Abstract
Somatic chromosome spreads from maize (Zea mays L.) plants containing B-A translocation chromosomes undergoing the chromosome type breakage–fusion–bridge cycle were examined by FISH. The size and type of extra chromosomes varied among cells of the same individual. A collection of minichromosomes derived from the chromosome type breakage–fusion–bridge cycle was examined for the presence of stable dicentric chromosomes. Six of 23 chromosomes in the collection contained two regions with DNA sequences typical of centromeres. Functional analysis and immunolabeling of CENH3, the centromere-specific histone H3 variant, revealed only one functional centromere per chromosome, despite the duplicate centromere sequences. One plant was found with an inactive B centromere that had been translocated to the short arm of chromosome 9. The translocated centromere region appeared identical to that of a normal B chromosome. The inactivation of the centromeres was stable for at least four generations. By using dicentrics from dispensable chromosomes, centromere inactivation was found to be quite common under these circumstances.
Keywords: B chromosome, breakage–fusion–bridge cycle, centromeres
Chromosomal rearrangements or de novo centromere formation can produce two linked centromeres that will migrate separately to newly forming daughter cells, forming a chromatin bridge that will break. The broken ends may subsequently fuse, reforming a dicentric chromosome, albeit with some of the intervening chromatin missing. This process is called the breakage–fusion–bridge (BFB) cycle (1, 2). When stable dicentric chromosomes have been recovered, they are functionally monocentric. In dicentric chromosomes with well separated centromeres, stabilization occurs by the poorly understood phenomenon of centromere inactivation (3). Stability can also be achieved if the centromeres are very close together and form only one heterochromatic block (3). To date, no examples of an inactivated plant centromere have been reported.
Extra chromosomes, called B chromosomes, have been identified in diverse taxa, (4) including maize (5). The presence of these chromosomes has little obvious effect on the phenotype of a plant that harbors them, yet they persist by taking advantage of the cellular mechanisms responsible for faithful chromosome replication and transmission. Because these chromosomes are entirely dispensable but contain the essential components required for efficient transmission through mitosis and meiosis, they provide an excellent model to study centromeres.
Reports of the chromosome type BFB cycle describe the fate of a translocation chromosome involving the B chromosome and a variant of chromosome 9, which contains an inverted duplication of its short arm (6, 7). The duplicated section of 9S can fold back and recombine with itself during meiosis I, creating a chromosome that will be cleaved during anaphase II, because it contains two centromeres. The B9-Dp9 chromosome undergoes nondisjunction at the second pollen mitosis, and two broken chromosomes can be delivered to the zygote, initiating the chromosome type BFB cycle (6). Because this chromosome is dispensable and moves independently from the intact chromosome 9, it provides a method to track the progress of a dicentric chromosome throughout the life cycle and to recover the resulting chromosomes.
We describe six independent stable dicentric chromosomes resulting from this process, including a chromosome in which the B centromere has been transferred to the short arm of chromosome 9. In all cases, only one centromere is functional. By using a dispensable chromosome, it was possible to demonstrate that centromere inactivation can be quite common and can occur in plants.
Results
Cytological Examination of Somatic Metaphase Chromosomes in Root Tips Undergoing the BFB Cycle.
A previous study of the BFB cycle involving the B9-Dp9 chromosome examined cells during telophase for double bridges, which indicate that the chromosome type BFB cycle is active (6). In about one third of the plants, minichromosomes were observed in meiotic samples (6). Subsequent work involved assembling a collection of these minichromosomes, which varied in size, transmission rate, and type of DNA elements that were present (7). Fig. 1 illustrates the hypothesized events leading to minichromosome formation.
Fig. 1.
The B9-Dp9 chromosome initiates the BFB cycle after crossing over with itself. (A) The duplicated portion of B9-Dp9 pairs with itself during meiosis I, and a crossover creates a dicentric chromosome and an acentric fragment. (B) During meiosis II, the sister centromeres migrate to opposite poles, and the resulting chromatin bridge is broken. (C) A chromosome with a broken end is delivered to the microsporocyte. (D) In the microsporocyte, the broken chromosome is replicated and fuses with itself. The centromeres migrate to opposite poles during the first pollen mitotic division, breaking the chromosome. (E) After the first pollen division, the broken chromosome is replicated in the generative cell. Because the B centromere usually undergoes nondisjunction during the second pollen division, both centromeres move to one pole. (F) In the subsequent somatic division after fertilization, the dicentric chromosome is replicated, and the centromeres could move to the poles independently of one another. If two centromeres on the same chromatid move to opposite poles, a double bridge is formed that might break at different locations. The broken ends of the chromosomes in each daughter cell can fuse, leading to another round of the chromosome type BFB cycle. As this process continues, chromosomes can be stabilized by inactivation of one of the centromeres or by healing of a broken end by addition of telomeres, leading to a variety of minichromosomes.
FISH using the B chromosome-specific element ZmBs and the 180-bp knob heterochromatin-repeat probes was performed on somatic chromosome spreads from root tips resulting from a hybrid between a tester stock and a male containing one copy of the B9-Dp9 and two copies of the reciprocal 9-B chromosome to visualize the effects of the BFB cycle. The ZmBs probe allows the centromere of the B chromosome to be identified, and the 180-bp knob repeat hybridizes to a location near the B centromere and to the knob normally found near the tip of chromosome 9 (8–10).
Some of the progeny contained intact B9-Dp9 chromosomes that have passed through meiosis without experiencing a crossover in the duplicated region (Fig. 2A). All chromosome spreads examined from this type of root tip contained two B9-Dp9 chromosomes. In other root tips, the number and appearance of chromosomes varied from cell to cell, indicative of the BFB cycle (Fig. 2 B–D). Large and small dicentric chromosomes were observed as well as telocentric chromosomes and small chromatin fragments. Up to four chromosomes with ZmBs signals were present in a single cell, all being telocentric. In some cells, two distinct dicentric chromosomes were observed (Fig. 2C). In root tips collected from 3-week-old seedlings, only very small fragments and larger telocentric chromosomes, but no dicentric chromosomes, were found.
Fig. 2.

BFB cycle in root tips. A plant containing one B9-Dp9 and two 9-B translocation chromosomes was crossed as a male to a yg2, bz1 tester line. The resulting kernels were germinated, chromosome spreads prepared from the root tips, and FISH performed by using the B chromosome-specific ZmBs probe (green) and the 180-bp knob heterochromatin probe (red). Lack of crossing over in the duplicated region, followed by nondisjunction during the second pollen mitosis, results in two intact B9-Dp9 chromosomes as in A. (B–D) Different cells from a single root tip vary in the size and number of extra chromosomes. The arrowhead indicates the ZmBs signal at the tip of the 9-B chromosome, and arrows indicate the ZmBs signal of the centromeric region.
Dicentric Minichromosomes Resulting from the BFB Cycle.
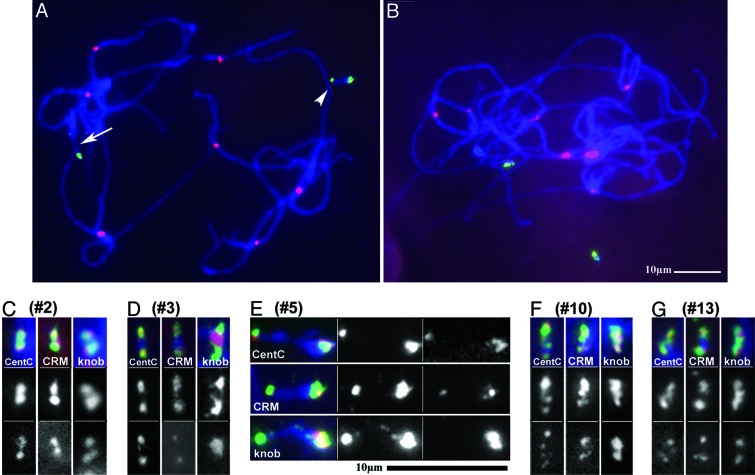
Because the BFB cycle can be stopped by inactivation of one centromere, the collection of minichromosomes (7) was screened for stable dicentrics. Chromosomes at the pachytene stage of meiosis are much more extended than at somatic metaphase; therefore, meiotic samples were collected and hybridized with FISH probes against the centromeric elements ZmBs, CentC, and CRM as well as the 180-bp knob repeat. Of the 23 chromosomes examined (including 9 previously unreported cases), 5 were identified that contained two sites where the ZmBs signal colocalized with CentC and CRM (minichromosomes 2, 3, 5, 10, and 13 (Fig. 3; and see Fig. 6, which is published as supporting information on the PNAS web site). An additional stable dicentric chromosome was identified that contained a translocated B centromere on chromosome 9S (discussed below).
Fig. 3.
Pachytene FISH of dicentric minichromosomes. (A) A pachytene spread with an intact B chromosome (indicated by the arrow) and minichromosome 5 hybridized with ZmBs (green) and CRM (red). The arrowhead indicates the active centromere of minichromosome 5. (B) A pachytene spread with two unpaired copies of minichromosome 10 hybridized with ZmBs (green) and CRM (red) illustrates the relative size of the minichromosomes. (C–G) Minichromosomes 2, 3, 5, 10, and 13, respectively, are depicted from pachytene spreads. Chromosomes were hybridized with ZmBs (green) and CentC, CRM, or the 180-bp knob repeat (red). The gray values for the probes are also displayed, first ZmBs and, second, the indicated repetitive probe (CentC, CRM, or the 180-bp knob repeat).
By comparing the FISH signal patterns of the minichromosomes to those of an intact B centromeric region (see Fig. 7, which is published as supporting information on the PNAS web site), some of the rearrangements leading to the formation of the minichromosomes can be determined. The centromeric region of the B chromosome contains two blocks of ZmBs that are megabases in length and that flank a 700-kb “core” consisting of interspersed CentC, CRM, and ZmBs elements. This core contains the functional kinetochore-binding domain and is the site of CENH3 localization (9, 11). Adjacent to the block of ZmBs on the long arm, there is a stretch of 180-bp knob repeat, followed by another block of ZmBs intermixed with CentC and CRM (8, 9). The short arm of chromosome 9 has a site of strong hybridization to the 180-bp knob repeat, which appears as two large signals on the B9-Dp9 chromosome (Fig. 2A).
At the cytological level, minichromosomes 2, 3, and 13 contained two identical B centromere segments (Fig. 3). This type of structure is one predicted outcome of the BFB process (Fig. 1) and might result when the breaks in both chromatids of the double bridge occur in the same location. ZmBs, CentC, CRM, and the 180-bp knob repeat hybridize to multiple sites in minichromosome 10, indicating that a complex series of rearrangements formed this minichromosome.
The ZmBs hybridization signals on minichromosome 5 were well separated, and both colocalized with CentC and CRM signals (Fig. 3). Only one of the two regions of centromeric-element hybridization also contained 180-bp knob-repeat signals and the intensity of the CentC/CRM signal was greater at this site than the other. The lack of 180-bp knob repeat at that centromere suggests that, as two active centromeres pulled the chromatin, a break occurred very near to one of the B centromeres, removing the block of 180-bp knob repeat, two blocks of ZmBs, and a portion of the 700-kb core domain. The resulting fragment contained little more than the centromere. This small fragment was healed by attachment to another chromatid fragment that resulted from a break further out on the chromosome arm.
Minichromosomes with Two Centromeric Regions Contain a Single Functional Centromere.
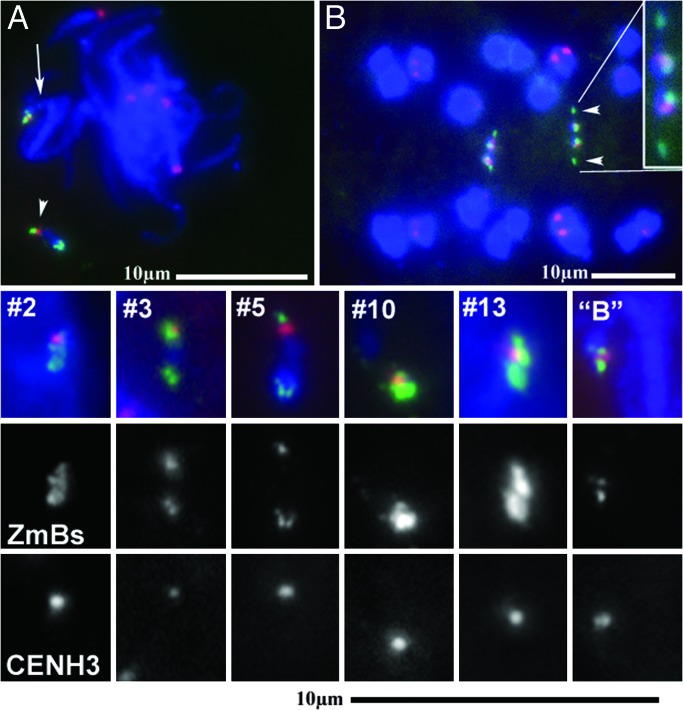
Antibodies against CenH3, the centromeric H3 histone variant, were used to label cytological preparations containing the stable dicentric minichromosomes to confirm that only one centromere remained active. In each case, a single site of localization was observed (Fig. 4). For minichromosomes 2, 3, and 13, which contain two identical centromere regions, it was not possible to distinguish which of the centromeric regions contained the functional centromere. In minichromosome 5, the region with no 180-bp knob labeling and less hybridization signal to the centromeric element probes is the region that was labeled by anti-CenH3 antibodies (Fig. 4). During anaphase, the smaller region of centromeric elements was stretched toward the two poles and leads the remainder of the chromosome (Fig. 4). The smaller site of centromeric elements was the sole site of CenH3 recruitment in five different plants observed over two generations, which indicated that the location of the active centromere is stable. In minichromosome 10, CenH3 occurs at the smaller site of ZmBs hybridization (Fig. 4).
Fig. 4.
Localization of the active centromeres of the minichromosomes. (A) Meiotic samples containing minichromosomes 5 (indicated with an arrowhead) and an intact B chromosome (indicated by an arrow) with immunolabeled CENH3 (red) hybridized with ZmBs (green). (B) Anaphase I cell spread, containing two unpaired copies of minichromsome 5 hybridized with ZmBs (green) and the 180-bp knob repeat (red). The smaller sites of ZmBs hybridization (indicated with arrowheads in one homologue) move toward the poles. The figures below show CENH3 immunolabeling (red) and ZmBs (green) hybridization of the minichromosomes and an intact B chromosome. Only one site of CENH3 labeling is observed per chromosome. The gray values for ZmBs and CENH3 are presented below the merged image.
In minichromosomes 3, 10, and 13, FISH showed a distinct site of CentC and CRM located between two blocks of ZmBs hybridization (Fig. 4), similar to the pattern seen on an intact B centromere (9, 11). The CenH3 labeling also appeared at this position in the minichromosomes and the intact B centromere. In minichromosome 5, the larger region of centromeric-element hybridization shows a similar pattern of element distribution as the intact B centromere, but this region does not function as a centromere (Fig. 4 A and B). Instead, the CenH3 labeling is immediately adjacent to the other, smaller area of ZmBs (Fig. 4A). CentC and CRM elements also hybridize to this site (Fig. 3E). Thus, CenH3 appears to be associated with the same interspersion of CentC, CRM, and ZmBs as occurs in the intact B centromere. Therefore, inactivation of one centromere did not affect the positioning of the remaining active centromere.
Origin of an A-B Dicentric Translocation Chromosome in Maize.
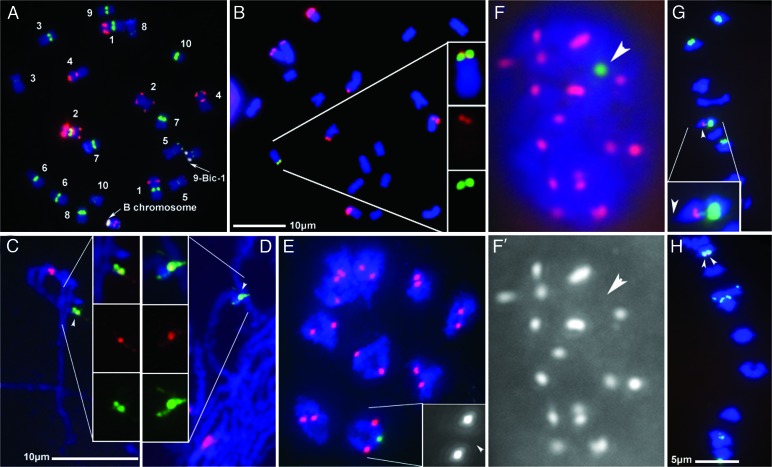
During a screen for additional minichromosomes, a large chromosome was discovered that contained strong ZmBs and intermediate 180-bp knob repeat signals at the tip of its short arm. Application of a mixture of FISH probes that allows the maize karyotype to be identified (10) indicated that this chromosome contained two centromeric regions, one from a B chromosome and the other from chromosome 9. Hereafter, this chromosome is referred to as 9-B inactive centromere-1 (9-Bic-1). CentC and CRM signals colocalized with the ZmBs signal at the tip of 9-Bic-1 (Fig. 5A and B; and see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 5.
Cytological analysis of the translocation chromosome 9-Bic-1. For all figures, samples from a 9-Bic-1/+ heterozygote plant were examined. (A) Somatic chromosome spreads containing an intact B chromosome hybridized with ZmBs (white) and a mixture of probes that allow the chromosomes to be identified. The probes include CentC (green), “TAG” microsatellite (red), 180-bp knob repeat (blue), NOR (green), 5S rRNA (yellow), subtelomeric repeat 4–12-1 (green), and a 1.1 subtelomeric repeat (red). The chromosomes were counterstained with DAPI (blue). The arrows designate the B centromeric regions. (B) Knob signal (red) is observed adjacent to the ZmBs signal (green). (C) Pachytene chromosomes hybridized with ZmBs (green) and CRM (red). (D) Pachytene chromosomes hybridized with ZmBs (green) and CentC (red). (E) Diakinesis chromosomes with immunolabeled CENH3 (red) hybridized with ZmBs (green). The arrowhead in the inset indicates the location of the ZmBs signal in an enlarged image showing only the CENH3 signal. (F) A mitotic interphase nucleus with immunolabeled CENH3 (red) hybridized with ZmBs (green). The CENH3 signal alone is shown in (F′), with an arrowhead indicating the location of ZmBs. (G and H) Anaphase chromosomes hybridized with ZmBs (green) and the 180-bp knob repeat (red). Arrowheads indicate the ZmBs signal. The ZmBs repeat is not leading to the poles. (Inset) The arrowhead indicates the portion of chromosome 9 that is leading. In G, the presence of ZmBs signal on both separating chromosomes results from a crossover. The scale for A is the same as for B, C–E are the same scale, and G and H are the same scale.
Self-pollination of plants carrying this chromosome resulted in progeny containing two copies of 9-Bic-1. Plants homozygous for this chromosome were albino and died at the seedling stage, most probably because of removal of the very distal part of chromosome 9S including the white deficiency (wd) gene. Kernels on the ears resulting from self-pollination were all recessive bronze1 (bz1) and dominant Colored1 (C1) (see Fig. 9, which is published as supporting information on the PNAS web site). Because the dominant C1 allele was present in all kernels (including homozygotes for 9-Bic-1), it must be present on the 9-Bic-1 chromosome. This information allowed the breakpoint of the translocation to be placed on the genetic map between wd and c1. The original B-9Dp9 chromosome contained the dominant Bz1 allele, suggesting that 9-Bic-1 was formed by placement of a B centromeric region onto the recessive bz1 tester chromosome. This formation could have occurred through a meiotic recombination between the bz1 tester chromosome and a dicentric minichromosome (see Fig. 10, which is published as supporting information on the PNAS web site). Pairing between 9-Bic-1 and chromosome 9 occurred along most of the chromosomes during meiosis (Fig. 5 C and D). The missing portion on 9S is not critical for gametophyte function, because 9-Bic-1 was transmitted through both the male and female flowers.
Pachytene FISH Analysis of the Translocated B Centromere of 9-Bic-1.
The 9-Bic-1 chromosome was labeled with FISH probes for CentC, CRM, ZmBs, and the 180-bp knob repeat to compare the patterns of hybridization to the intact B chromosome. The ZmBs probe hybridized to three blocks, two located immediately adjacent to each other (and that appeared as a single large block in most spreads), and the third was separated by a short distance. The CentC and CRM signal appeared at a site located between the closely adjoined ZmBs signals with an additional weak signal colocalizing with the third block of ZmBs in some spreads (Fig. 5 C and D). This pattern is the same as an intact B centromeric region (9, 11). Because of the close proximity of the large knob on the normal chromosome 9 that was paired with 9-Bic-1, it was not possible to observe the 180-bp knob-repeat signal on the translocated B region of 9-Bic-1. However, the 180-bp-knob signal is revealed in somatic preparations in close proximity to the ZmBs signal (Fig. 5B). Thus, the translocated B centromere contained all of the same elements as the progenitor B centromere and, at the cytological level, appeared unaltered.
The Translocated B Centromere Is Inactive.
Interphase root-tip nuclei and meiotic samples containing 9-Bic-1 were labeled with antibodies against CENH3 and hybridized with ZmBs. CENH3 signal did not colocalize with ZmBs (Fig. 5 E and F). At anaphase, the ZmBs signal lagged behind the centromere from chromosome 9 (Fig. 5 G and H), indicating that it did not form a functional kinetochore or bind microtubules. In addition to the site of microtubule attachment, centromeric regions serve as the site of sister-chromatid cohesion. During mitotic metaphase, the portions of the chromosomes that remain bound are marked by phosphorylation of the serine-10 residue of the canonical H3 histone protein (12, 13). Mitotic-chromosome spreads containing 9-Bic-1 were labeled by using antibodies against this chromatin modification (pSer10-H3). Only the chromosome 9 centromeric region was labeled (see Fig. 11, which is published as supporting information on the PNAS web site). Additionally, ZmBs signals on sister chromatids of 9-Bic-1 separated at mitotic metaphase, whereas the centromeric regions from chromosome 9 remain attached (Figs. 5 A and B and 8). The 9-Bic-1 chromosome has been maintained for more than four generations, and >100 root tips have been examined as kernels were scored for the 9-Bic-1 chromosome during routine classification. No evidence of chromosome instability that would arise from reactivation of the translocated B centromere has been observed. Taken together, this evidence indicates that the translocated B centromere region located on 9S is stably inactive for kinetochore formation and sister-chromatid cohesion typical of centromeres at metaphase.
Discussion
Many studies have been conducted in plants to examine the passage of dicentric chromosomes through cell division, but, to our knowledge, there are no reports of inactivated plant centromeres. In other species, inactivation of one centromere has allowed the recovery of dicentric chromosomes, leading to the conclusion that the mere presence of particular DNA elements is not sufficient to organize a centromere (14). The previous inability to observe inactive plant centromeres may have been because of the difficulty in recovering chromosomes with large deficiencies, which would be selected against in the gametophytic generation. The approach described in this study, namely, to use the maize B chromosome to study dicentrics, takes advantage of the dispensable nature of the maize B chromosome. Any chromosomes that result, including the stabilized dicentric chromosomes, segregate independently of the other chromosomes and can be recovered for study.
One third of the plants containing a B9-Dp9 chromosome undergoing the BFB have a minichromosome observed in meiosis (6). In the remaining plants, the B9-Dp9 chromosome is completely lost during development. This loss could occur as dicentric chromosomes lag behind other chromosomes and are excluded from both daughter nuclei. Nondisjunction at any cell division could also result in loss of the additional chromosome in one cell lineage and the gain of a chromosome in another. Some metaphase cells of root tips from B9-Dp9 progeny undergoing the BFB cycle were observed with two dicentric chromosomes or with three or four telocentric chromosomes (Fig. 2), suggesting that nondisjunction has occurred during at least two mitotic divisions subsequent to dicentric formation in meiosis. Normal maize B chromosomes regularly fail to disjoin at the second pollen division, placing two copies of the B chromosome into one sperm and none into the other (15). Also, nondisjunction occurs at a low level in the first pollen division (16) in tapetal cells (17) and in endosperm cells (18). Nondisjunction of a rye B chromosome results when the chromosome lags behind the other chromosomes at the metaphase plate (19). Frequent nondisjunction of the dicentric B9-Dp9 chromosome during development may result from the tendency of functionally dicentric chromosomes to lag or from the action of the B chromosome nondisjunction mechanism.
Every round of the BFB cycle presents an opportunity for the chromosome to be stabilized by healing the ends of the chromosome or by inactivating a centromere. Six of the 23 recovered chromosomes contained inactivated centromeres. Thus, although centromere inactivation is not as frequent as end healing in stabilizing the dicentric chromosomes, centromere inactivation cannot be considered an extremely rare event. Except for 9-Bic-1, the dicentric chromosomes were very small, indicating that centromere inactivation occurred after most of the chromatin from the B chromosomes and the duplicated portion of 9S had already been lost, suggesting that smaller dicentric chromosomes are more prone to centromere inactivation. Such inactivation might result by nondisjunction in which one centromere fails to attach to the spindle and does not recover activity in subsequent mitoses.
Previously, a collection of translocation chromosomes containing broken or misdivided maize B centromeres was created and analyzed (11, 20, 21). The misdivisions occur primarily in the 700-kb core, and some of the resulting centromere derivatives retain only a small portion of their functional chromatin. Despite losing most of the centromeric sequences, these derivatives are able to form functional centromeres, demonstrating that even a small portion of the regular DNA elements of the B centromere is sufficient for centromere function.
Both the active and inactive centromeres of the minichromosomes and the translocated portion of chromosome 9-Bic-1 all appear to retain at least a portion of the functional core of the B centromere, and, in many cases, the core region appears intact cytologically. Therefore, the inactivation of the B centromere of 9-Bic-1 and the inactive centromeres on the minichromosomes are not because of a lack of suitable sequences to form a centromere. When the two centromeric regions could be distinguished, as in minichromosome 5 and chromosome 9-Bic-1, the inactivation of an intact B centromere was shown to be stable over several generations.
Because the inactivated centromere contains all DNA elements found in a functional centromere, it can be concluded that primary sequence alone is not sufficient for centromere maintenance in plants. Recovery of barley chromosomes lacking any detectable centromeric elements is complementary evidence that DNA sequence is not necessary for centromere formation (22). Similarities in structure and a shared lack of sequence-dependence in plants and animals suggest a common mechanism for centromere identity in the two kingdoms.
Methods
Plant Materials.
Lines containing minichromosomes 2, 3, 5, 10, 13, and 23 were selected from the progeny of plants that were hemizygous for the B9-Dp9 chromosome together with two 9-B chromosomes (6). Individual plants from each line were scored cytologically for the presence of a B chromosome-specific element, ZmBs (23), and then grown in the greenhouse or field. Immature tassels were fixed in ethanol/acetic acid (3:1, vol/vol) on ice for 2 h, and transferred to 70% ethanol and stored at −20°C.
FISH and Immunostaining.
Probes were prepared as described in ref. 10, except that 10 units of polymerase I was used in the nick translation reaction. Chromosome preparation, FISH, image capturing, and image processing were performed as described in ref. 10, with the following modifications. Digested root tips were washed and broken in 70% ethanol. The cells were rinsed in 100% ethanol and resuspended in 100% acetic acid before application to slides. The probe mixture (4 ng/ul of each probe in 2× SSC and 1× TE buffer) was heated for 5 minutes at 95°C and then cooled on ice before applying to slides. Probe and chromosomes were denatured together by heating at 100°C for 5 minutes. Tissue preparation and immunolabeling was performed as described in refs. 9 and 12.
Supplementary Material
Acknowledgments
We thank M. Sivaguru and B. Hardiman at the Molecular Cytology Core, Life Sciences Center, for their help with image acquisition. This work was supported by National Science Foundation Grants DBI0421671 and DBI0423898, U.S. Department of Agriculture Grant 2002-01280, and a University of Missouri Life Sciences Fellowship (to J.C.L.).
Abbreviations
- BFB
breakage–fusion–bridge
- 9-Bic-1
9-B inactive centromere-1
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McClintock B. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClintock B. Proc. Natl. Acad. Sci. USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan B. A., Willard H. F. Nat. Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 4.Jones G. H., Rees H. B Chromosomes. London: Academic; 1982. [Google Scholar]
- 5.Longley A. E. J. Agric. Res. 1927;35:796. [Google Scholar]
- 6.Zheng Y. Z., Roseman R. R., Carlson W. R. Genetics. 1999;153:1435–1444. doi: 10.1093/genetics/153.3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato A., Zheng Y. Z., Auger D. L., Phelps-Durr T., Bauer M. J., Lamb J. C., Birchler J. A. Cytogenet. Genome Res. 2005;109:156–165. doi: 10.1159/000082395. [DOI] [PubMed] [Google Scholar]
- 8.Hsu F. C., Wang C. J., Chen C. M., Hu H. Y., Chen C. C. Genetics. 2003;164:1087–1097. doi: 10.1093/genetics/164.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb J. C., Kato A., Birchler J. A. Chromosoma. 2005;113:337–349. doi: 10.1007/s00412-004-0319-z. [DOI] [PubMed] [Google Scholar]
- 10.Kato A., Lamb J. C., Birchler J. A. Proc. Natl. Acad. Sci. USA. 2004;101:13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin W., Lamb J. C., Vega J. M., Dawe R. K., Birchler J. A., Jiang J. Plant Cell. 2005;17:1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaszas E., Cande W. Z. J. Cell Sci. 2000;113:3217–3226. doi: 10.1242/jcs.113.18.3217. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder-Reiter E., Houben A., Wanner G. Chromosome Res. 2003;11:585–596. doi: 10.1023/a:1024952801846. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan B. A., Blower M. D., Karpen G. H. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 15.Roman H. Genetics. 1947;32:391–409. doi: 10.1093/genetics/32.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusche M. L., Mogensen H. L., Shi L., Keim P., Rougier M., Chaboud A., Dumas C. Genetics. 1997;147:1915–1921. doi: 10.1093/genetics/147.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiavarino A. M., Rosato M., Manzanero S., Jimenez G., Gonzalez-Sanchez M., Puertas M. J. Genetics. 2000;155:889–897. doi: 10.1093/genetics/155.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfenito M. R., Birchler J. A. Maydica. 1990;35:359–366. [Google Scholar]
- 19.Jones N., Houben A. Trends Plant Sci. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- 20.Kaszas E., Birchler J. A. EMBO J. 1996;15:5246–5255. [PMC free article] [PubMed] [Google Scholar]
- 21.Kaszas E., Birchler J. A. Genetics. 1998;150:1683–1692. doi: 10.1093/genetics/150.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasuda S., Hudakova S., Schubert I., Houben A., Endo T. R. Proc. Natl. Acad. Sci. USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfenito M. R., Birchler J. A. Genetics. 1993;135:589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.