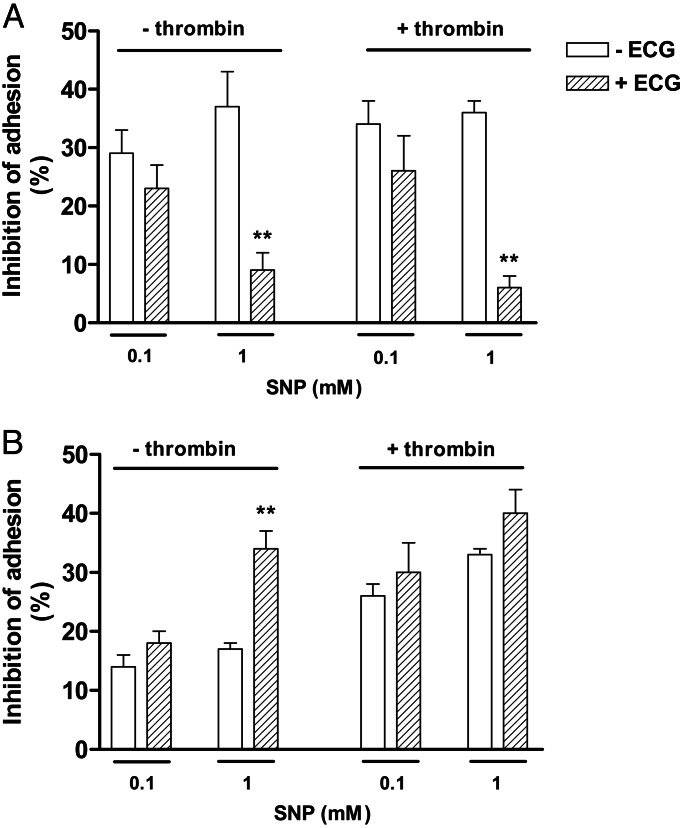
Pretreatment of platelets with ECG (5 μM) alone did not significantly affect the platelet adhesion at 15 and at 60 min (data not shown). In addition, at 15 min, ECG (5 μM) failed to modify the inhibitory responses of 0.1 mM SNP in both nonactivated and thrombin-activated platelets, whereas at 1 mM SNP, the inhibitory effect was significantly reduced by ECG (Fig. 4A). At 60 min, ECG did not significantly affect the inhibition of platelet adhesion by SNP in any condition used, except in nonactivated platelets at 1 mM SNP. In this case, ECG significantly potentiated platelet adhesion inhibition (Fig. 4B).
Fig. 4.
Effect of ECG on inhibition of platelet adhesion by SNP at 15 (A) and 60 (B) min. Nonactivated or thrombin (50 milliunits/ml)-activated platelets (1.2 × 108 platelets/ml) were incubated with SNP (0.1 and 1 mM) for either 15 min or 60 min in the presence of ECG (5 μM). Results represent the mean ± SEM (n = 3). ∗∗, P < 0.01 compared to the adhesion in absence of ECG.
Western Blotting Analysis of Nitrotyrosine.
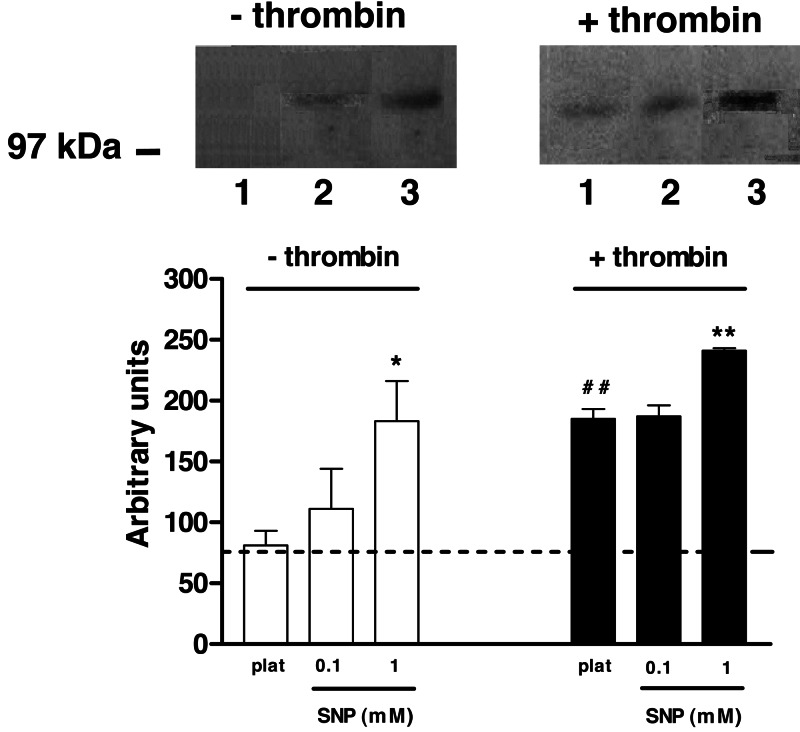
Western blot analysis of nonactivated platelets (6 × 106 platelets/ml) did not detect any protein band reacting with anti-nitrotyrosine antibodies (Fig. 5). Incubation of platelets with 0.1 mM SNP for 15 min did not reveal any nitrated protein, but when platelets were treated with 1 mM SNP, a single tyrosine-nitrated protein with an apparent molecular mass of ≈105 kDa was detected (Fig. 5). Activation of platelets by thrombin alone also led to the appearance of a nitrated band similar to SNP (1 mM) in nonactivated platelets. Detection of nitrated protein was prevented when the monoclonal antibody against nitrotyrosine was preincubated with free nitrotyrosine, confirming the specificity of nitrotyrosine immunodetection (data not shown). Exposure of thrombin-stimulated platelets to 0.1 mM SNP did not affect the intensity of immunoreactive protein band, but treatment with 1 mM SNP for 15 min enhanced the signal by 30%, as demonstrated by densitometry analysis (Fig. 5). The nitration of the detected 105-kDa band was transient, because platelets samples treated with SNP (0.1 and 1 mM) for 60 min did not show any tyrosine-nitrated protein.
Fig. 5.
Detection of nitrated protein in human platelets. Lines 1, 2, and 3 represent untreated platelets, and platelets incubated with SNP at 0.1 and 1 mM, respectively. Position of molecular marker (in kDa) is indicated on the left. (Lower) The graph shows densitometric analysis of nitrated protein bands in platelets from three different individuals. Dashed line represents background density. ∗, P < 0.05 and ∗∗, P < 0.01 compared with respective control. ##, P < 0.01 compared with nonactivated platelets.
Purification and Identification of Nitrated Protein.
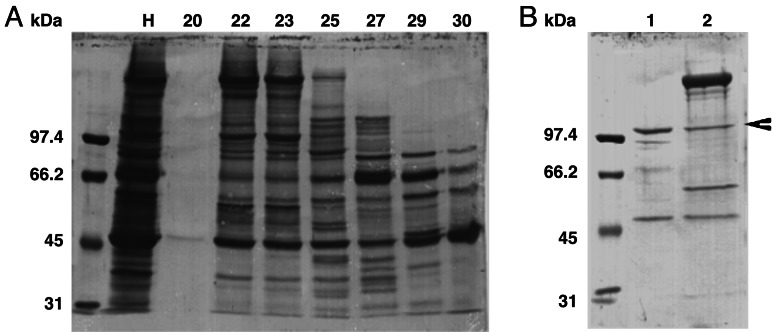
Fig. 6A shows the protein profile of the fractions obtained from Sephacryl S-200 HR column elution. Fractions 22 and 23, containing nitrated 105-kDa protein, were pooled and loaded onto a hydroxyapatite column. The elution of hydroxyapatite column with 100 and 200 mM Na2HPO4 resulted in substantial purification of the tyrosine-nitrated protein (Fig. 6B). Using capillary liquid chromatography/electrospray ionization-MS/MS, the mass and sequence of 18 alkylated tryptic peptides, obtained from the SDS gel band containing nitrated protein, were determined. The nitrated protein was identified as human α-Actinin 1 cytoskeletal isoform (P12814).
Fig. 6.
Purification of nitrated protein in washed activated platelets. Coomassie-stained SDS-polyacrylamide gel of proteins contained in fractions from elution of Sephacryl S-200 HR column (A) and hydroxyapatite column (B). Western blotting analysis using monoclonal anti-nitrotyrosine antibody indicated the presence of a nitrated band in fractions 22 and 23, which were pooled and loaded onto a hydroxyapatite column. Numbers 1 and 2 represent the bound proteins eluted with 100 and 200 mM sodium phosphate. The arrowhead indicates the protein band excised and analyzed by using capillary liquid chromatography/electrospray ionization-MS/MS. Line H represents the proteins contained in the total material before chromatography (30 μg of protein per line). Position of the molecular marker (in kDa) is indicated on the left.
Discussion
In this work, we have demonstrated that the NO-donor SNP inhibits platelet adhesion to immobilized fibrinogen mostly through cGMP-dependent mechanisms. However, cGMP-independent mechanisms, possibly through α-actinin nitration, account for the inhibition of platelet adhesion in conditions of short-time incubation (15 min) and high SNP concentration (1 mM).
The compound ODQ inhibits NO-stimulated soluble guanylyl cyclase activity and has been extensively used to study the function of the NO-cGMP transduction pathway. This inhibitory effect of ODQ is due to changes in the oxidation state of the haem moiety without adverse effects on the catalytic activity of the enzyme (21). Therefore, we have used ODQ to explore the cGMP-dependent mechanisms mediating the inhibitory effect of SNP on platelet adhesion in fibrinogen-coated plates. At a concentration used in our study, ODQ completely abolished the SNP-induced cGMP elevation, irrespective of the SNP concentration used (0.1 and 1.0 mM) or activation state of the platelets. Additionally, ODQ markedly reversed the inhibition of platelet adhesion by SNP in most of our experimental conditions, reinforcing that cGMP plays a major role mediating this phenomenon. However, our findings that ODQ failed to affect the inhibition of platelet adhesion at 15 min when SNP was used at a concentration of 1 mM indicate that a cGMP-independent mechanism takes place under this experimental condition. cGMP-independent mechanisms have also been proposed to mediate the inhibition of platelet aggregation in response to NO donors (10, 11). In our study, we have further investigated the cGMP-independent mechanisms that take place in SNP (1 mM)-treated platelets in conditions of short-time incubation (15 min). In vivo concentrations of NO can range from 5 nM (minimum amount for sGC activation) to 4 μM (postcerebral ischemic injury) (22, 23). In addition, in human platelets exposed to 10 μM SNP, the rate of spontaneous NO release is 0.04 ± 0.001 nmol/min (24). Thus, in our study, the incubation of SNP (1 mM) with platelets would generate amounts of NO found in physiological and/or pathological conditions.
Superoxide anion (O2−) has been previously shown to activate human platelets (25, 26). In agreement with these studies, our results showed that, in absence of SNP, the O2− scavenger SOD significantly prevented the adhesion of nonactivated and thrombin-activated platelets to fibrinogen, further supporting a role for O2− as a platelet activator. On the other hand, in the presence of SNP (1 mM, 15 min), SOD markedly reversed the inhibitory effect of this NO donor independently of the platelet activation state. It is well established that, in presence of SOD, O2− can be converted to hydrogen peroxide (H2O2). A previous study showed that H2O2 has no effect on platelet adhesion to immobilized fibrinogen, even at 1 mM concentration after 30 or 60 min of incubation (19). This finding suggests that in our study, H2O2 does not mediate the inhibitory effect of 15 min of treatment with 1 mM SNP on platelet adhesion. Hydrogen peroxide may also interact with O2− to form hydroxyl radical (OH•), which is reactive with a variety of cellular components and may be toxic for the cell. However, both the MTT and the acid phosphatase activity assays did not reveal any toxic effect of SNP, thus excluding that inhibition of platelet adhesion by SNP reflects OH• formation and cell death.
The O2− is relatively unreactive in comparison with many other radicals, but biological systems can convert it into other more reactive species, such as peroxynitrite (ONOO−). This molecule is derived from the extremely fast reaction with NO and is described as a potent nitrating agent. Only few reports described nitrated proteins in platelets. In one study, 3-nitrotyrosine was detected in nonactivated platelets (27). In another study, vasodilator-stimulated phosphoprotein and its phosphorylated form was mildly nitrated in nonactivated platelets, and this nitration significantly increased after activation with collagen (28). In our work, a significant nitration of a protein with an apparent molecular mass of ≈105 kDa was revealed in platelets after incubation with high concentration of SNP alone (1 mM) at 15 min, an effect further enhanced in thrombin-activated platelets. In functional assays, inhibition of platelet adhesion by high concentration of SNP at 15 min was reversed by ECG, a polyphenolic antioxidant and potent ONOO− scavenger known to prevent tyrosine nitration (29). This finding suggests that the ONOO− formation and, subsequently, tyrosine nitration have an important role in limiting the platelet activation. Interestingly, platelet activation by thrombin alone (in absence of SNP) also led to the appearance of a nitrated protein that may reflect the enhancement by thrombin of O2− production through mitochondrial oxidation or NADPH oxidase. Of interest, peroxynitrite has been shown to inhibit human platelet aggregation induced by collagen (30, 31) or arachidonic acid (32).
The nitrated protein was identified as α-actinin 1 by sequencing of 18 peptides from tryptic digestion. α-Actinin 1 is a cytoskeletal isoform member of actin-cross-linking proteins family. This protein is associated into reorganized actin cable networks in platelets during activation (33). Additionally, α-actinin may form a bridge between actin and the cytoplasmic domain of integrin (proteins that act as receptors for the extracellular matrix) at focal contacts (34).
In our study, at 60 min, no nitrated band could be detected in the platelets in any experimental condition used. Therefore, our data clearly show that the increase in platelet nitration by SNP above basal levels is detected only in conditions where the inhibition of platelet adhesion is reversed by SOD or ECG and unaffected by ODQ. The mechanism determining the loss of nitration at 60 min is unclear, but the decrease of protein nitration in nonactivated platelets has been described (35). Denitration processes, which may occur by enzymatic or chemical reaction, have also been reported in other tissues (36–38).
In conclusion, our results demonstrated that SNP, at high concentration and short incubation times, inhibits platelet adhesion by cGMP-independent mechanisms, involving O2− generation and α-actinin nitration. Several reports suggest that nitration prevents phosphorylation of specific tyrosine residues (35, 39–43). α-Actinin is closely associated with both transmembrane adhesion receptors and cytoskeletal proteins and, thus, may be an attractive regulatory target. Recently, exposure of human ventricular myocytes to peroxynitrite has been shown to produce α-actinin nitration and alterations in its conformation (44). α-Actinin is phosphorylated in position 12 by the focal adhesion kinase, which is closely linked to platelet spreading (45). Therefore, we predict that the nitration of this protein interferes with its phosphorylation and contributes consequently to the inhibitory role of NO on platelet adhesion.
Materials and Methods
Washed Platelet Preparation.
Normal human blood from healthy volunteers who had not received any medication within the previous 10 days was collected in 1:9 (vol/vol) 12.4 mM sodium citrate/13 mM citric acid/11 mM glucose and centrifuged (200 × g, 15 min). Five milliliters of platelet-rich plasma were added to 7 ml of washing buffer (140 mM NaCl/0.5 mM KCl/12 mM trisodium citrate/10 mM glucose/12.5 mM saccharose, pH 6), and centrifuged (800 × g, 10 min). The pellet was resuspensed in washing buffer, and the procedure was repeated once. Finally, the platelets were gently suspended in Krebs solution (118 mM NaCl/25 mM NaHCO3/1.2 mM KH2PO4/1.7 mM MgSO4/5.6 mM glucose, pH 7.4). The platelet number was adjusted to 1.2 × 108 platelets/ml in presence of 1 mM CaCl2.
Adhesion Assay.
Adhesion assay was carried out according to Bellavite et al. (20). Briefly, the 96-well microtiter plates were coated (overnight at 4°C) by adding 50 μl per well of human fibrinogen (50 μg/ml). Before use, the wells were washed twice with Krebs solution. The nonspecific adhesion was blocked by incubation of wells with 1% BSA (1 h, 37°C). At the end of incubation, the plate was washed, and platelet suspension (50 μl containing 6 × 106 platelets) was added to each well. The plate was carefully washed twice with Krebs solution (200 μl per well) to remove unattached platelets. Adherent platelets were quantified through the measurement of acid phosphatase activity. Wells containing adherent platelets were incubated with 150 μl per well of acid phosphatase substrate solution (0.1 M citrate buffer, pH 5.4, containing 5 mM p-nitrophenyl phosphate and 0.1% Triton X-100), and after 1 h of incubation at room temperature, the reaction was stopped and the color was developed by the addition of 100 μl of 2 M NaOH. The p-nitrophenol produced in reaction was measured with a microplate reader at 405 nm. The percentage of adherent cells was calculated on the basis of a standard curve obtained with known numbers of platelets. All experiments were performed in triplicate. Cell toxicity was estimated by using the tetrazolium salt reduction test (MTT assay) after exposure to NO donor (46).
Extraction and Measurement of cGMP.
Platelets (1.2 × 108 platelets/ml) were incubated with the phosphodiesterase inhibitor 3-isobutyl-l-methyl-xanthine (2 mM) for 15 min. Nonactivated or thrombin-activated platelets were incubated with SNP (0.1 or 1.0 mM) for 15 or 60 min. The reaction was interrupted by the addition of cold-acidified absolute ethanol (67%, vol/vol), and samples were vigorously agitated for 30 s. Cell samples were centrifuged (4,000 × g, 30 min at 4°C). Supernatants were dried at 55–60°C under a stream of nitrogen. cGMP was measured by using a kit from Cayman Chemical (Ann Arbor, MI).
Western Blotting for 3-Nitrotyrosine.
Washed platelets (6 × 106 platelets/ml) were incubated (37°C) for 15 or 60 min with SNP (0.1 or 1 mM) in the presence or absence of thrombin (50 milliunits/ml). The platelet suspension (1 ml) was centrifuged at 10,000 × g for 10 min at 4°C. The pellet was suspended in 40 μl of dissociating buffer (50 mM Tris·HCl, pH 6.8/2% SDS/10% glycerol/2% 2-mercaptoetanol/0.1% bromofenol blue). Samples (40 μl per well) and 1 μg per well of nitrated BSA were loaded on SDS/PAGE (7.0%). After separation, proteins were electrophoretically transferred to poly(vinylidene difluoride) (PVDF) membranes and blocked for 30 min at room temperature in solution containing 5% of nonfat milk in Tris-buffered saline-Tween (TBS-T; 20 mM Tris·HCl, pH 7.2/0.3 M NaCl with 0.1% Tween-20). The membrane was incubated overnight in 5% nonfat milk containing a 1:2,000 dilution of mouse monoclonal anti-nitrotyrosine antibodies (Upstate Biotechnology) at 4°C. After washing with TBS-T, immureactive proteins were detected by using horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biosciences) and enhanced chemiluminescence. Control experiments for nitrotyrosine immunoreactivity were performed by preincubating (30 min, room temperature) the anti-nitrotyrosine antibody with 4 mM free 3-nitrotyrosine before incubation with PVDF membrane.
Purification and Identification of the Nitrated Protein.
Resting or thrombin-activated (50 milliunits/ml) platelet samples (1 × 109 platelets/ml) were collected in the presence of protease inhibitors (10 μg/ml soybean trypsin inhibitor/64 μM benzamidine/0.005 unit/ml aprotinin/25 μM leupeptin/15 μM pepstatin A/8 μM antipain/200 μM PMSF) and immediately frozen in liquid nitrogen. The samples were sonicated for 10 min at 4°C and centrifuged at 10,000 × g for 5 min at 4°C. The supernatant was collected and centrifuged at 100,000 × g for 1 h at 4°C. The samples were stored at –80°C until further processing.
Proteins from platelets high-speed supernatant were precipitated with 60% of ammonium sulfate for 18 h at 4°C with agitation. The suspension was centrifuged at 12,000 × g (4°C) for 10 min, and the pellet was suspended in 15 ml of 10 mM sodium phosphate buffer (pH 7.2). The Sephacryl S-200 HR column (1 × 100 cm; Amersham Pharmacia Biosciences) was equilibrated with 10 mM sodium phosphate buffer (pH 7.2) containing 0.3 M NaCl, and then 1 ml of platelet supernatant was loaded and chromatographed in the same buffer at a flow rate of 10 ml/h. Collected fractions were analyzed by Western blotting with the monoclonal anti-nitrotyrosine antibody. Fractions containing nitrated protein were pooled and loaded onto a hydroxyapatite column (1 × 2 cm; Bio-Rad) equilibrated with 10 mM sodium phosphate buffer (pH 7.2). Bound proteins were eluted with a step gradient of increasing sodium phosphate concentration (50, 100, 150, and 200 mM) at a flow rate of 6 ml/h, and the fractions analyzed by Western blotting. The SDS gel band containing the purified nitrated protein was stained with Coomassie blue and excised by using a razor blade. The excised gel band was destained, and the protein was reduced, alkylated with iodoacetamide, and digested with trypsin (47). The resulting alkylated tryptic peptides were extracted, dried, and analyzed by using capillary liquid chromatography/electrospray ionization-MS/MS in data-dependent acquisition mode. The tandem MS data were processed by using proteinlynx software (Micromass, Manchester, U.K.) and the Mascot MS/MS Ion Search tool (www.matrixscience.com).
Statistical Analysis.
The results of adhesion and viability assays are expressed as means ± SEM, and the statistical significance between groups was determined by using the Bonferroni test for ANOVA. Where appropriate, Student’s t test was used to compare specific groups. Significance was established at P < 0.05.
Acknowledgments
S.M. was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo, Brazil.
Abbreviations
- cGMP
cyclic GMP
- ECG
epigallocatechin gallate
- SNP
sodium nitroprusside
- SOD
superoxide dismutase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Dopheide S. M., Yap C. L., Jackson S. P. Clin. Exp. Pharmacol. Physiol. 2001;28:355–363. doi: 10.1046/j.1440-1681.2001.03468.x. [DOI] [PubMed] [Google Scholar]
- 2.Niiya K., Hodson E., Bader R., Byers-Ward V., Koziol J. A., Plow E. F., Ruggeri Z. M. Blood. 1987;70:475–483. [PubMed] [Google Scholar]
- 3.Savage B., Shattil S. J., Ruggeri Z. M. J. Biol. Chem. 1992;267:11300–11306. [PubMed] [Google Scholar]
- 4.Nolte C., Eigentaler M., Horstrup K., Honig-Liedl P., Walter U. Biochem. Pharmacol. 1994;48:1569–1575. doi: 10.1016/0006-2952(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 5.Noack E., Feelisch M. Basic Res. Cardiol. 1991;2:37–50. doi: 10.1007/978-3-642-72461-9_5. [DOI] [PubMed] [Google Scholar]
- 6.Anfossi G., Russo I., Massucco P., Mattiello L., Balbo A., Cavalot F., Trovati M. Thromb. Res. 2001;102:319–330. doi: 10.1016/s0049-3848(01)00240-7. [DOI] [PubMed] [Google Scholar]
- 7.Chintala M. S., Bernardino V., Chiu P. J. J. Pharmacol. Exp. Ther. 1994;271:1203–1208. [PubMed] [Google Scholar]
- 8.Polanowska-Grabowska R., Gear A. R. Blood. 1994;83:2508–2515. [PubMed] [Google Scholar]
- 9.Wu C.-C., Ko F.-N., Teng C.-M. Biochem. Biophys. Res. Commun. 1997;231:412–416. doi: 10.1006/bbrc.1996.5998. [DOI] [PubMed] [Google Scholar]
- 10.Brune B., Lapetina E. G. J. Biol. Chem. 1989;264:8455–8458. [PubMed] [Google Scholar]
- 11.Tsikas D., Ikie M., Tewes K. S., Raida M., Frölich J. C. FEBS Lett. 1999;442:162–166. doi: 10.1016/s0014-5793(98)01633-0. [DOI] [PubMed] [Google Scholar]
- 12.Greenacre S. A. B., Ischiropoulos H. Free Radical Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 13.Souza J. M., Daikhin E., Yudkoff M., Raman C. S., Ischiropoulos H. Arch. Biochem. Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard-Fillion B., Souza J. M., Friel T., Jiang G. C., Vrana K., Sharov V., Barron L., Schoneich C., Quijano C., Alvarez B., et al. J. Biol. Chem. 2001;276:46017–46023. doi: 10.1074/jbc.M105564200. [DOI] [PubMed] [Google Scholar]
- 15.Zou M., Martin C., Ullrich V. Biol. Chem. 1997;378:707–713. doi: 10.1515/bchm.1997.378.7.707. [DOI] [PubMed] [Google Scholar]
- 16.Yamakura F., Taka H., Fujimura T., Murayama K. J. Biol. Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 17.Turko I. V., Marcondes S., Murad F. Am. J. Physiol. 2001;281:H2289–H2294. doi: 10.1152/ajpheart.2001.281.6.H2289. [DOI] [PubMed] [Google Scholar]
- 18.Miller M. R., Hanspal I. S., Hadoke P. W., Newby D. E., Rossi A. G., Webb D. J., Megson I. L. Cardiovasc. Res. 2003;57:853–860. doi: 10.1016/s0008-6363(02)00779-4. [DOI] [PubMed] [Google Scholar]
- 19.Belisario M. A., Tafuri S., Domenico C., Squillacioti C., Morte R. D., Lucisano A., Staiano N. Biochim. Biophys. Acta. 2000;1495:183–193. doi: 10.1016/s0167-4889(99)00160-3. [DOI] [PubMed] [Google Scholar]
- 20.Bellavite P., Andrioli G., Guzzo P., Arigliano P., Chirumbolo S., Manzato F., Santonastaso C. Anal. Biochem. 1994;216:444–450. doi: 10.1006/abio.1994.1066. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y., Brandish P. E., DiValentin M., Schelvis J. P., Babcock G. T., Marletta M. A. Biochemistry. 2000;39:10848–10854. doi: 10.1021/bi9929296. [DOI] [PubMed] [Google Scholar]
- 22.Malinski T., Bailey F., Zhang Z. G., Chopp M. J. Cereb. Blood Flow Metab. 1993;13:355–358. doi: 10.1038/jcbfm.1993.48. [DOI] [PubMed] [Google Scholar]
- 23.Hanafy K., Krumenacker J. S., Murad F. Med. Sci. Monit. 2001;7:801–819. [PubMed] [Google Scholar]
- 24.Gordge M. P., Hothersall J. S., Noronha-Dutra A. A. Br. J. Pharmacol. 1998;124:141–148. doi: 10.1038/sj.bjp.0701821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvemini D., De Nucci G., Sneddon J. M., Vane J. R. Br. J. Pharmacol. 1989;97:1145–1150. doi: 10.1111/j.1476-5381.1989.tb12572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krotz F., Sohn H. Y., Gloe T., Zahler S., Riexinger T., Schiele T. M., Becker B. F., Theisen K., Klauss V., Pohl U. Blood. 2002;100:917–924. doi: 10.1182/blood.v100.3.917. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H., Balazy M. Nitric Oxide. 1998;2:350–359. doi: 10.1006/niox.1998.0196. [DOI] [PubMed] [Google Scholar]
- 28.Naseem K. M., Low S. Y., Sabetkar M., Bradley N. J., Khan J., Jacobs M., Bruckdorfer K. R. FEBS Lett. 2000;473:119–122. doi: 10.1016/s0014-5793(00)01490-3. [DOI] [PubMed] [Google Scholar]
- 29.Fiala E. S., Sodum R. S., Bhattacharya M., Li H. Experientia. 1996;52:922–926. doi: 10.1007/BF01938881. [DOI] [PubMed] [Google Scholar]
- 30.Moro M. A., Darley-Usmar V. M., Goodwin D. A., Read N. G., Zamora-Pinto R., Feelisch M., Radomski M. W., Moncada S. Proc. Natl. Acad. Sci. USA. 1994;91:6702–6706. doi: 10.1073/pnas.91.14.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown A. S., Moro M. A., Masse J. M., Cramer E. M., Radomski M., Darley-Usmar V. Cardiovasc. Res. 1998;40:380–388. doi: 10.1016/s0008-6363(98)00182-5. [DOI] [PubMed] [Google Scholar]
- 32.Boulos C., Jiang H., Balazy M. J. Pharmacol. Exp. Ther. 2000;293:222–229. [PubMed] [Google Scholar]
- 33.Tanaka K., Itoh K. J. Struct. Biol. 1998;124:13–41. doi: 10.1006/jsbi.1998.4051. [DOI] [PubMed] [Google Scholar]
- 34.Otey C. A., Pavalko F. M., Burridge K. J. Cell. Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabetkar M., Low S. Y., Naseem K. M., Bruckdorfer K. R. Free Radical Biol. Med. 2002;33:728–736. doi: 10.1016/s0891-5849(02)00890-0. [DOI] [PubMed] [Google Scholar]
- 36.Kamisaki Y., Wada K., Bian K., Balabanli B., Davis K., Martin E., Behbod F., Lee Y. C., Murad F. Proc. Natl. Acad. Sci. USA. 1998;95:11584–11589. doi: 10.1073/pnas.95.20.11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irie Y., Saeki M., Kamisaki Y., Martín E., Murad F. Proc. Natl. Acad. Sci. USA. 2003;100:5634–5639. doi: 10.1073/pnas.1131756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koeck T., Fu X., Hazen S. L., Crabb J. W., Stuehr D. J., Aulak K. S. J. Biol. Chem. 2004;279:27257–27262. doi: 10.1074/jbc.M401586200. [DOI] [PubMed] [Google Scholar]
- 39.Gow A. J., Duran D., Malcolm S., Ischiropoulos H. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 40.Kong S.-K., Yim M. B., Stadtman E. R., Chock P. B. Proc. Natl. Acad. Sci. USA. 1996;93:3377–3382. doi: 10.1073/pnas.93.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman D. K., Hoffman S., Kotamraju S., Zhao T., Wakim B., Kalyanaraman B., Newman P. J. Biochem. Biophys. Res. Commun. 2002;296:1171–1179. doi: 10.1016/s0006-291x(02)02060-0. [DOI] [PubMed] [Google Scholar]
- 42.Monteiro H. P. Free Radical Biol. Med. 2002;33:765–773. doi: 10.1016/s0891-5849(02)00893-6. [DOI] [PubMed] [Google Scholar]
- 43.Casoni F., Basso M., Massignan T., Gianazza E., Cheroni C., Salmona M., Bendotti C., Bonetto V. J. Biol. Chem. 2005;280:16295–16304. doi: 10.1074/jbc.M413111200. [DOI] [PubMed] [Google Scholar]
- 44.Borbély A., Tóth A., Édes I., Virág L., Papp J. G., Varró A., Paulus W. J., Velden J., Stienen G. J. M., Papp Z. Cardiovasc. Res. 2005;67:225–233. doi: 10.1016/j.cardiores.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 45.Izaguirre G., Aguirre L., Hu Y-P., Lee H. Y., Schlaepfer D. D., Aneskievich B. J., Haimovich B. J. Biol. Chem. 2001;276:28676–28685. doi: 10.1074/jbc.M101678200. [DOI] [PubMed] [Google Scholar]
- 46.Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 47.Hellman U., Wernstedt C., Gonez J., Heldin C. H. Anal. Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]