Abstract
Recent evidence suggests that tumor necrosis factor α (TNFα) signaling in vascular cells can have antiatherogenic consequences, but the mechanisms are poorly understood. TNFα is released by free cholesterol-loaded apoptotic macrophages, and the clearance of these cells by phagocytic macrophages may help to limit plaque development. Macrophage cholesterol uptake induces ATP-binding cassette (ABC) transporter ABCA1 promoting cholesterol efflux to apolipoprotein A-I and reducing atherosclerosis. We show that TNFα induces ABCA1 mRNA and protein in control and cholesterol-loaded macrophages and enhances cholesterol efflux to apolipoprotein A-I. The induction of ABCA1 by TNFα is reduced by 65% in IκB kinase β-deficient macrophages and by 30% in p38α-deficient macrophages, but not in jun kinase 1 (JNK1)- or JNK2-deficient macrophages. To evaluate the potential pathophysiological significance of these observations, we fed TNFα-secreting free cholesterol-loaded apoptotic macrophages to a healthy macrophage monolayer (phagocytes). ABCA1 mRNA and protein were markedly induced in the phagocytes, a response that was mediated both by TNFα signaling and by liver X receptor activation. Thus, TNFα signals primarily through NF-κB to induce ABCA1 expression in macrophages. In atherosclerotic plaques, this process may help phagocytic macrophages to efflux excess lipids derived from the ingestion of cholesterol-rich apoptotic corpses.
Keywords: atherosclerosis, cytokine, ATP-binding cassette transporter
ABCA1 belongs to the ATP-binding cassette (ABC) transporter superfamily (1, 2) and promotes efflux of cholesterol and phospholipids from cellular membranes to apolipoprotein A-I (apoA-I) (3). In macrophages, oxysterol-activated liver X receptor (LXR) (4) and retinoid X receptor form a heterodimer that binds to a direct repeat 4 sequence (5, 6) located in the proximal promoter of the ABCA1 gene, resulting in increased gene transcription and increased cholesterol and phospholipid efflux to apoA-I (7). Bone marrow transplantation studies have shown that the expression of ABCA1 in macrophage foam cells has antiatherogenic consequences (8, 9).
Atherosclerosis represents an inflammatory reaction in the arterial wall, initiated by the retention of lipoprotein lipids (10, 11). One of the most studied inflammatory cytokines is tumor necrosis factor α (TNFα), which is active in both human and rodent atherosclerotic plaques (10, 12). Many of the inflammatory properties of TNFα suggest that TNFα signaling is proatherogenic (13). Paradoxically, other studies have shown that signaling by means of the TNFα receptor I (p55) may have an overall atheroprotective effect (14) and moreover that TNFα signaling through NF-κB in macrophages and vascular smooth muscle cells may be antiatherogenic (14–17). However, the mechanisms of atheroprotective effects of TNFα signaling in macrophages are not well understood. In this work, we show that TNFα induces ABCA1 through NF-κB in macrophages and in phagocytes ingesting apoptotic cells, revealing a previously undescribed antiatherogenic mechanism of TNFα signaling in macrophages.
Results
TNFα Induces ABCA1 mRNA and Protein Expression in Mouse Peritoneal Macrophages.
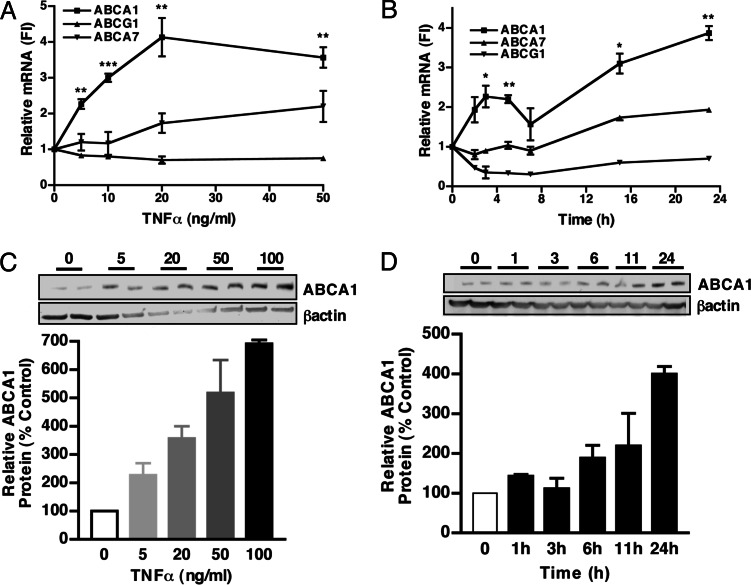
We first investigated whether TNFα can alter expression of different ABC transporters potentially involved in lipid efflux from cells, i.e., ABCA1, ABCA7 (18), and ABCG1 (19, 20). We treated mouse peritoneal macrophages for 24 h with increasing doses of TNFα (0–50 ng/ml). As shown in Fig. 1A, TNFα up-regulated ABCA1 mRNA levels in a concentration-dependent manner up to 4-fold (50 ng/ml) (P < 0.01). ABCA7 was slightly increased by TNFα, but only at higher doses (20–50 ng/ml). In contrast, ABCG1 mRNA was repressed by TNFα treatment. No signs of cellular apoptosis or necrosis were detected by TUNEL or other assays even at the highest dose (data not shown), as expected because TNFα does not usually induce apoptosis unless NF-κB signaling is impaired (21).
Fig. 1.
TNFα regulates ABC transporter expression in mouse peritoneal macrophages. Thioglycollate-elicited macrophages were treated with increasing concentrations of TNFα (0–50 and 0–100 ng/ml, respectively) in DMEM containing 10% FBS for 24 h (A and C) or with TNFα (10 ng/ml) (B and D) for the indicated times. (A and B) RNA extracts were prepared from cultured cells, and mRNA levels were analyzed by Taqman real-time PCR, as described in Materials and Methods. The average copy numbers of ABCA1, ABCG1, or ABCA7 were normalized to housekeeping gene expression. Results are expressed as fold induction (FI) as compared with untreated controls (control = 1 arbitrary unit) ± SEM. ∗∗∗, P < 0.001; ∗∗, P < 0.01; ∗, P < 0.05. (C and D) Protein extracts were prepared. (Lower) The ratio of ABCA1 to β-actin is expressed as a percent of untreated controls. In each experiment, treatments were performed in duplicate, and values shown represent the average of two different cell preparations. Upper shows a Western blot and is representative of one experiment.
Fig. 1B shows the time course of the response of ABCA1, ABCA7, and ABCG1 mRNAs to TNFα (10 ng/ml). ABCA1 mRNA was increased by ≈2.5-fold (P < 0.05) at 2–6 h and by ≈4-fold at 16–24 h (P < 0.01). ABCA7 was slightly increased by TNFα at later time points (2-fold; P < 0.05; 24 h), whereas ABCG1 mRNA was repressed (0–24 h). A similar induction of ABCA1 by TNFα was observed in bone-marrow-derived macrophages cultured in the presence of macrophage-colony stimulating factor (see below) and in human THP-1 macrophages (data not shown). In similar experiments, we monitored the levels of ABCA1 protein (Fig. 1 C and D). Treatment of mouse peritoneal macrophages with TNFα resulted in an increase in ABCA1 protein in a dose- and time-dependent manner, with a 4-fold induction at 50 ng/ml (Fig. 1C) and at 24 h (dose = 50 ng/ml) (Fig. 1D). These changes were comparable in magnitude with the increases in ABCA1 mRNA (Fig. 1).
Mechanisms of the Induction of ABCA1 by TNFα.
TNFα is known to induce the expression of genes via a number of different signaling pathways, notably the NF-κB pathway (22, 23) and various mitogen-activated protein kinase (MAPK) signaling pathways (24, 25). We initially used inhibitors to assess the role of these pathways in the induction of ABCA1 by TNFα, as shown in Fig. 6, which is published as supporting information on the PNAS web site. We used different inhibitors of NF-κB signaling. The most common heterodimer of NF-κB consists of p50 and p65 subunits. SN50, an inhibitor of the nuclear translocation of the p50 subunit of NF-κB, caused a significant increase (60%; P < 0.05) in the induction of ABCA1 by TNFα, whereas the control peptide SN50M had no effect (Fig. 6). MG-132 and CAPE reduced or eliminated this response by 80% (P < 0.01) and 35% (P < 0.01), respectively. (Fig. 6) These experiments could indicate differential roles of p65 and p50 in the induction of ABCA1. However, we must consider that p65 and p50 inhibitors may have nonspecific effects; thus, we cannot be sure whether they truly have differential roles. We also used inhibitors to evaluate signaling via the MAPK pathways i.e., extracellular signal-regulated kinase (ERK), jun kinase (JNK), and p38-MAPK pathways (Fig. 6). Whereas ERK and JNK inhibitors had no effect, the p38-MAPK inhibitor SB202180 caused a 35% reduction (P < 0.01) in the TNFα response. Thus, the inhibitor experiments suggest a possible involvement of NF-κB and p38-MAPK signaling pathway in the induction of ABCA1 by TNFα.
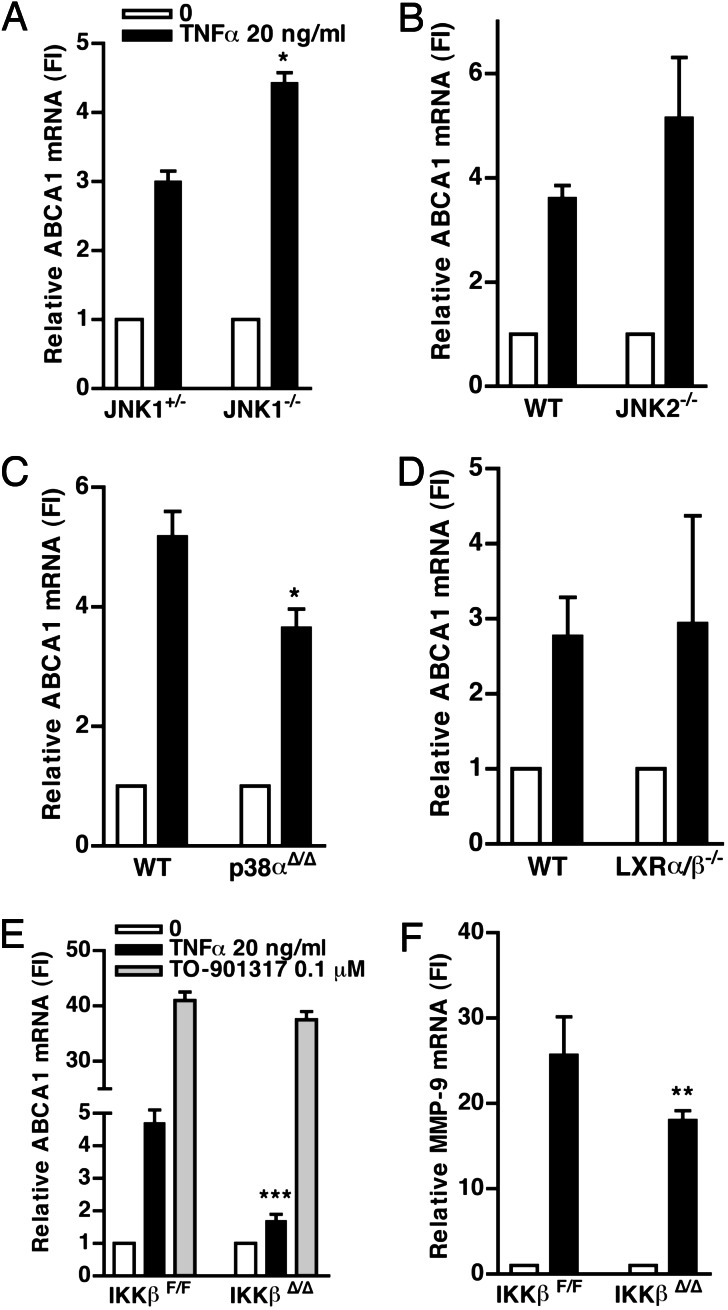
To more clearly define the signaling pathways involved in this response, we next carried out experiments using macrophages from mice deficient in key molecules involved in the different signaling pathways. TNFα induction of ABCA1 was slightly increased in macrophages from JNK1−/− (P < 0.05) or JNK2−/− (not significant) mice (Fig. 2A and B). Although there may be redundancy in the JNK1 and JNK2 signaling pathways (26) together with the inhibitor data (Fig. 6), this result suggests that JNK signaling does not contribute in a positive fashion to the induction of ABCA1 by TNFα. To assess the p38-MAPK signaling pathway, we used macrophages deficient in p38α. As shown in Fig. 2C, the increase of ABCA1 by TNFα was reduced by 30% (P < 0.05) in the p38α-deficient macrophages as compared with the wild-type (WT) control.
Fig. 2.
NF-κB and p38-MAPK, but not JNK, mediate the increase of ABCA1 mRNA by TNFα. (A, B, E, and F) Bone-marrow-derived-macrophages from JNK1−/− (A), JNK2−/− (B), and IKKβF/F and IKKβΔ/Δ (E and F) were cultured in DMEM containing 10% FBS and macrophage-colony stimulating factor (20 ng/ml) for 5–9 days, before addition of TNFα (20 ng/ml). (C and D) Peritoneal macrophages from WT, p38αΔ/Δ (C), or LXRα/β−/− (D) mice were cultured as in Fig. 1 and treated with TNFα (20 ng/ml). RNA was extracted 5 h later, and ABCA1 or MMP-9 mRNA was analyzed by Taqman and corrected for housekeeping genes as described in Fig. 1. Results are expressed as fold induction (FI) compared with the control (no treatment). ∗∗∗, P = 0.003; ∗∗, P = 0.01; ∗, P < 0.05. Each graph represents two or three different cell preparations, except for p38, which was conducted in one cell preparation. All experiments were performed in triplicate wells.
TNFα induction of ABCA1 was well preserved in LXRα/β−/− macrophages (Fig. 2D), indicating that TNFα does not act via LXR to induce ABCA1. IKKβ phosphorylates IκBα, leading to its ubiquitination and proteasomal degradation and releases p65 for nuclear translocation (22). The induction of ABCA1 by TNFα was markedly reduced (by 65%) in IKKβ−/− macrophages (Fig. 2E). In comparison, the response of the well known NF-κB target gene metalloprotease-9 (MMP-9) to TNFα was reduced by 35% (P = 0.01) in the IKKβ−/− cells (Fig. 2F). In contrast, induction of ABCA1 by the LXR activator TO-901317 (abbreviated TO-1317) was intact in these cells, ruling out a nonspecific effect on the ABCA1 promoter. The data shown in Fig. 2 were obtained at the 5-h time point. A separate set of experiments carried out 12 h after addition of TNFα showed that the induction of ABCA1 by TNFα was reduced by 80% at 12 h (data not shown). Given that the deletion of IKKβ is only ≈75% efficient (27), these experiments indicate a principal role of NF-κB signaling in the induction of ABCA1 by TNFα and a secondary role of p38-MAPK signaling in this response.
Analysis of the ABCA1 promoter by matinspector (Genomatix Software, Munich) suggested potential NF-κB binding sites in the proximal promoter region. However, TNFα did not induce a response after transfection of the human ABCA1 1-Kb promoter-luciferase in RAW macrophages or in primary mouse peritoneal macrophages (data not shown). This finding could indicate that there is a NF-κB binding site(s) elsewhere in the gene or that the effects of NF-κB are indirectly mediated.
TNFα Induces ABCA1 and Cholesterol Efflux in Cholesterol-Loaded Macrophages.
We next determined whether induction of ABCA1 by TNFα would occur under conditions of relevance to atherogenesis. Thus, we examined the effects of TNFα in macrophages that had been cholesterol loaded with acetyl-low density lipoprotein (AcLDL) or treated with the synthetic LXR activator (TO-1317).
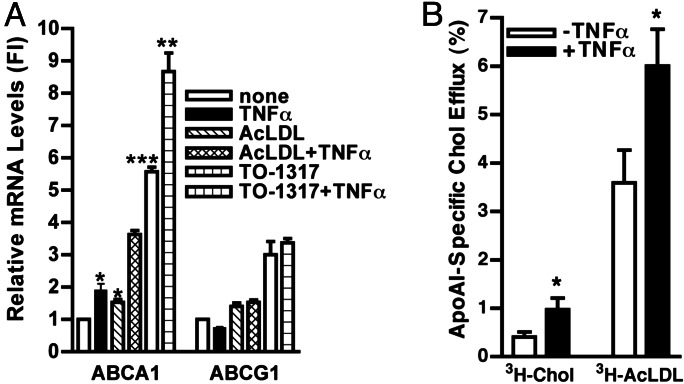
Peritoneal macrophages were treated with submaximal doses of TNFα and TO-1317 (0.1 μM) or AcLDL (50 μg/ml) simultaneously for 24 h. As shown in Fig. 3A, the increase of ABCA1 mRNA resulting from treatment with TNFα and AcLDL (3.6-fold; P < 0.0001) or TNFα and TO-1317 (8.6-fold; P < 0.001) was at least additive, as compared with TNFα alone (1.9-fold), AcLDL alone (1.5-fold), or TO-1317 alone (5.6-fold). ABCG1 mRNA also was induced by AcLDL (1.4-fold) or TO-1317 (3.0-fold). However, TNFα or TNFα in combination with TO-1317 or with AcLDL had no additional effect on ABCG1 mRNA. TNFα and AcLDL, and TNFα and TO-1317, increased ABCA1 protein in a more than additive manner as well (data not shown). The mechanism of the apparent cooperation between LXR and TNFα (NF-κB) signaling in the induction of ABCA1 is unknown.
Fig. 3.
Effect of TNFα, AcLDL, and LXR activator on the induction of ABCA1 mRNA and effect of TNFα on cholesterol efflux. Thioglycollate-elicited macrophages were cultured as described in Fig. 1. (A) An LXR agonist (TO-1317) (0.1 μM) or AcLDL (50 μg/ml) and TNFα (20 ng/ml) were added to the cells simultaneously in DMEM containing 10% lipoprotein-deficient serum (LPDS). ABCA1 and ABCG1 mRNA were analyzed as described in Materials and Methods. Results are expressed as fold induction (FI) compared with untreated controls ± SEM. ∗∗∗, P < 0.0001; ∗∗, P < 0.001; ∗, P < 0.01. (B) Cells were labeled in DMEM and 10% LPDS containing [3H]cholesterol or [3H]AcLDL, with or without TNFα (50 ng/ml), for 18 h. Cholesterol efflux was calculated as described in Materials and Methods. Results are expressed as a percent of cholesterol efflux ± SEM. Cholesterol efflux to apoA-I was measured in triplicate or quadruplicate, and experiments were performed in three different cell preparations. ∗, P < 0.05
To assess cholesterol efflux, peritoneal macrophages were loaded with free-cholesterol ([3H]cholesterol) or cholesterol incorporated into AcLDL ([3H]AcLDL) overnight in the presence or absence of TNFα (50 ng/ml), and the efflux of cholesterol to apoA-I was measured during a subsequent 4-h incubation (Fig. 3B). As reported in ref. 28, AcLDL loading resulted in a higher level of cholesterol efflux (shown in Fig. 3B, open bars) than loading with cholesterol. TNFα increased cholesterol efflux by 2.5-fold (P < 0.05) when macrophages were loaded with [3H]cholesterol and 1.7-fold (P < 0.05) after loading with [3H]AcLDL. These studies show that ABCA1 induced by TNFα is functional in terms of cholesterol efflux, both in basal and cholesterol-loaded cells.
Increase of ABCA1 During Phagocytosis of Apoptotic Cells Is Inhibited by Neutralizing Anti-TNFα Antibodies (Abs).
Macrophages undergo apoptosis in atherosclerotic plaques, in response to loading with free cholesterol or oxidized lipids (12, 29). Apoptotic macrophages may be taken up by other phagocytic macrophages or, if not cleared in this way, may undergo postapoptotic necrosis, inciting an inflammatory response (30–32). Free cholesterol-induced macrophage apoptosis involves the unfolded protein response and leads to MAPK activation and release of inflammatory cytokines, notably TNFα (33). To further explore the potential physiological relevance of our observations, we next asked whether ABCA1-induction by TNFα could occur during macrophage phagocytosis of TNFα-secreting, free cholesterol-induced apoptotic macrophages (FC-AMs). In this model of phagocytosis, mouse peritoneal macrophages are loaded with free-cholesterol for 18 h [using 100 μg/ml AcLDL in the presence of acyl-CoA:cholesterol acyltransferase (ACAT) inhibitor] to induce apoptosis, the FC-AMs are then fed to a monolayer of healthy macrophages (phagocytes) for a time period, and the monolayer is washed and analyzed for mRNA or protein responses. The phagocytes have been shown to ingest only apoptotic cells with high efficiency, and the washing procedure removes all uningested apoptotic and nonapoptotic cells from the monolayer (54). Moreover, the phagocytes show evidence of NF-κB activation, possibly in response to TNFα synthesized by the FC-AMs or the phagocytes themselves (33).
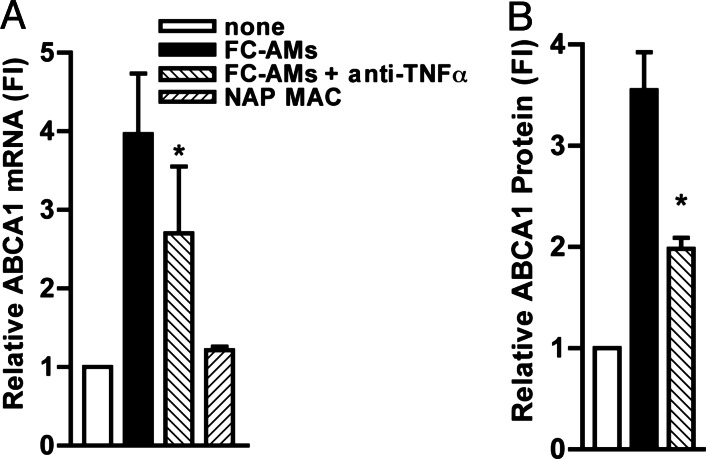
Peritoneal macrophages were incubated with the FC-AMs, in the presence of neutralizing anti-TNFα or control Abs for 8 h. In this model of phagocytosis, uptake of apoptotic cells occurs rapidly (as early as 15 min) and the phagocytic index is high (up to 40%). However, preliminary time-course studies indicated that the induction of ABCA1 and the effect of TNFα Abs required more prolonged incubation, and thus phagocytes were examined after 8 h of incubation. ABCA1 mRNA in the phagocytes was increased by ≈4-fold upon incubation with apoptotic cells, and this increase was suppressed by ≈40% (P < 0.05) in the presence of anti-TNFα Abs (Fig. 4A). This reduction of the response by TNFα Abs was reproduced in four separate experiments. A control IgG Ab for anti-TNFα had no effect on the induction of ABCA1 in phagocytes (data not shown). In a control experiment, we showed that ABCA1 was not increased in macrophages that were fed with nonapoptotic macrophages (Fig. 4A). Fig. 4B shows that ABCA1 protein also was increased (3.5-fold) in phagocytes after incubation with FC-AMs, and this increase was suppressed by ≈60% (P < 0.02) by neutralizing anti-TNFα Abs. These results show a marked induction of ABCA1 mRNA and protein in phagocytes after ingestion of FC-AMs and demonstrate that a major part of this response can be attributed to TNFα. It is unlikely that apoptosis of phagocytes themselves contributed to the induction of ABCA1 because Feng and Tabas (34) have shown that FC-induced apoptosis in macrophages markedly reduced ABCA1 protein levels.
Fig. 4.
Neutralizing anti-TNFα Abs suppress the induction of ABCA1 that occurs during phagocytosis of apoptotic cells by peritoneal macrophages. Peritoneal macrophages were incubated with FC-AMs (A and B) or with nonapoptotic macrophages (NAP-MAC) as a control (A) with or without anti-TNFα Ab (10 μg/ml), phagocytes were washed vigorously to remove any bound but undigested apoptotic cells, and RNA extracts (A) or protein extracts (B) were prepared from the phagocytes 8 h later. mRNA data were normalized to housekeeping genes, and protein data were normalized to β-actin levels and are expressed as fold induction (FI) as compared with the controls that did not receive FC-AMs or NAP-MAC (control = 1 arbitrary unit). ∗, P < 0.05. The graph represents the average of three experiments conducted in three different cell preparations.
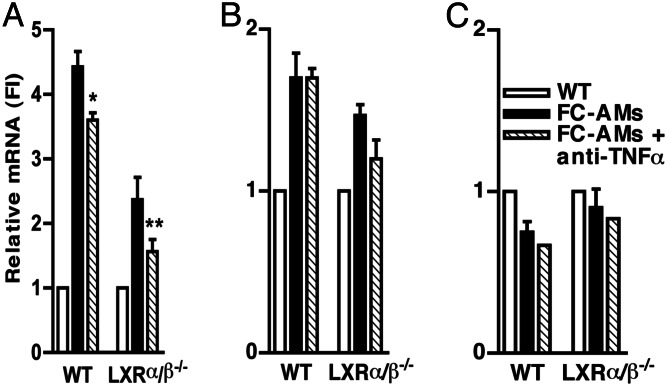
ABCA1 Induction in Phagocytes Is Decreased in LXR α/β−/− Mice: Joint Role of TNFα and LXR in Induction of ABCA1 in Phagocytes.
Because the phagocytes are ingesting a cholesterol-rich meal, it seemed likely that LXR induction of ABCA1 also could be involved in the response. To assess the specific role of LXR, we carried out similar experiments using phagocytes from LXRα/β−/− mice, in which the sterol induction of ABCA1 is abolished (35). WT or LXRα/β−/− macrophage monolayers were incubated with WT apoptotic cells for 8 h, the phagocytes were harvested, and RNA was extracted. The increase of ABCA1 mRNA by apoptotic cells was diminished by 60% in LXRα/β−/− phagocytes compared with control WT littermates (Fig. 5A). The mRNA of ABCG1, another LXR target gene, was modestly increased by the FC-AMs (Fig. 5B). This effect was reduced in LXRα/β−/− cells. ABCA7, which is not regulated by LXR ligands (18) and only slightly induced by TNFα (Fig. 1B), was not induced by FC-AMs in either WT or LXRα/β−/− cells (Fig. 5C). These data show that ABCA1 induction in phagocytes is partially mediated by LXR. Nonetheless, there was considerable ABCA1 expression remaining in the LXR-deficient mice in the presence of apoptotic cells. We hypothesized that the remaining induction of ABCA1 mRNA levels was due to the effect of TNFα. To test this hypothesis, we investigated the effect of neutralizing anti-TNFα Abs on ABCA1 in LXRα/β−/− phagocytes compared with control WT littermates (Fig. 5). Neutralizing TNFα Abs added to phagocytes in the presence of apoptotic cells decreased ABCA1 mRNA by a similar amount in both WT and LXRα/β−/− mice cells and reduced ABCA1 mRNA almost to the control level in the LXRα/β−/− cells. In contrast, ABCG1 and ABCA7 mRNA levels were not significantly changed by neutralizing anti-TNFα Abs. The small residual response in LXRα/β−/− cells treated with TNFα Abs could represent incomplete neutralization of TNFα or other factors inducing ABCA1 in this model of phagocytosis. These experiments indicate a major role for both TNFα and LXR in the induction of ABCA1 in phagocytes.
Fig. 5.
ABCA1-induction is decreased in LXRα/β−/− phagocytes: joint role of TNFα and LXR in induction of ABCA1 in phagocytes. Thioglycollate-elicited peritoneal macrophages were isolated from LXRα/β−/− mice and their control WT littermates and cultured as described in Materials and Methods. Macrophage monolayers (WT or LXRα/β−/−) were incubated with WT FC-AMs in the presence or absence of anti-TNFα Ab (10 μg/ml), and RNA extracts were performed, as described in Fig. 4. ABCA1 (A), ABCG1 (B), and ABCA7 (C) mRNA were analyzed by real-time quantitative PCR, and data are shown normalized for housekeeping genes. ∗, P < 0.05; ∗∗, P = 0.056. The experiment was conducted in two different cell preparations; the graphs show the average of triplicates of wells of one representative experiment.
Discussion
TNFα acting via NF-κB induces ABCA1 expression in macrophages. Macrophage ABCA1 expression is markedly up-regulated during phagocytosis of cholesterol-induced apoptotic cells, reflecting both the effects of TNFα and LXR activation in the phagocytes. These studies show an important link between inflammatory signals and lipid efflux pathways in macrophages as they phagocytose apoptotic cells. It is likely that the induction of ABCA1 by TNFα and LXR facilitates the resolution of inflammatory lesions and represents a beneficial response of the phagocyte in the context of atherosclerosis. The findings are consistent with the emerging view that NF-κB signaling is involved both in the formation and in the resolution of inflammatory lesions (36). There was also a lesser role of p38-MAPK in the induction of ABCA1 by TNFα. p38-MAPK may contribute to NF-κB activation through phosphorylation of p65 (37) or indirectly through phosphorylation and activation of other factors that synergize with NF-κB (38–40).
The induction of ABCA1 by TNFα may be relevant to atherosclerotic plaques in which apoptotic free cholesterol-loaded macrophages may be cleared by healthy macrophages (41, 42). ABCA1 and ABCG1 both could contribute to the efflux of cholesterol from the phagocytes after ingestion of the cholesterol-enriched apoptotic corpses. ABCG1 promotes cholesterol efflux to high-density lipoprotein (HDL) but has minimal effects on phospholipid efflux (20). In contrast, ABCA1 enhances both cholesterol and phospholipid efflux from macrophages to lipid-poor apolipoproteins. In addition to efflux of cholesterol and native phospholipids, the induction of ABCA1 could help the phagocyte to deal with oxidant stress and reactive oxygen species generation, subsequent to the ingestion of oxidized lipids in phagocytes. Macrophages play an important role in the clearance of apoptotic corpses that contain large amounts of oxidized phospholipids (43), and ABCA1 has been suggested to increase efflux of oxidized lipids from cells to apoA-I (44) limiting the formation and the nucleation of minimally oxidized low-density lipoprotein (LDL) (45), thereby potentially protecting against atherosclerosis
More generally, our study shows a positive interaction between inflammation and lipid metabolism. The cooperation of TNFα and LXR in the induction of ABCA1 contrasts with the well documented antiinflammatory transrepression of a number of TNFα target genes by LXR activation (46–48). LXR activation inhibits lipopolysaccharide (LPS)- or TNFα-activated MMP-9 in macrophages (46). However, unlike ABCA1 these genes are not direct targets of LXR. In an elegant study, Castrillo et al. (47) also showed that LPS inhibits LXR-induced ABCA1 gene expression in macrophages, in a process that involves TLR4 and IRF-3 mediated signaling events. These pathways are not activated by TNFα, which may explain the different responses to LPS and TNFα. This result illustrates the different responses of ABCA1 in a pathologic process involving LPS compared with its physiologic induction in phagocytes ingesting apoptotic cells.
Our studies likely have relevance to the role of TNFα in atherogenesis. Some recent studies have suggested that TNFα signaling via NF-κB may be atheroprotective (14–16). Although some other studies have suggested a proatherogenic role of TNFα signaling, this role may depend on the cell type involved as well as the specific receptors and signaling pathways downstream of TNFα. Thus, there seems to be abundant evidence for a proatherogenic role of TNFα signaling in endothelial cells (49, 50). In contrast, TNFα signaling via NF-κB in macrophages and smooth muscle cells may have antiatherogenic consequences. Idel et al. (17) have shown that an atheroprotective locus on chromosome 10 in the C57Bl6 mouse contains the A20 gene, which is involved in down-modulating NF-κB signaling. Compared with the FVB variant, an amino acid change in the B6 A20 gene is associated with more sustained NF-κB signaling in response to TNFα in vascular smooth muscle cells. By using a macrophage-specific deletion of IKKβ similar to that used in the current study (Fig. 2), Kanters et al. (16) showed that inhibition of NF-κB activation in macrophages increases atherosclerosis in LDL-receptor−/− mice. By using the same strategy to achieve macrophage-specific knockout of NF-κB signaling, we show that TNFα induction of ABCA1 is virtually abolished in these cells. Together, this information suggests that TNFα-induction of macrophage ABCA1 by NF-κB is likely to be antiatherogenic.
Materials and Methods
Materials.
The LXR activator, TO-1317, was obtained from Sigma. Tritium-labeled cholesterol ([1,2-3H]cholesterol) was purchased from PerkinElmer Life Sciences. Rabbit anti-ABCA1 Ab was from Novus Biologicals (Littleton, CO); mouse anti-β-actin, horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit Abs were from Sigma. Recombinant mouse TNFα, recombinant mouse macrophage-colony stimulating factor, goat anti-mouse TNFα Ab, and control IgG were from R & D Systems. AcLDL was from Biomedical Technologies (Stoughton, MA). ApoA-I was from Biodesign International (Kennebunkport, ME).
Animals and Cell Culture.
Male C57BL/6J mice were purchased from The Jackson Laboratory. LXRα/β−/− mice and their control WT littermates were kindly provided by David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas) (51). Peritoneal macrophages were obtained from mice injected intraperitoneally with thioglycollate (52). Bone-marrow-derived macrophages were isolated and cultured in DMEM/10% FBS supplemented with macrophage-colony stimulating factor (20 ng/ml) and antibiotics for 5–9 days before the experiment. IKKβF/F and IKKβΔ/Δ, JNK1± and JNK1−/− bone-marrow-derived macrophages are described in ref. 27. Macrophages deficient in p38α (p38αΔ/Δ) were obtained from p38αflox/flox (p38αF/F) mice crossed with LysMCre/C57BL6J mice (53).
Cholesterol Efflux.
Macrophages were incubated with AcLDL and [3H]cholesterol in the presence or absence or TNFα (50 ng/ml), then incubated with apoA-I in the presence or absence of TNFα for 4 h. The cells were lysed in 0.5 ml of 0.1 M sodium hydroxyde containing 0.1% SDS at room temperature, and the radioactivity in the cell lysates and medium was quantified. To obtain apoA-I-specific efflux, radioactivity in apoA-I-free medium was subtracted. Cholesterol efflux was calculated as the percent of radioactivity released from cells into the medium relative to the total radioactivity in cells and medium.
RNA Analysis.
Total RNA was isolated from mouse peritoneal macrophages by using the RNeasy Minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Real-time quantitative PCR assays were performed as described in ref. 52. All samples were analyzed for β-actin, 36B4 expression, or 18S RNA in the same run as ABCA1, ABCG1, ABCA7, or MMP-9. The sequences of the probes and primers can be provided by the authors upon request.
Western Blot Analysis.
Protein extracts were prepared in modified radioimmunoprecipitation assay buffer as described in ref. 52. Equal amounts of protein (15 μg) were fractioned on a 4–15% gradient SDS/polyacrylamide gel, then transferred to a nitrocellulose membrane, incubated with anti-ABCA1 and anti-β-actin Abs, and processed with horseradish peroxidase-conjugated secondary Ab using SuperSignal West Pico Chemiluminescent substrate (Pierce). The band intensity was analyzed with imagequant (Amersham Pharmacia). Data were normalized for β-actin expression.
Generation of Apoptotic Cells.
Con A or methyl-BSA elicited macrophages from female C57BL/6J mice were cultured in DMEM containing 10% FBS (Mediatech, Washington, DC), antibiotics, and 20% L cell-conditioned medium. The cells were incubated with 100 μg/ml AcLDL and 10 μg/ml acyl-CoA:cholesterol acyltransferase (ACAT) inhibitor 58035 (Sigma) for 16–20 h to induce early apoptosis as described in ref. 33. Apoptosis was confirmed by Annexin V staining. In a typical experiment, 30–40% of the population was apoptotic with <5% necrosis. These FC-AMs were collected, resuspended into X-VIVO10 medium (BioWhittaker), and added to the phagocytes. The ratio of phagocytes to apoptotic cells was ≈1:5.
Phagocytosis of Apoptotic Cells.
The phagocytes were incubated with the FC-AMs or control nonapoptotic macrophages for 8 h in the presence or absence of 10 μg of anti-TNFα Ab or control IgG Ab in X-VIVO10 medium. Phagocytes then were washed vigorously with the medium to remove any bound but undigested apoptotic cells, and RNA or protein was extracted from the phagocytes, as described earlier.
In preliminary experiments, the interaction of fluorescently labeled FC-AMs with phagocytes was examined by confocal microscopy. We found that ingestion of FC-AMs by the phagocytes occurred as early as 15 min after incubation (54).
Statistics.
All results are expressed as the means ± SEM unless otherwise mentioned. Statistical significance was determined by using the Student t test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. David Mangelsdorf and Angie Bookout (Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas) for providing LXRα/β−/− mice and Drs. Yibin Wang (University of California, Los Angeles) and Tracie DeVries-Seimon (Columbia University) for providing the p38αΔ/Δ mice. This work was supported by National Institutes of Health Grants HL22682, HL57560, and HL75662 and by Dr. Saal van Zwanenbergstichting (The Netherlands).
Abbreviations
- ABCA1
ATP-binding cassette-A1
- AcLDL
acetyl-low density lipoprotein
- apoA-I
apolipoprotein A-I
- FC-AM
free cholesterol-induced apoptotic macrophage
- IKK
IκB kinase
- JNK
jun kinase
- LXR
liver X receptor
- MAPK
mitogen-activated protein kinase
- TO-1317
TO-901317.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Broccardo C., Luciani M., Chimini G. Biochim. Biophys. Acta. 1999;1461:395–404. doi: 10.1016/s0005-2736(99)00170-4. [DOI] [PubMed] [Google Scholar]
- 2.Schmitz G., Langmann T. Curr. Opin. Lipidol. 2001;12:129–140. doi: 10.1097/00041433-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Wang N., Silver D. L., Costet P., Tall A. R. J. Biol. Chem. 2000;275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- 4.Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 5.Willy P. J., Umesono K., Ong E. S., Evans R. M., Heyman R. A., Mangelsdorf D. J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 6.Teboul M., Enmark E., Li Q., Wikstrom A. C., Pelto-Huikko M., Gustafsson J. A. Proc. Natl. Acad. Sci. USA. 1995;92:2096–2100. doi: 10.1073/pnas.92.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costet P., Luo Y., Wang N., Tall A. R. J. Biol. Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 8.Francone O. L., Royer L., Boucher G., Haghpassand M., Freeman A., Brees D., Aiello R. J. Arterioscler. Thromb. Vasc. Biol. 2005;25:1198–1205. doi: 10.1161/01.ATV.0000166522.69552.99. [DOI] [PubMed] [Google Scholar]
- 9.van Eck M., Bos I. S., Kaminski W. E., Orso E., Rothe G., Twisk J., Bottcher A., Van Amersfoort E. S., Christiansen-Weber T. A., Fung-Leung W. P., et al. Proc. Natl. Acad. Sci. USA. 2002;99:6298–6303. doi: 10.1073/pnas.092327399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Libby P., Ridker P. M., Maseri A. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 11.Williams K. J., Tabas I. N. Engl. J. Med. 1999;340:1928. (lett.), and author reply (1999) 340, 1929. [PubMed] [Google Scholar]
- 12.Glass C. K., Witztum J. L. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 13.Monaco C., Andreakos E., Kiriakidis S., Mauri C., Bicknell C., Foxwell B., Cheshire N., Paleolog E., Feldmann M. Proc. Natl. Acad. Sci. USA. 2004;101:5634–5639. doi: 10.1073/pnas.0401060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreyer S. A., Peschon J. J., LeBoeuf R. C. J. Biol. Chem. 1996;271:26174–26178. doi: 10.1074/jbc.271.42.26174. [DOI] [PubMed] [Google Scholar]
- 15.Brand K., Page S., Rogler G., Bartsch A., Brandl R., Knuechel R., Page M., Kaltschmidt C., Baeuerle P. A., Neumeier D. J. Clin. Invest. 1996;97:1715–1722. doi: 10.1172/JCI118598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanters E., Pasparakis M., Gijbels M. J., Vergouwe M. N., Partouns-Hendriks I., Fijneman R. J., Clausen B. E., Forster I., Kockx M. M., et al. J. Clin. Invest. 2003;112:1176–1185. doi: 10.1172/JCI18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idel S., Dansky H. M., Breslow J. L. Proc. Natl. Acad. Sci. USA. 2003;100:14235–14240. doi: 10.1073/pnas.1835672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N., Lan D., Gerbod-Giannone M., Linsel-Nitschke P., Jehle A. W., Chen W., Martinez L. O., Tall A. R. J. Biol. Chem. 2003;278:42906–42912. doi: 10.1074/jbc.M307831200. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy M. A., Barrera G. C., Nakamura K., Baldan A., Tarr P., Fishbein M. C., Frank J., Francone O. L., Edwards P. A. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang N., Lan D., Chen W., Matsuura F., Tall A. R. Proc. Natl. Acad. Sci. USA. 2004;101:9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin M., Lin A. Nat. Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 22.Karin M., Ben-Neriah Y. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 23.Leung T. H., Hoffmann A., Baltimore D. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Moon S. K., Cha B. Y., Kim C. H. J. Cell Physiol. 2004;198:417–427. doi: 10.1002/jcp.10435. [DOI] [PubMed] [Google Scholar]
- 25.Wajant H., Pfizenmaier K., Scheurich P. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 26.Kuan C. Y., Yang D. D., Samanta Roy D. R., Davis R. J., Rakic P., Flavell R. A. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 27.Karin M., Greten F. R. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y., Hao M., Luo Y., Liang C. P., Silver D. L., Cheng C., Maxfield F. R., Tall A. R. J. Biol. Chem. 2003;278:5813–5820. doi: 10.1074/jbc.M208687200. [DOI] [PubMed] [Google Scholar]
- 29.Libby P., Aikawa M., Schonbeck U. Biochim. Biophys. Acta. 2000;1529:299–309. doi: 10.1016/s1388-1981(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 30.Ball R. Y., Stowers E. C., Burton J. H., Cary N. R., Skepper J. N., Mitchinson M. J. Atherosclerosis. 1995;114:45–54. doi: 10.1016/0021-9150(94)05463-s. [DOI] [PubMed] [Google Scholar]
- 31.Aikawa M., Libby P. Can. J. Cardiol. 2004;20:631–634. [PubMed] [Google Scholar]
- 32.Fan J., Watanabe T. J. Atheroscler. Thromb. 2003;10:63–71. doi: 10.5551/jat.10.63. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Schwabe R. F., Devries-Seimon T., Yao P. M., Gerbod-Giannone M. C., Tall A. R., Davis R. J., Flavell R., Brenner D. A., Tabas I. J. Biol. Chem. 2005;280:21763–21772. doi: 10.1074/jbc.M501759200. [DOI] [PubMed] [Google Scholar]
- 34.Feng B., Tabas I. J. Biol. Chem. 2002;277:43271–43280. doi: 10.1074/jbc.M207532200. [DOI] [PubMed] [Google Scholar]
- 35.Repa J. J., Turley S. D., Lobaccaro J. A., Medina J., Li L., Lustig K., Shan B., Heyman R. A., Dietschy J. M., Mangelsdorf D. J. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence T., Gilroy D. W., Colville-Nash P. R., Willoughby D. A. Nat. Med. 2001;7:1291–1297. doi: 10.1038/nm1201-1291. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S., Karin M. Cell. 2002;109(Suppl.):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 38.Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. J. Biol. Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 39.Okazaki T., Sakon S., Sasazuki T., Sakurai H., Doi T., Yagita H., Okumura K., Nakano H. Biochem. Biophys. Res. Commun. 2003;300:807–812. doi: 10.1016/s0006-291x(02)02932-7. [DOI] [PubMed] [Google Scholar]
- 40.Vega M. I., Huerta-Yepaz S., Garban H., Jazirehi A., Emmanouilides C., Bonavida B. Oncogene. 2004;23:3530–3540. doi: 10.1038/sj.onc.1207336. [DOI] [PubMed] [Google Scholar]
- 41.Savill J. J. Leukocyte Biol. 1997;61:375–380. doi: 10.1002/jlb.61.4.375. [DOI] [PubMed] [Google Scholar]
- 42.Henson P. M., Bratton D. L., Fadok V. A. Curr. Biol. 2001;11:R795–R805. doi: 10.1016/s0960-9822(01)00474-2. [DOI] [PubMed] [Google Scholar]
- 43.Chang M. K., Binder C. J., Miller Y. I., Subbanagounder G., Silverman G. J., Berliner J. A., Witztum J. L. J. Exp. Med. 2004;200:1359–1370. doi: 10.1084/jem.20031763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navab M., Ananthramaiah G. M., Reddy S. T., Van Lenten B. J., Ansell B. J., Fonarow G. C., Vahabzadeh K., Hama S., Hough G., Kamranpour N., et al. J. Lipid Res. 2004;45:993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Reddy S. T., Hama S., Ng C., Grijalva V., Navab M., Fogelman A. M. Arterioscler. Thromb. Vasc. Biol. 2002;22:1877–1883. doi: 10.1161/01.atv.0000035700.82829.2a. [DOI] [PubMed] [Google Scholar]
- 46.Castrillo A., Joseph S. B., Marathe C., Mangelsdorf D. J., Tontonoz P. J. Biol. Chem. 2003;278:10443–10449. doi: 10.1074/jbc.M213071200. [DOI] [PubMed] [Google Scholar]
- 47.Castrillo A., Joseph S. B., Vaidya S. A., Haberland M., Fogelman A. M., Cheng G., Tontonoz P. Mol. Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- 48.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. Nat. Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 49.Nakashima Y., Raines E. W., Plump A. S., Breslow J. L., Ross R. Arterioscler. Thromb. Vasc. Biol. 1998;18:842–851. doi: 10.1161/01.atv.18.5.842. [DOI] [PubMed] [Google Scholar]
- 50.Hajra L., Evans A. I., Chen M., Hyduk S. J., Collins T., Cybulsky M. I. Proc. Natl. Acad. Sci. USA. 2000;97:9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 52.Costet P., Lalanne F., Gerbod-Giannone M. C., Molina J. R., Fu X., Lund E. G., Gudas L. J., Tall A. R. Mol. Cell. Biol. 2003;23:7756–7766. doi: 10.1128/MCB.23.21.7756-7766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devries-Seimon T., Li Y., Yao P. M., Stone E., Wang Y., Davis R. J., Flavell R., Tabas I. J. Cell Biol. 2005;171:61–73. doi: 10.1083/jcb.200502078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Gerbod-Giannone M. C., Seitz H., Cui D., Thorp E., Tall A. R., Matsushima G. K., Tabas I. J. Biol. Chem. 2005 doi: 10.1074/jbc.M510579200. December 27, www.jbc.org/cgi/reprint/m510579200v1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.