Abstract
Many bacterial toxins act on conserved components of essential host-signaling pathways. One consequence of this conservation is that genetic model organisms such as Drosophila melanogaster can be used for analyzing the mechanism of toxin action. In this study, we characterize the activities of two anthrax virulence factors, lethal factor (LF) and edema factor, in transgenic Drosophila. LF is a zinc metalloprotease that cleaves and inactivates most human mitogen-activated protein kinase (MAPK) kinases (MAPKKs). We found that LF similarly cleaves the Drosophila MAPK kinases Hemipterous (Hep) and Licorne in vitro. Consistent with these observations, expression of LF in Drosophila inhibited the Hep/c-Jun N-terminal kinase pathway during embryonic dorsal closure and the related process of adult thoracic closure. Epistasis experiments confirmed that LF acts at the level of Hep. We also found that LF inhibits Ras/MAPK signaling during wing development and that LF acts upstream of MAPK and downstream of Raf, consistent with LF acting at the level of Dsor. In addition, we found that edema factor, a potent adenylate cyclase, inhibits the hh pathway during wing development, consistent with the known role of cAMP-dependent PKA in suppressing the Hedgehog response. These results demonstrate that anthrax toxins function in Drosophila as they do in mammalian cells and open the way to using Drosophila as a multicellular host system for studying the in vivo function of diverse toxins and virulence factors.
Keywords: mitogen-activated protein kinase kinase, c-Jun terminal kinase, downstream of raf1, development, cAMP-dependent PKA
Over the past two decades, the fruit fly Drosophila melanogaster has proven to be a powerful model system for deciphering genetic networks controlling the development and physiology of metazoan organisms. More recently, advances in reverse genetics in conjunction with the completion of genome sequencing have stimulated interest in using Drosophila for functional analysis of many human disease genes that are conserved between humans and insects (1–3). Drosophila also played a pioneering role in the discovery of the innate immune system and its regulation by the Toll pathway (4). In contrast, relatively little use has been made of Drosophila in analyzing the activity of virulence factors produced by pathogens. Because bacteria often achieve infectivity through the secretion of toxins that target well conserved components of signaling pathways or other essential cellular networks, there is a large untapped potential for exploiting Drosophila in the analysis of toxin–host interactions. In this preliminary proof-of-principle study, we examine the activities of two well characterized anthrax toxins in transgenic Drosophila and compare the in vivo effects of these toxins to their known effects in human cells.
Anthrax is caused by Bacillus anthracis, a Gram-positive bacterium that infects primarily herbivores and occasionally humans. B. anthracis secretes three exotoxins [lethal factor (LF), edema factor (EF), and protective antigen (PA)] that are required for its virulence. Anthrax toxins belong to the A/B subfamily of exotoxins, in which the B subunit (PA) binds to a host membrane component and promotes the entry of catalytic A subunits (LF and EF) into host cells (5, 6). PA binds to the human cell-surface receptors tumor endothelial marker 8 or capillary morphogenesis protein 2 (7, 8), two related, widely expressed transmembrane proteins of unknown function (9). After cleavage by furin proteases, PA becomes activated and forms a heptameric prepore, which binds three molecules of EF, LF, or a combination of both, after which the complex undergoes endocytosis. A pH drop in endocytic vesicles triggers a conformational change in the PA ring, leading to translocation of EF and LF into the cytosol (5). LF is a zinc metalloprotease that cleaves six of the seven known human mitogen-activated protein kinase (MAPK) kinases (MAPKKs) in their N-terminal proline-rich regulatory domain, which prevents them from binding to their substrates and thereby inhibits phosphorylation and activation of downstream MAPKs (10–12). EF, the second catalytic anthrax toxin, is a Ca2+/calmodulin-dependent adenylate cyclase with a specific activity ≈1,000-fold higher than that of endogenous mammalian counterparts (13). Because bacteria lack calmodulin, EF becomes active only after entering host eukaryotic cells, in which it causes an unregulated rise in cAMP levels.
Both LF and EF play a central role in anthrax pathogenesis, as demonstrated by the greatly reduced infectivity of B. anthracis strains lacking either toxin (14). In addition, the isolated toxins can cause death (LF) or edema (EF) when coinjected with PA. The best characterized cellular response to LF is in macrophages, which undergo programmed cell death and lysis after LF exposure (15, 16). There is also evidence that LF induces defects in permeability of the vascular endothelium, which, in combination with cytokines produced by dying macrophages, may contribute to the shock-like death of animals exposed to LF. The cellular basis for EF action is less well characterized than that of LF, but it has been reported that EF blocks phagocytosis in monocytes (17), impairs the function of dendritic cells (18), and inhibits antigen presentation to T cells (19). In addition, a recent report examining the systemic effects of EF reveals that this toxin causes severe tissue damage and multiple organ failure followed by rapid death in mice (20).
In this report, we analyze the effect of LF and EF expressed in various tissues during Drosophila development. We found that LF cleaves the Drosophila MAPKKs Hemipterous (Hep) and Licorne (Lic) (21, 22) in vitro. Phenotypic analysis of LF expression during embryonic and imaginal development reveals that LF inhibits the activities of the MAPKKs Hep and Downstream-of-Raf1 (Dsor1) (23). These LF-induced phenotypes were not observed with a mutant version of LF lacking its catalytic site (11), consistent with LF functioning by proteolysis of target proteins. Expression of EF causes phenotypes in the wing similar to those caused by loss-of-function hedgehog (hh) pathway mutations, consistent with the known effect of cAMP-dependent PKA in inhibiting Hedgehog (Hh) signaling (24, 25). These results validate Drosophila as a multicellular model organism for analyzing the function of anthrax toxins and possibly other bacterial or viral virulence factors targeting unknown host proteins.
Results
Drosophila MAPKKs Are Cleaved by LF.
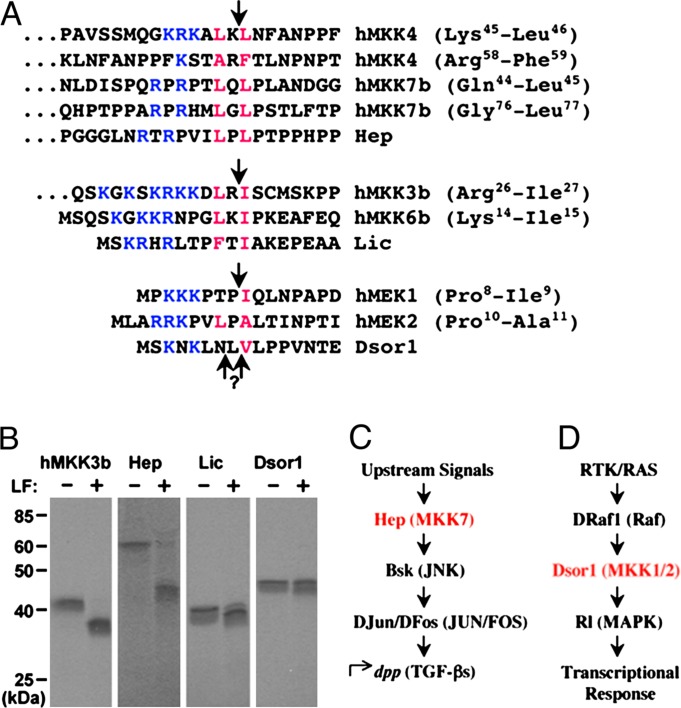
LF is a zinc metalloprotease that cleaves within the N terminus of six different human MAPKKs (10–12). A consensus amino acid N-terminal sequence was derived consisting of a few basic residues and a hydrophobic residue preceding the LF cleavage site, followed immediately by another hydrophobic residue (Fig. 1A). We searched for this LF recognition motif within Drosophila MAPKKs and found matches in the N-terminal domains of all four known MAPKKs. We focused on the three MAPKKs for which mutant alleles have been characterized (Hep, Dsor1, and Lic; see Fig. 1A). To test whether they can be cleaved by LF, Hep, Dsor1, and Lic proteins were synthesized and radiolabeled in vitro. Human MAPKK3b protein was used as a positive control, because it has been shown to serve as a substrate for LF under our in vitro cleavage conditions (15). Incubation of Hep with LF generated a smaller product of ≈44 kDa (Fig. 1B), a size consistent with cleavage of full-length Hep by LF (Fig. 1B). As the predicted LF cleavage site in Lic is only 11 aa downstream from its N terminus, we observed only a slight reduction in the size of Lic (≈1–2 kDa) upon incubation with LF (Fig. 1B). Although we did not detect a change in the mobility of Dsor1 by LF under these conditions, it is possible that cleavage takes place too close to the N terminus (only 6 or 8 aa away) to be detected by this assay. In vivo analysis of LF function is consistent with this possibility, because the activities of both Dsor and Hep are inhibited by LF (see below). Alternatively, cleaved Dsor might migrate more slowly on SDS/PAGE than expected, or our in vitro cleavage conditions may not be optimal for this MAPKK.
Fig. 1.
In vitro cleavage of Drosophila MAPKKs by anthrax LF. (A) A multiple protein sequence alignment of known LF cleavage sites in human MAPKKs and cognate sequences in Drosophila MAPKKs. Conserved residues defining the cleavage motif are denoted in color (blue, basic residues; red, hydrophobic residues). LF cleavage sites are indicated by arrows. Amino acid residues flanking known cleavage sites are in parentheses. (B) 35S-labeled MAPKK proteins were synthesized in vitro and incubated with LF (250 ng) for 1 h. MAPKK cleavage was analyzed by SDS/PAGE and autoradiography. (C) Schematic diagram of JNK pathway components in Drosophila and mammals (parentheses) examined in this study. (D) Schematic diagram of RTK/RAS pathway components. Mammalian homologs appear in parentheses.
LF Inhibits Hep/c-Jun N-Terminal Kinase (JNK) Signaling in Vivo.
We tested the in vivo activity of LF by expressing the toxin in specific cell types in Drosophila using the GAL4/UAS system (26). In these experiments, we crossed flies carrying a UAS-LF transgene to a second stock expressing the yeast GAL4 transactivator in one of a variety of patterns and examined progeny carrying both transgenes. We first verified that LF was expressed at detectable levels by preparing protein extracts from heat-induced adult flies carrying a UAS-LF transgene and a heat-inducible GAL4 source (hsGAL4>LF flies). SDS/PAGE analysis of these extracts followed by Western blotting with anti-LF antibodies confirmed that LF was expressed and detected as a single band with an apparent molecular mass of 85 kDa (data not shown).
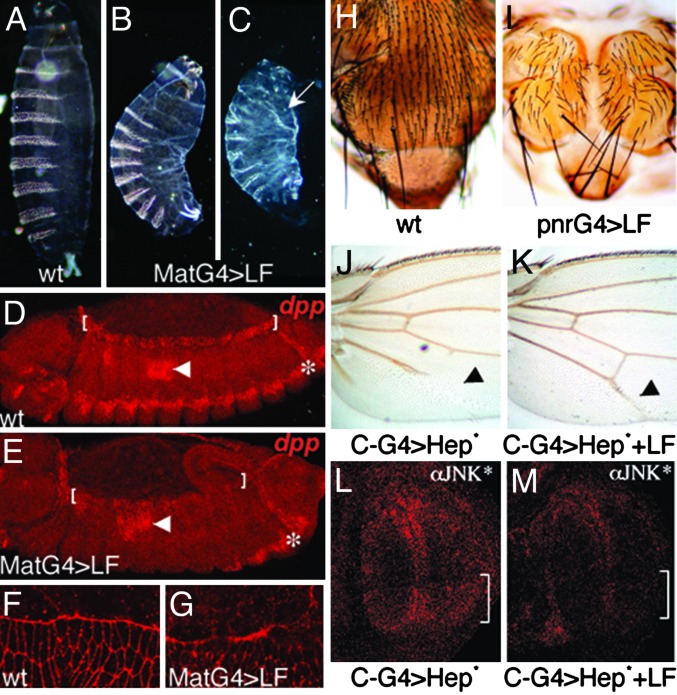
Next, we examined the in vivo effects of expressing LF during embryogenesis using various GAL4 drivers (such as teashirtGAL4, twistGAL4, actinGAL4, or MatGAL4) and observed nearly complete lethality. For further analysis, we focused on MatGAL4>LF embryos in which a maternal promoter provides strong ubiquitous expression of GAL4 in the embryo. Cuticle preparations of dead MatGAL4>LF embryos revealed a range of developmental defects. The least severely affected embryos had a U-shaped phenotype (Fig. 2B; compare with the wild-type embryo in Fig. 2A), which is typical of mutations interfering with dorsal closure, a process wherein lateral epithelial sheets move dorsally and fuse to cover extraembryonic cells known as the amnioserosa (27, 28). More severely affected embryos were greatly reduced in size, had dorsal holes in the cuticle, and lacked all head skeletal structures (Fig. 2C), which are hallmarks of more completely disrupted dorsal closure (21). Because Hep (21), the Drosophila ortholog of mammalian MAPKK7, is a key component of the JNK signaling pathway that controls dorsal closure (Fig. 1C), these defects may result from cleavage and inhibition of Hep by LF. To test this possibility, we assayed expression of the JNK pathway target gene decapentaplegic (dpp) in MatGAL4>LF embryos. Induction of dpp expression in leading-edge epithelial cells (Fig. 2D, brackets) depends on activity of the Hep/JNK pathway (29, 30). We observed a strong reduction of dpp expression in leading-edge cells of MatGAL4>LF embryos (Fig. 2E). In contrast, dpp expression was unaffected in other regions of the embryo such as the midgut (Fig. 2 D and E, arrowheads), consistent with previous evidence that dpp expression in these cells is independent of JNK regulation (29, 30). We also observed severe defects in the F-actin network in leading-edge cells of MatGAL4>LF embryos (Fig. 2G), which forms a regular line in wild-type embryos (Fig. 2F). Similar defects have been reported in bsk− embryos (31). Interestingly, a role in actin stress fibers formation was previously demonstrated for the mammalian JNK pathway (32).
Fig. 2.
LF inhibits Hep/JNK signaling in Drosophila. (A–C) Cuticle preparations of Drosophila wild-type (wt) (A) or MatGAL4>LF (B and C) embryos. LF-expressing embryos are similar to Hep/JNK pathway mutant embryos (21, 30). GAL4 drivers indicated in labels in this and subsequent figures are abbreviated as G4. Among embryos expressing high levels of LF that have dorsal closure defects the various phenotypic categories occur at the following respective frequencies (n = 54): U-shaped (46%), U-shaped with head cuticle defects and/or dorsal holes (42%), and severe cuticle phenotype (12%). In C the arrow indicates an anterior dorsal hole in the cuticle. (D and E) in situ hybridization of Drosophila embryos using a dpp antisense probe. Brackets indicate leading-edge cells, which express dpp in wild-type embryos (D), but fail to do so in MatGAL4>LF embryos (E). Arrows point to dpp-expressing midgut cells, and the asterisk indicates a lateral stripe of dpp expression, independent of JNK regulation. (F and G) High-magnification views of F-actin in embryos stained with Alexa Fluor 555-coupled phalloidin. Elongated leading-edge cells have a sharp and regular F-actin front in wild-type (wt) embryos (F), whereas in MatGAL4>LF embryos (G) the F-actin front is irregular and discontinuous. (H and I) Thoraxes dissected from wild-type (wt) (H) and pnrGAL4>LF (I) adults. LF-expressing individuals display a dorsal cleft phenotype typical of Hep viable mutants (33). (J and K) Proximal portions of adult wings. (J) A C-GAL4>Hep-CA (activated Hep) wing. (K) A C-GAL4>Hep-CA+LF wing, in which LF suppresses the phenotype induced by Hep-CA. (L and M) Third-instar imaginal discs stained with an anti-activated JNK antibody. (L) A C-GAL4>Hep-CA disc. Bracket indicates the posterior domain of GAL4 expression. (M) A C-GAL4>Hep-CA+LF disc showing reduced JNK activation caused by LF.
A process closely related to dorsal closure is also required to suture imaginal discs along the thoracic dorsal midline during pupal development, which likewise depends on Hep/JNK signaling (33, 34). We tested whether LF affects thorax closure by expressing LF under the control of the pannier (pnr) GAL4 driver in the dorsal region (notum) derived from the wing imaginal disc (35). This region elongates during pupariation to form the dorsal–medial region of the adult thorax. LF expression driven by pnrGAL4 caused a dorsal cleft phenotype (Fig. 2I; compare with wild-type thorax in Fig. 2H), phenocopying hypomorphic hep mutants (33). This phenotype was not observed when we expressed a form of LF with a mutated catalytic site (E720C) (11), consistent with dorsal cleft defects being caused by cleavage of a host target protein. We also tested whether LF could inhibit ectopic activation of the Hep/JNK pathway by coexpressing LF with a constitutively activated Hep (HepCA) (36). Weak expression of HepCA in the posterior region of the wing using the C-GAL4 driver led to a partial loss of the fifth longitudinal vein (L5) (Fig. 2J). When LF was coexpressed with HepCA, this phenotype was almost completely suppressed (Fig. 2K), indicating that LF acts in parallel to or downstream of Hep. In addition, we assayed the in situ activation pattern of Basket (Bsk) using a phosphospecific anti-D-JNK antibody (37) in C-GAL4>hepCA±LF discs. We found that expression of HepCA alone in the posterior region of wing discs led to elevated levels of activated Bsk (Fig. 2L, bracket) and that coexpression with LF eliminated this activation (Fig. 2M), providing evidence that LF acts upstream of Bsk. In aggregate, the data indicate that LF inhibits the JNK pathway in three distinct developmental settings, most likely by cleaving and inhibiting the Drosophila MAPKK Hep.
LF Inhibits the Dsor/MAPK Pathway in the Wing Disc.
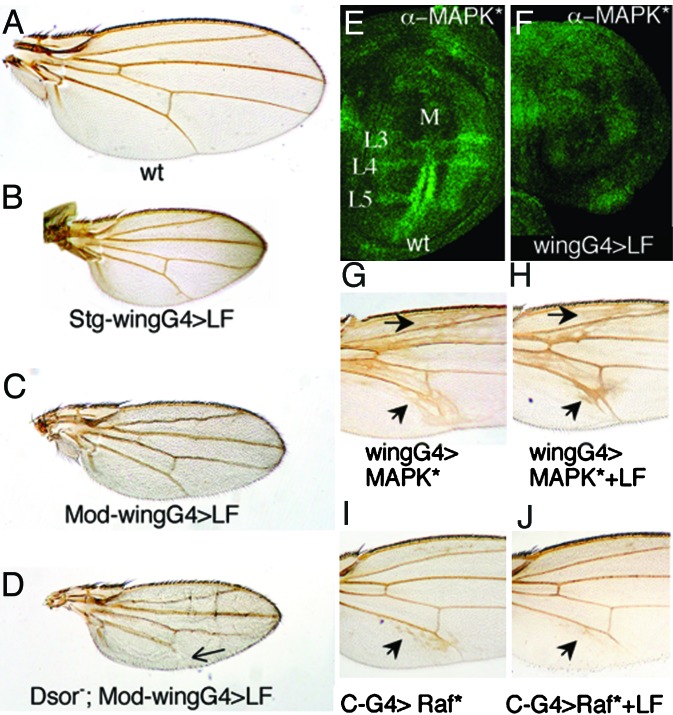
Dsor1, the Drosophila homolog of human hMEK1 and hMEK2, is a central component of the canonical RTK/RAS/Dsor/MAPK pathway (23) (Fig. 1D). Although we could not detect a mobility shift resulting from LF treatment of Dsor1 in our in vitro cleavage assay, sequence analysis indicated that Dsor1 should be a good substrate for LF. As mentioned previously, a possible explanation is that cleavage occurs but is too close to the N terminus of Dsor1 to be detected by SDS/PAGE analysis. To determine whether LF can inhibit Dsor1 in vivo, we expressed LF ubiquitously in the wing pouch using a wing-specific GAL4 driver (MS1096GAL4 = wingGAL4). The RTK/RAS/Dsor/MAPK pathway plays a dual role in the wing in promoting cell survival/proliferation (at basal levels of signaling) and vein differentiation (strong localized activation) (reviewed in ref. 38). When expressed at high levels in the wing disc, LF caused small scooped wings, which were elongated along the proximal–distal axis (Fig. 3 B and C). Similar phenotypes are caused by expressing dominant-negative (DN) forms of RTK/RAS/Dsor/MAPK pathway components, such as DN-EGF-R (39, 40), in line with LF inhibiting this pathway. Another function of the Dsor/MAPK pathway is to promote wing vein over intervein development, which depends in part on localized processing of membrane-bound EGF ligands by the Rhomboid protease (41, 42). Consistent with a role in inhibiting EGF-R signaling, strong expression of LF with the wingGAL4 can induce distal vein truncations, and moderate expression of LF, which has no vein phenotype on its own, also causes vein loss in heterozygous combinations with recessive vein-loss mutations such as rhove, vn/++, or kniri (data not shown). We tested whether heterozygous mutations in any of the four known Drosophila MAPKKs enhanced the effects of LF in the wing. We found that two loss-of-function Dsor1 alleles (Fig. 3D), but not mutations in hep, lic, or MAPKK4 (data not shown), resulted in a aggravated wingGAL4>LF phenotype consisting of more pronounced scooped wings and distal vein truncations (compare Fig. 3 D and A). This specific interaction of LF with Dsor1 supports the view that the observed LF wing phenotype results from reduced RTK/RAS/Dsor/MAPK signaling. We also assayed the effect of LF on the in situ activation level of MAPK (MAPK*) (43). In wild-type wing discs, detectable MAPK* is normally restricted to the presumptive margin and vein primordia (40, 43) (Fig. 3E). In wingGAL4>LF expressing discs, however, little, if any, MAPK* was observed (Fig. 3F), suggesting that LF inhibits the Dsor/MAPK pathway upstream of MAPK. Although the loss of MAPK* staining in vein primordia of LF-expressing wing discs appears to be more severe than the resulting adult wing phenotype, this discrepancy may reflect the difficulty in detecting low levels of MAPK* activation in situ. Alternatively, compensatory developmental mechanisms acting during pupal development may correct the earlier defects, as has been observed previously with vein-loss mutants (40). In accord with the MAPK* staining data, LF did not suppress the ectopic vein phenotype caused by UAS-rolledSem, an activated allele of the rolled MAPK (44) (compare Fig. 3 G and H). In contrast, LF did suppress the strong ectopic vein phenotypes caused by ectopic expression of rhomboid (41) (data not shown) or an activated form of Raf (26) (compare Fig. 3 I and J). We conclude that LF inhibits the RTK/RAS/Dsor/MAPK pathway downstream of Raf and upstream of MAPK. Consistent with LF cleaving and inactivating Dsor1, none of the various LF phenotypes were observed with a catalytically inactive mutant form of LF (11).
Fig. 3.
LF inhibits Dsor/MAPK signaling. (A–D) Adult wings. Stg-wingGAL4>LF refers to males with three copies of the UAS-LF construct driven by the MS-1096 ubiquitous wing driver at 29°C. The small wing vein phenotype (B) has nearly 100% penetrance, and ≈50% of these individuals also have vein truncations (not present in the wing shown in B). Mod-wingGAL4 refers to the MS1096GAL4 driving expression of three copies of the UAS-LF construct in females at 25°C (A). (D) The Dsor1 allele used is Dsor1rl. Arrow points at a truncated L5 vein. (E and F) Third-instar imaginal discs stained with an anti-di-phosphoMAPK antibody. (E) A wild-type (wt) disc, showing localized MAPK activation in the margin and vein primordial. (F) An MS1096>LF disc, in which LF suppresses MAPK activation. (G) An MS1096>UAS-Sem wing, in which the activated allele Sem of the rolled MAPK causes ectopic veins (arrows). This phenotype is not suppressed by coexpression of LF (H). (I and J) Medial portions of adult wings, showing ectopic veins (arrows) in C-GAL4>Rafgof wings (I) and suppression of this phenotype by LF (J).
EF Causes hh-Like Phenotypes in the Wing.
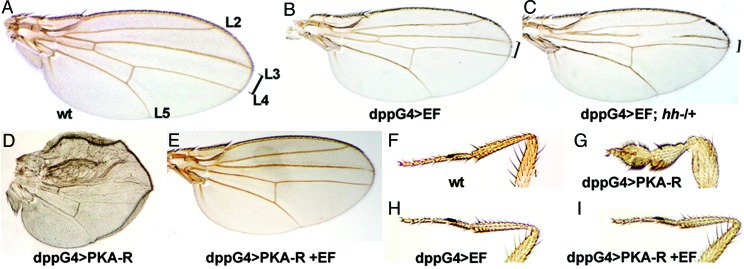
We also examined the effect of expressing EF in various cell types. We crossed a stock carrying a UAS-EF transgene to different GAL4 drivers and found that EF expression caused lethality with many of the drivers tested. We obtained surviving adult flies, however, when expressing EF under the control of the dppGAL4 driver, which activates gene expression in a stripe of six to eight cells between the presumptive L3 and L4 vein primordia in the wing imaginal disc. These dppGAL4>EF flies had a reduced spacing between the L3 and L4 veins and occasional truncations of the L3 vein (compare Fig. 4B with the wild-type wing shown in Fig. 4A). Similar phenotypes result from weak mutations in the Hh pathway, which specifies cell fates in the central region of the wing (45, 46). The Hh-like phenotype caused by EF is consistent with the known inhibitory role of cAMP-dependent PKA in this pathway. PKA phosphorylates the Cubitus interruptus transcription factor (Ci) and promotes its ubiquitination and proteolysis into a transcriptional repressor (CiR), which inhibits expression of hh target genes (reviewed in ref. 24). Elevated levels of cAMP induced by EF are therefore expected to inhibit hh signaling through activation of PKA. We tested whether EF was interfering with the Hh pathway by reducing the gene dose of hh in flies expressing EF and found that such dppGAL4>EF; hh/+ individuals exhibited a significant enhancement of the EF phenotype (Fig. 4C). This enhanced EF phenotype was not suppressed by coexpression of the antiapoptotic protein p35 (47) (data not shown), suggesting that EF causes patterning or growth defects rather than apoptosis. To confirm that EF acts on the Hh pathway via PKA, we expressed EF along with the regulatory subunit of PKA (PKAr) (48), which mediates the cAMP dependence of PKA activity (49). When expressed alone, PKAr causes strong phenotypes mimicking ectopic activation of Hh signaling. For example, dppGAL4>PKAr individuals die during late pupariation, and excavated adults show severe wing (Fig. 4D) and leg (Fig. 4G) patterning defects (compare with wild-type limbs in Fig. 4 A and F, respectively). Although EF expression caused no detectable phenotype on its own in the leg (Fig. 4H) and only a moderate Hh-like phenotype in the wing (Fig. 4B), it completely suppressed PKAr-induced lethality and the associated pupal phenotypes when coexpressed with PKAr (Fig. 4 E and I). These observations are compatible with the known effect of cAMP on the regulatory subunit of PKA, which binds to and suppresses the activity of the catalytic subunit of PKA (PKAc) under low concentrations of cAMP but dissociates from PKAc under high cAMP concentrations. We conclude that EF phenotypes in the wing are likely to be mediated by its adenylate cyclase activity.
Fig. 4.
EF inhibits PKA-dependent Hh signaling. (A–E) Adult wings. Brackets indicate the distance between longitudinal veins L3 and L4, which is reduced in dpp-discGAL4>EF wings (B). This phenotype is clearly enhanced in hh heterozygous mutants (C). (D) A dppGAL4>PKAr wing, in which expression of the regulatory subunit of PKA causes sublethality and severe patterning defects. This phenotype is almost completely suppressed by coexpression with EF (E). (F–I) Anterior legs of adult males. Expression of the regulatory subunit of PKA causes malformation of legs (G). This phenotype is also suppressed by coexpression with EF (I). EF expression with other GAL4 drivers causes lethality (apGAL4, ptcGAL4, and dppGAL4 at 25°C), strong composite wing phenotypes (71BGAL4, 1348GAL4, scaGAL4, MS1096GAL4, and vgGAL4), and small or rough eyes (eyeGAL4 and GMRGAL4, respectively). wt, wild type.
Discussion
B. anthracis and its virulence factors LF and EF have been intensively studied over the past few years, and, although much has been learned about the structure of the toxins and their mode of entry into host cells (5), the link between their biochemical activities and their physiological effects has remained elusive (6). In this study we examined the effects of anthrax toxins in the model genetic host, Drosophila. In the case of LF, we found that two Drosophila MAPKKs are cleaved in vitro by LF and that the in vivo function of two MAPKK pathways (Hep/Bsk = MAPKK7/JNK and Dsor/Rolled = MEK1,2/MAPK) can be blocked by expression of an LF transgene. Although we have not directly examined the effect of LF on p38 pathways, we observed strong cuticle phenotypes in embryos expressing high levels of LF (data not shown). These severe phenotypes are very similar to those of hep lic double mutants in which both JNK and p38 activation are blocked (22). In addition, as shown in Fig. 1B, Lic is a substrate for LF in vitro. Further analysis will be necessary to determine the in vivo effect of LF on Lic and MAPKK4.
EF toxin also causes expected patterning defects when expressed in developing wing imaginal discs. The EF phenotypes are similar to those caused by mutations reducing Hh signaling and are likely to be mediated by cAMP-dependent PKA, which suppresses Hh signaling (46). These initial studies validate Drosophila as a genetic multicellular model host for analyzing the function of the LF and EF anthrax toxins.
We note that, while there is strong evidence for LF acting at least in part by cleaving and inactivating MAPKK targets, this protease may also have other targets contributing to its lethal effects (6). Another important question is how LF and EF toxins cooperate to achieve optimal virulence in the host. Recent reports indicate that EF and LF can act in either opposing or synergistic fashions depending on the cellular context (18, 50). In preliminary experiments, we have observed other phenotypes caused by expression of LF and EF in various cell types in addition to the expected phenotypes reported here. This study therefore provides a starting point for analyzing potentially novel effects of LF and EF and may lead to the identification of new targets mediating cooperative effects of these two toxins.
B. anthracis is not known to infect hosts other than mammals. Consistent with this observation, we found no homolog of anthrax toxin receptors tumor endothelial marker 8 and capillary morphogenesis protein 2 encoded by the Drosophila genome, suggesting that Drosophila is not a suitable model for infection by anthrax. This is also likely to be true for many human pathogens, which have evolved to infect mammals via multiple sequential events, including host recognition, adherence, induction of virulence genes, virulence factor delivery, or evasion of host defenses. In some cases, however, it has been possible to infect Drosophila with human pathogens, such as Vibrio cholerae (51), Pseudomonas aeruginosa (52), or Staphylococcus aureus (53). In contrast to infection with a pathogenic organism, expression of a single virulence factor, which affects only a limited set of conserved host targets, is more likely to produce a specific and interpretable response. Because many pathogens act on specific protein targets that have been highly conserved in Drosophila, we anticipate that Drosophila will become a widely used in vivo system for the analysis of bacterial toxins or viral virulence factors with unknown activities or unidentified targets. In addition, toxins such as LF that have multiple host target proteins may be used to simultaneously reduce or eliminate the activities of several related proteins that perform overlapping functions. Thus, pharmacogenetic strategies can complement classic loss-of-function genetics in cases where multiple genes carry out related functions.
Materials and Methods
In Vitro Cleavage of Drosophila MAPKK Proteins with LF.
Hep, Dsor1 and Lic proteins were synthesized and 35S-labeled by incubating their cDNA plasmids with TnT reticulocyte lysates (Promega). For the in vitro cleavage assay, 35S-labeled MAPKK proteins were incubated with 250 ng of LF in 25 μl of assay buffer (25 mM potassium phosphate, pH 7.4/20 mM NaCl/0.1 mM calcium chloride/0.1 mM zinc chloride/10 mM magnesium chloride/1 mM DTT/10% glycerol) for 1 h. Samples were analyzed by SDS/PAGE and autoradiography.
Construction of UAS-LF and UAS-EF Plasmids.
LF and EF sequences were amplified by high-fidelity PCR (Extensor PCR Master Mix 1, ABgene catalog no. 0792/A) from plasmids pLF7 (Lf-wt), pSJ121F (Lf-E720C), and pSE42 (EF), which were kindly provided by Steve Leppla (National Institutes of Health, Bethesda), by using the following primers: LF sense, 5′-CATAGCGGCCGCGGCGCCAGCATGGCGGGCGGTCATGGTGATGTA-3′; LF antisense, 5′-GAGCTCTAGATTATGAGGTAATAATGAACTTAATC3′; EF sense, 5′-GATCCTCGAGGGCGCCAGCATGAATGAACATTACACTGAGAGT-3′; EF antisense, 5′-CAGCTCTAGATTATTTTTCATCAATAATTTTTTGG-3′. The bacterial signal sequences comprising the first 33 aa of the LF and EF proteins were not included and were replaced by a methionine preceded by an optimal Drosophila Kozak's sequence (GCCAGC). PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen) and checked for proper sequence. LF was then inserted into the pUASt vector. EF was inserted into a modified version of the pUWT vector (54) (details available on request), which has an FLP-excisable white+ cassette that prevents leaky expression of the transgene.
Drosophila Stocks and Crosses.
All crosses were grown under standard conditions at 25°C, except for dppGAL4>EF combinations, which were raised at room temperature. For expression of LF we used either a UAS-LF2X/FM7 stock or a UAS-LF3X/FM7 stock. The GAL4 driver lines used included MatGAL4 (55), pnrGAL4 (35), wingGAL4 known as MS1096GAL4 (56), C-GAL4 (57), and dpp-discGAL4 (dppGAL4). Dsor1rl and Dsor1LH110 alleles were kindly provided by N. Perrimon (Harvard Medical School), and UAS-Sem was provided by Ilaria Rebay (University of Chicago, Chicago). UAS-HepCA, Hepr75, HepG0107, and MAPKK4e01485 stocks were obtained from the Bloomington Stock Center (Bloomington, IN). For expression of EF we used a UAS<w+>EF UAS-FLP/TM6 stock, and for epistasis experiments we crossed this dppGAL4 UAS<w+>EF UAS-FLP/TM6 stock to hh2/TM3, hh21/TM3 stocks (obtained from Bloomington Stock Center) and to a UAS-PKAr stock (kindly provided by Dan Kalderon, Columbia University, New York).
In Situ Hybridization and Immunofluorescence.
In situ hybridization on embryos using a digoxigenin (DIG) dpp antisense probe was performed as described (58). The final detection steps were carried out with a sheep anti-DIG antibody, followed by a donkey anti-sheep Alexa Fluor 555-coupled antibody (Invitrogen/Molecular Probes). We detected in situ activation of Bsk (JNK) by using a primary rabbit anti-active JNK pAb (1/500) (Promega, catalog no. V7931) followed by incubation with an Alexa Fluor 555 goat anti-rabbit secondary antibody (Invitrogen/Molecular Probes). In situ detection of Rolled activation (MAPK*) was performed by using a primary mouse anti-diphosphoMAPK mAb (1/200) (Sigma, catalog no. M9692) followed by an Alexa Fluor 488 chicken anti-mouse secondary antibody (1/500, Invitrogen/Molecular Probes). F-actin was detected by using Alexa Fluor 555-phalloidin (Invitrogen/Molecular Probes).
Acknowledgments
We thank Steve Leppla for kindly providing EF, LF, and mutant LF cDNAs and John Collier and members of the E.B. laboratory for helpful discussions. This work was supported by National Institutes of Health Grants R21 AI60976 (to E.B.) and R01 AIOG1712 (to M.K.).
Abbreviations
- LF
lethal factor
- EF
edema factor
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- Hep
Hemipterous
- HepCA
constitutively activated Hep
- Bsk
Basket
- Dsor1
Downstream-of-Raf1
- PA
protective antigen
- Lic
Licorne
- JNK
c-Jun N-terminal kinase
- Hh
Hedgehog
- PKAr
regulatory subunit of PKA
See Commentary on page 3013 .
Footnotes
Conflict of interest statement: E.B. holds shares in NovaScape Sciences.
References
- 1.Reiter L. T., Potocki L., Chien S., Gribskov M., Bier E. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bier E. Nat. Rev. Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 3.Bier E., McGinnis W. In: Molecular Basis of Inborn Errors of Development. Epstein C. J., Erikson R. P., Wynshaw-Boris A., editors. New York: Oxford Univ. Press; 2004. pp. 25–45. [Google Scholar]
- 4.Beutler B. Mol. Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Collier R. J., Young J. A. Annu. Rev. Cell Dev. Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 6.Mourez M. Rev. Physiol. Biochem. Pharmacol. 2004;152:135–164. doi: 10.1007/s10254-004-0028-2. [DOI] [PubMed] [Google Scholar]
- 7.Bradley K. A., Young J. A. Biochem. Pharmacol. 2003;65:309–314. doi: 10.1016/s0006-2952(02)01455-7. [DOI] [PubMed] [Google Scholar]
- 8.Scobie H. M., Rainey G. J., Bradley K. A., Young J. A. Proc. Natl. Acad. Sci. USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonuccelli G., Sotgia F., Frank P. G., Williams T. M., de Almeida C. J., Tanowitz H. B., Scherer P. E., Hotchkiss K. A., Terman B. I., Rollman B., et al. Am. J. Physiol. 2005;288:C1402–C1410. doi: 10.1152/ajpcell.00582.2004. [DOI] [PubMed] [Google Scholar]
- 10.Vitale G., Bernardi L., Napolitani G., Mock M., Montecucco C. Biochem. J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 11.Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., Vande Woude G. F. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 12.Chopra A. P., Boone S. A., Liang X., Duesbery N. S. J. Biol. Chem. 2003;278:9402–9406. doi: 10.1074/jbc.M211262200. [DOI] [PubMed] [Google Scholar]
- 13.Leppla S. H. Proc. Natl. Acad. Sci. USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezard C., Berche P., Mock M. Infect. Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J. M., Greten F. R., Li Z. W., Karin M. Science. 2002;297:2048–2051. doi: 10.1126/science.1073163. [DOI] [PubMed] [Google Scholar]
- 16.Friedlander A. M. J. Biol. Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 17.O’Brien J., Friedlander A., Dreier T., Ezzell J., Leppla S. Infect. Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tournier J. N., Quesnel-Hellmann A., Mathieu J., Montecucco C., Tang W. J., Mock M., Vidal D. R., Goossens P. L. J. Immunol. 2005;174:4934–4941. doi: 10.4049/jimmunol.174.8.4934. [DOI] [PubMed] [Google Scholar]
- 19.Paccani S. R., Tonello F., Ghittoni R., Natale M., Muraro L., D’Elios M. M., Tang W. J., Montecucco C., Baldari C. T. J. Exp. Med. 2005;201:325–331. doi: 10.1084/jem.20041557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firoved A. M., Miller G. F., Moayeri M., Kakkar R., Shen Y., Wiggins J. F., McNally E. M., Tang W. J., Leppla S. H. Am. J. Pathol. 2005;167:1309–1320. doi: 10.1016/S0002-9440(10)61218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glise B., Bourbon H., Noselli S. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 22.Suzanne M., Irie K., Glise B., Agnes F., Mori E., Matsumoto K., Noselli S. Genes Dev. 1999;13:1464–1474. doi: 10.1101/gad.13.11.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuda L., Inoue Y. H., Yoo M. A., Mizuno M., Hata M., Lim Y. M., Adachi-Yamada T., Ryo H., Masamune Y., Nishida Y. Cell. 1993;72:407–414. doi: 10.1016/0092-8674(93)90117-9. [DOI] [PubMed] [Google Scholar]
- 24.Nybakken K., Perrimon N. Curr. Opin. Genet. Dev. 2002;12:503–511. doi: 10.1016/s0959-437x(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 25.Jiang J., Struhl G. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- 26.Brand A. H., Perrimon N. Development (Cambridge, U.K.) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 27.Martin P., Parkhurst S. M. Development (Cambridge, U.K.) 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y., Karin M. Trends Cell Biol. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Hou X. S., Goldstein E. S., Perrimon N. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- 30.Glise B., Noselli S. Genes Dev. 1997;11:1738–1747. doi: 10.1101/gad.11.13.1738. [DOI] [PubMed] [Google Scholar]
- 31.Ricos M. G., Harden N., Sem K. P., Lim L., Chia W. J. Cell Sci. 1999;112:1225–1235. doi: 10.1242/jcs.112.8.1225. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Wang W., Hayashi Y., Jester J. V., Birk D. E., Gao M., Liu C. Y., Kao W. W., Karin M., Xia Y. EMBO J. 2003;22:4443–4454. doi: 10.1093/emboj/cdg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agnes F., Suzanne M., Noselli S. Development (Cambridge, U.K.) 1999;126:5453–5462. doi: 10.1242/dev.126.23.5453. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Blanco E., Pastor-Pareja J. C., Garcia-Bellido A. Proc. Natl. Acad. Sci. USA. 2000;97:7888–7893. doi: 10.1073/pnas.97.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calleja M., Moreno E., Pelaz S., Morata G. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 36.Adachi-Yamada T., Fujimura-Kamada K., Nishida Y., Matsumoto K. Nature. 1999;400:166–169. doi: 10.1038/22112. [DOI] [PubMed] [Google Scholar]
- 37.Lee S. B., Cho K. S., Kim E., Chung J. Development (Cambridge, U.K.) 2003;130:4001–4010. doi: 10.1242/dev.00595. [DOI] [PubMed] [Google Scholar]
- 38.Bier E. BioEssays. 1998;20:189–194. doi: 10.1002/(SICI)1521-1878(199803)20:3<189::AID-BIES1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Buff E., Carmena A., Gisselbrecht S., Jimenez F., Michelson A. M. Development (Cambridge, U.K.) 1998;125:2075–2086. doi: 10.1242/dev.125.11.2075. [DOI] [PubMed] [Google Scholar]
- 40.Guichard A., Biehs B., Sturtevant M. A., Wickline L., Chacko J., Howard K., Bier E. Development (Cambridge, U.K.) 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- 41.Sturtevant M. A., Roark M., Bier E. Genes Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- 42.Urban S., Lee J. R., Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 43.Gabay L., Seger R., Shilo B. Z. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 44.Brunner D., Oellers N., Szabad J., Biggs W. H., III, Zipursky S. L., Hafen E. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 45.Vervoort M. BioEssays. 2000;22:460–468. doi: 10.1002/(SICI)1521-1878(200005)22:5<460::AID-BIES8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 46.Lum L., Beachy P. A. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 47.Hay B. A., Wolff T., Rubin G. M. Development (Cambridge, U.K.) 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 48.Kiger J. A., Jr., Eklund J. L., Younger S. H., O’Kane C. J. Genetics. 1999;152:281–290. doi: 10.1093/genetics/152.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W., Ohlmeyer J. T., Lane M. E., Kalderon D. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- 50.Park J. M., Greten F. R., Wong A., Westrick R. J., Arthur J. S., Otsu K., Hoffmann A., Montminy M., Karin M. Immunity. 2005;23:319–329. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Blow N. S., Salomon R. N., Garrity K., Reveillaud I., Kopin A., Jackson F. R., Watnick P. I. PLoS Pathog. 2005;1:e8. doi: 10.1371/journal.ppat.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avet-Rochex A., Bergeret E., Attree I., Meister M., Fauvarque M. O. Cell Microbiol. 2005;7:799–810. doi: 10.1111/j.1462-5822.2005.00512.x. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Lara J., Needham A. J., Foster S. J. FEMS Immunol. Med. Microbiol. 2005;43:311–323. doi: 10.1016/j.femsim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Keller A., Sweeney S. T., Zars T., O’Kane C. J., Heisenberg M. J. Neurobiol. 2002;50:221–233. doi: 10.1002/neu.10029. [DOI] [PubMed] [Google Scholar]
- 55.Bossing T., Barros C. S., Brand A. H. Genesis. 2002;34:123–126. doi: 10.1002/gene.10145. [DOI] [PubMed] [Google Scholar]
- 56.Capdevila J., Guerrero I. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lunde K., Trimble J. L., Guichard A., Guss K. A., Nauber U., Bier E. Development (Cambridge, U.K.) 2003;130:235–248. doi: 10.1242/dev.00207. [DOI] [PubMed] [Google Scholar]
- 58.Kosman D., Mizutani C. M., Lemons D., Cox W. G., McGinnis W., Bier E. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]