Abstract
The androgen receptor not only mediates prostate development but also serves as a key regulator of primary prostatic cancer growth. Although initially responsive to selective androgen receptor modulators (SARMs), which cause recruitment of the nuclear receptor–corepressor (N-CoR) complex, resistance invariably occurs, perhaps in response to inflammatory signals. Here we report that dismissal of nuclear receptor–corepressor complexes by specific signals or androgen receptor overexpression results in recruitment of many of the cohorts of coactivator complexes that permits SARMs and natural ligands to function as agonists. SARM-bound androgen receptors appear to exhibit failure to recruit specific components of the coactivators generally bound by liganded nuclear receptors, including cAMP response element-binding protein (CBP)/p300 or coactivator-associated arginine methyltransferase 1 (CARM1) to the SARM-bound androgen receptor, although still causing transcriptional activation of androgen receptor target genes. SARM-bound androgen receptors use distinct LXXLL (L, leucine; X, any amino acid) helices in the p160 nuclear receptor interaction domains that may impose selective allosteric effects, providing a component of the molecular basis of differential responses to different classes of ligands by androgen receptor.
Keywords: corepressor, nuclear receptor–corepressor, methyltransferase
The ability of many nuclear receptors to bind a variety of ligands, including antagonists that cause either a failure to respond or markedly diminished gene activation, raises intriguing issues about the molecular mechanism by which quantitative responses to ligands is regulated. One opportunity to investigate this issue is provided by androgen receptor, which can bind both ligands acting as strong agonists or can act to inhibit activation events.
The effects of androgens are mediated through the androgen receptor, a member of the nuclear receptor family functioning as a ligand-inducible transcription factor that regulates the expression of target genes having an androgen response element (1). Analogous to other members of nuclear receptor superfamily, the androgen receptor contains a DNA-binding domain and a C-terminal ligand-binding domain functioning as ligand-dependent nuclear receptors with activation functions 1 and 2 (AF-1 and AF-2) transcription activation domains (2–4). The integrity of the C-terminal AF-2 domain and recruitment of many cofactors including histone acetyltransferase (HAT)-containing proteins and the TRIP/DRIP/ARC complex are important for the transcriptional regulation. The AF-2 domain of nuclear receptors binds to p160 cofactors at the LXXLL motif (L, leucine; X, any amino acid), and these motifs were shown to confer helical conformation (5–12). The structural basis of estrogen receptor and coactivator recognition revealed that each of the ligands induces a different conformation of helix 12, and various antagonists block AF-2 function by disrupting the topography of the AF-2 structure (11, 13–18). The binding of antagonist did not trigger AF-2 activity, whereas the binding of agonist did (19). A series of structural analyses of the ligand-binding domains of multiple nuclear receptors and the demonstration of altered conformations of the AF2 helix-and-loop in estrogen receptor on binding of distinct ligands (11, 14–17) provides a context for further investigation. The androgen receptor not only mediates prostate development but also serves as a key regulator of primary prostatic cancer growth (20).
Histone acetylation has emerged as a fundamental component of the control of gene activation so that various HATs stimulate transcription through acetylation of histones and other factors (reviewed in refs. 21–24). In addition to various coactivator proteins that have HAT activity, coactivator-associated arginine methyltransferase 1 (CARM1) was identified as the binding protein to the C-terminal region of p160 coactivators by using a yeast two-hybrid system (25). The activation domain 1 of p160 coactivators binds CBP/p300, and activation domain 2 of p160 coactivators binds CARM1 after propagating activating signals (25). The presence of both protein methyltransferase and transcriptional coactivator activities in CARM1 suggests that methylation of histones or other proteins may play a role in transcriptional activation. However, a large series of enzymatic activities have been associated with coactivator complexes, including HATs, “histone” methyltransferases, ubiquitylation complexes, phosphatases, and protein kinases (reviewed in ref. 23).
Conversely, histone deacetylases (HDACs) antagonize HAT activity and represses transcription. Thus, investigation of active repression of gene expression by unliganded nuclear receptors has led to the identification of the nuclear receptor–corepressor (N-CoR) (26) and a closely related silencing mediator for retinoic acid and thyroid hormone receptor (SMRT) (27, 28). N-CoR and SMRT contain two nuclear receptor interaction domains and at least three repression domains that could transfer their active repression function by recruiting HDACs and other corepressors. Retinoic acid receptor (RAR) and thyroid hormone receptors (T3Rs) regulate both ligand-dependent activation and active repression by unliganded nuclear receptors (reviewed in ref. 23). Estrogen receptor α (ERα) can interact with N-CoR in the presence of antagonist such as 4-hydroxytamoxifen (4-OHT) (29–31). Based on the genetic analysis, N-CoR proves to be required for repression by T3R and RAR and for the function of ER antagonists. N-CoR is present in many complexes, including TBL1/TBLR1/HDAC3/GPS2 (31–35), TAB2/HDAC3 (36), Sin3/HDAC1/HDAC 2 complexes (37–39), and complexes containing SWI/SNF-related proteins.
Sequences initially referred to as corepressor/nuclear receptor (CoRNR) box (40) or LXXX IXXX I/L motif (38, 41) appear to bind in the hydrophobic pocket that is occupied by the coactivator LXXLL helical motifs on binding to ligand, but conformational effects of ligand binding inhibit the binding of corepressors, implicating the molecular mechanism for ligand-dependent displacement of the corepressor complex.
Based on the roles of N-CoRs in mediating repressive actions of specific nuclear receptors, including ER on binding of antagonists (29–31, 42, 43), it becomes important to explore the potential roles of N-CoRs as required components for therapeutic effectiveness of selective androgen receptor (AR) modulators (SARMs), which function as antagonists in the specific tissue and promoter context.
In this article, we provide evidence that inflammatory signals such as IL-1β cause determination of the N-CoR/TAB2 complex to convert androgen antagonists to agonists by permitting the recruitment of coactivators to AR target gene promoters, resulting in SARM-dependent activation of AR target genes less effective than that achieved by agonist binding, reflecting a usage of distinct LXXLL-helical motifs in recruitment of p160 factors in response to SARMs or agonists, which in turn causes an allosteric effect that determines that CBP/P300 and CARM1 are no longer recruited in response to SARMs. These findings provide evidence for the molecular basis of the distinctions in quantitative responses in gene activation induced by agonists versus SARMs in the presence of signals that cause removal of the N-CoR complex.
Results
IL-1β Converts Androgen Antagonists to Function as Agonists.
The interaction between N-CoR and AR in the presence of SARMs, which is regulated by several signal transduction pathways, provides an opportunity for modulating tumor progression and drug resistance. Proinflammatory cytokines, such as IL-1β, cause nuclear export of the N-CoR/TAB2 complex, resulting in derepression of the N-CoR-mediated repression of NF-κB p50 homodimer gene targets in the nucleus (36). Treatment with IL-1β caused cyproterone acetate (CPA) or bicalutamide to function as an agonist of AR (Fig. 1A).
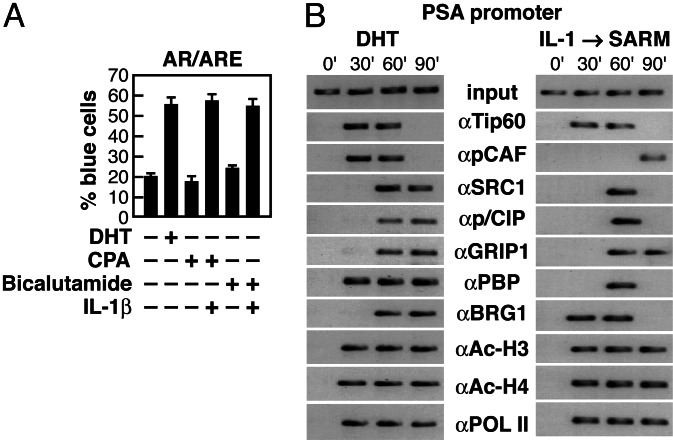
Fig. 1.
Evaluation of the recruitment of coactivators to AR-dependent promoter in response to 5α-dihydrotestosterone (DHT) and SARMs. (A) Pretreatment of cells with IL-1β abolished CPA or bicalutamide-mediated repression of a reporter containing androgen receptor response element (ARE). (B) ChIP assay to monitor the occupancy of prostate-specific antigen (PSA) promoter by Tip60, pCAF, SRC1, p/CIP, GRIP1, PBP, BRG1, acetylated histones H3 and H4, and RNA polymerase II (POL II) at the indicated times (min) after treatment with either DHT or IL-1β and SARM. α, Antibody. Soluble chromatin was prepared from LNCaP prostate cancer cells treated with either DHT for 1 h or IL-1β and CPA for 1 h.
Evaluation of the Roles of Coactivators in 5α-Dihydrotestosterone (DHT) and SARM-Dependent Activation.
Having demonstrated that SARMs function as agonists in response to IL-1β, we asked in this case whether the transcription machinery used in response to SARMs is identical to that used by native activating ligand, especially because the magnitude of transcriptional activation is quite diminished (≈3- to 5-fold) in response to SARMs. We therefore compared the potential ligand-specific difference in the temporal patterns of recruitment of various coactivator proteins with the AR-dependent transcription complex, and the relationship of these events to the state of histone acetylation on the PSA promoter. We found that many components of various coactivator complexes, including p160 factors (p/CIP, GRIP1, and SRC-1), pCAF, Tip60, PBP [a coactivator of peroxisome proliferator-activated receptor (PPAR)], thyroid hormone receptors, and Sp1 complexes, were recruited to the PSA promoter by both DHT-bound AR and by SARM-bound AR in IL-1β-treated cells (Fig. 1B). However, it appears that SARM-bound AR recruited certain coactivators only transiently or at a relatively later stage. Despite the ligands used, recruitment of many complexes including p160 members and a chromatin remodeling component such as Brg1, is a later event (Fig. 1B).
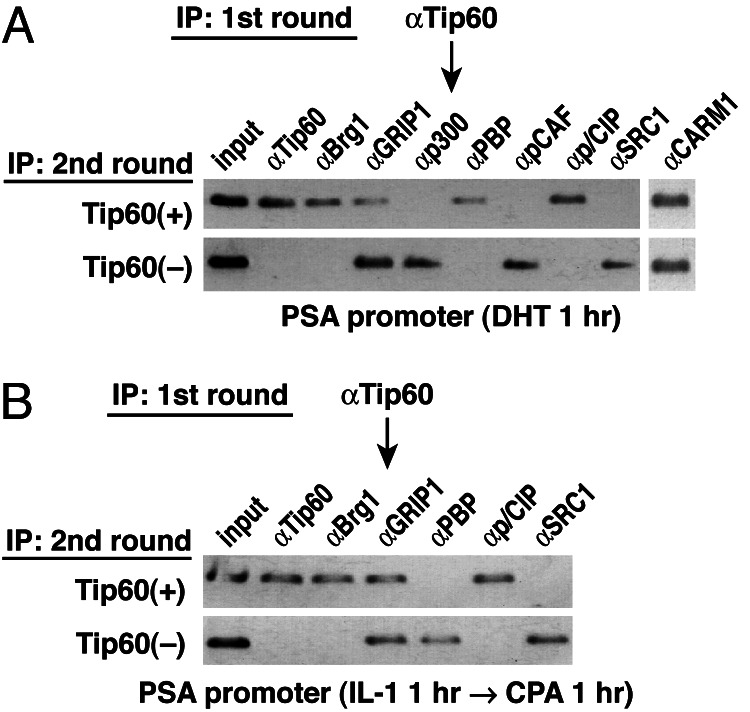
To investigate whether cohorts of coactivator complexes are simultaneously or selectively recruited to the identical PSA promoters, we performed sequential chromatin immunoprecipitation (ChIP) experiments in the presence of either DHT- or IL-1β/SARM-treated cells (Fig. 2A and B), by using αTip60 IgG for this two-step ChIP. Interestingly, however, we found that specific HAT-containing complexes, including CBP/p300, p/CAF, and SRG1, were not corecruited with Tip60 to the PSA promoter (Fig. 2). In the presence of IL-1β and SARM, Tip60 was also corecruited with BRG1, p/CIP, and GRIP1 complexes, but again not with other HAT-containing coactivator complexes, e.g., SRC-1 (Fig. 2). These experiments suggest that an ordered recruitment of distinct combinations of HAT-containing coactivator complexes (e.g., Tip60, CBP/p300, p/CAF, and SRC-1) occurs in ligand-dependent transcriptional activation of the PSA promoter. Corecruitment of one of several possible HAT-containing coactivator complexes may be sufficient to achieve transcriptional activation of the PSA promoter.
Fig. 2.
Two-step ChIP test of co-occupancy of coactivators. Cells were treated with DHT (1 h) (A) or with CPA (1 h) after IL-1 for 1 h (B). Aliquots were first immunoprecipitated with IgG against Tip60 (first IP). The bound materials and supernatant were collected and reimmunoprecipitated with IgGs against Tip60, Brg1, GRIP1, p300, PBP, pCAF, pCIP, SRC1, or CARM1.
Failure to Recruit CBP/p300 Coactivators and CARM1 Methyltransferase in SARM-Dependent Activation in IL-1β-Treated Cells.
Although many coactivator complexes are recruited both by DHT and by SARMs, we did identify a striking difference in coactivator recruitment by SARMs compared with DHT. Intriguingly, agonistic activity of SARMs in the presence of IL-1β was achieved in the absence of the recruitment of CBP/p300 or CARM1 (Fig. 3A), both of which are recruited to the C-terminal domain of the p160 proteins (25, 44). Furthermore, DHT-dependent, but not SARM-dependent, activation of the PSA promoter was correlated with methylation of histone H3 at R17 and R26, but not histone H4 at R3, indicating that CARM1-mediated methylation at R17 and R26 of histone H3 takes place during DHT-dependent transcriptional activation events but does not occur in response to SARMs (Fig. 3A). The acetylation of histone H3 at K9 and K14 was observed both in DHT- and in SARM-mediated activation, which correlates with a transcriptionally active modification pattern (data not shown).
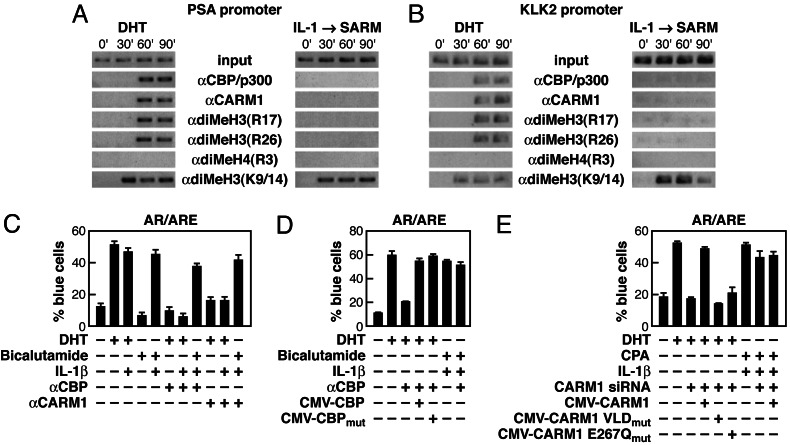
Fig. 3.
CBP/p300 and CARM1 are required for DHT but not SARM-dependent activation. (A) ChIP analysis of CBP/p300, CARM1, and other histone-modifying factors on PSA promoter after treatment with either DHT or IL-1β and SARM. (B) KLK2 promoter, another AR-responsive promoter, was examined after challenging with SARM and IL-1β, revealing that CBP/p300 were not recruited on KLK2 promoter. (C) Injection of anti-CBP IgG blocked the DHT-dependent activation of a reporter containing ARE. (D) Plasmid rescue experiments were performed as indicated after microinjection of anti-CBP IgG. (E) The function of CARM1 was assessed in cells injected with the ARE reporter. Rescue experiments with CARM1 expression plasmids confirmed a requirement for the functional methyltransferase activity.
To determine whether the failure to recruit CBP/p300 and CARM1 was unique to the PSA promoter in LNCaP cells, we examined their potential recruitment to other AR-dependent promoters such as KLK2 and found a failure of recruitment of CBP/p300 and CARM1 in the presence of IL-1β and SARM to the promoter and absence of methylation at R17 and R26 of H3 (Fig. 3B). Furthermore, failure of CBP/p300 and CARM1 recruitment in response to CPA or bicalutamide-mediated stimulation was also observed in LNCaP cells expressing high levels of AR, where SARMs display agonist activity (45).
The differential recruitment of CBP/p300 and CARM1 by DHT versus SARMs was unexpected and raised the possibility that these proteins may have nonessential functions on DHT-activated promoters. However, microinjection of specific αCBP/p300 IgGs or small interfering RNAs (siRNAs) against CBP/p300 markedly inhibited DHT-dependent activation but exerted no effect on the activation by IL-1β and SARMs (Fig. 3C and data not shown). Surprisingly, plasmids expressing either the functional CBP or mutant CBP with impaired HAT enzymatic function were able to rescue the activation function of DHT, indicating that the functional HAT enzymatic activity of CBP was not required for the DHT-dependent activation (Fig. 3D). Because histone H3/4 acetylation is observed for agonists and SARM/IL-1β-mediated activation, other HAT-containing complexes, particularly those containing Tip60, p/CAF, and/or p160 members must exert histone acetylation functions for SARM-dependent gene activation. Indeed, the HAT activities of both p/CAF and Tip60 were shown to be required for both DHT and SARM-dependent activation events by using the single-cell nuclear microinjection assay (data not shown).
Similarly, microinjection of specific antibodies or siRNA against CARM1 blocked DHT-dependent activation without any effect on SARM-dependent activation in IL-1β-treated cells (Fig. 3C and data not shown). In each case, injection of plasmids expressing functional CARM1 from a heterologous species, and thus resistant to the siRNA used, reversed the effect of CARM1 siRNA. However, unlike CBP/p300, mutant CARM1 with impaired enzymatic function (26) did not rescue the effects of CARM1 siRNA, suggesting that functional arginine methyltransferase activity is required for the DHT-dependent activation (Fig. 3E), consistent with the observed methylation of histone H3 at R17 and R26 (Fig. 3 A and B).
Differential Mode of p160 Factors Is Required for the DHT Versus SARM-Dependent Activation.
These findings raise the issue of the molecular mechanisms underlying differential recruitment of CBP/p300 and CARM1 by DHT versus SARMs. Because CBP/p300 and CARM1 are recruited by p160 cofactors (26), we investigated whether failure of their recruitment might reflect a difference in mode of p160 cofactor recruitment in response to SARMs. This possible mechanism was investigated by using specific anti-SRC-1 blocking IgG, which largely abrogates activation function of AR bound to agonist (Fig. 4A). Overexpression of wild-type SRC-1 restored AR agonist actions to both DHT and SARMs of IL-1β-pretreated cells (Fig. 4A). We therefore evaluated the requirement for the three LXXLL helices (LXDs) of the nuclear receptor interaction domains (6, 12). LXD1, as well as LXD2, was selectively required for the DHT-mediated activation, whereas LXD3, as well as LXD2, was specifically required for activation of SARMs (Fig. 4 B and C). Thus, it appears that, because DHT and SARMs induce distinct AR conformations of the AF-2 helices, the result is preferential binding to LXD1 and LXD3 of SRC-1, respectively. We postulate that these different binding interactions alter the conformation of SRC-1 and, hence, the subsequent assembly of the full transcription complex.
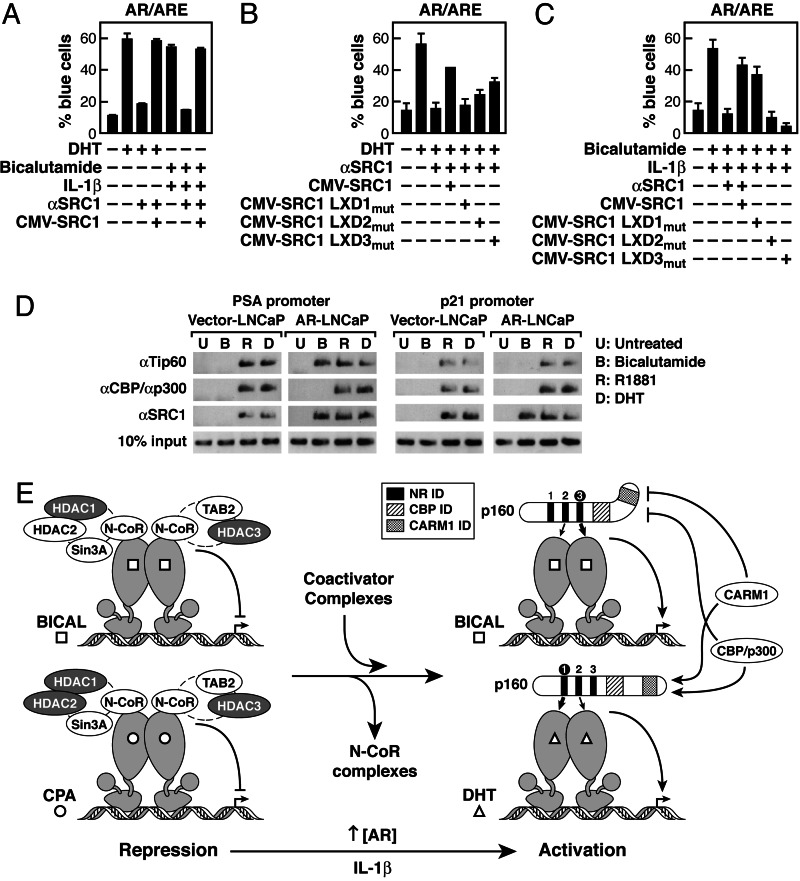
Fig. 4.
Differential mode of p160 factors for the DHT- versus SARM-dependent activation. (A) Injection of anti-SRC-1 IgG blocked both the DHT-dependent and bicalutamide/IL-1β-dependent activation of a reporter containing ARE. (B and C) After microinjection of anti-SRC-1 IgG, each LXXLL mutant of SRC-1 plasmid rescued the activation in the presence of DHT (B) or bicalutamide and Il-1β (C). Effects of mutating each LXXLL motif in SRC1 on DHT-dependent and bicalutamide/IL-1β-dependent activation are shown. (D) LNCaP cells stably infected with the AR virus or the control vector were generated and were starved for 5 days and then challenged with bicalutamide (10 μM), R1881 (100 pM), or DHT (10 nM). After 1 h of treatment, ChIP assay was performed with various IgGs. (E) Schematic representation of the action of coactivator use in SARMs-dependent gene activation.
Consistent with the role of N-CoR in SARM actions, AR levels are consistently up-regulated in hormone-refractory prostate cancer, causing a failure of N-CoR recruitment by SARMs (45), further supporting the hypothesis that N-CoR is a key determinant of SARM function. In the AR-overexpressed prostate cancer cell line, a failure of recruitment of CBP/p300 or CARM1 was again observed in response to CPA or bicalutamide-mediated stimulation, whereas all other coactivators examined, such as SRC-1, were recruited to the AR target gene promoters, PSA and p21 (Fig. 4D and data not shown). Thus, whether dismissal of N-CoR complexes occurs in response to specific signals or to AR overexpression, the specificity of coactivator complex recruitment in response to agonist or antagonists is distinct for different target genes.
Discussion
Molecular Mechanisms of Differential Gene Activation by DHT and SARMs.
In the absence of N-CoR, SARMs activate AR target genes, although not as robustly as native agonists, permitting clarification of molecular mechanisms that regulate transcriptional response to distinct ligands. Recruitment of most coactivator complexes was conserved and independent of ligand; however, a striking difference observed in SARM-dependent activation was that neither CBP/p300 nor CARM1 was recruited, nor were they required for activation. This result potentially provides, at least in part, the explanation of why SARMs are somewhat less effective than native agonists in levels of gene activation, even with corepressor dismissal. Because CARM1 is required by p160 factors, we investigated the potential differential modes of p160 factor recruitment in response to SARMs. Indeed, there were ligand-specific differences in the usage the three LXDs of the nuclear receptor interaction domains of p160 factors. LXD1 and LXD2 in SRC-1 are required for DHT-mediated activation, whereas LXD3 and LXD2 in SRC-1 are required for the activation of SARMs. Because recruitment of p160 factors by agonists and SARMs involves use of distinct LXXLL helical motifs in the nuclear receptor interaction domain, an allosteric mechanism can be suggested to account for the observation that the p160-dependent recruitment of CARM1, as well as CBP/p300, does not occur in CPA/bicalutamide-dependent activation of gene targets either in the presence of IL-1β or with AR overexpression.
Although neither CARM1-dependent methylation of histones nor CBP/p300 are required for AR-dependent gene activation with antagonist, these factors are required for full agonist-dependent gene activation. Because RNA profiling experiments show that there is a much decreased quantitative response in bicalutamide-dependent transcription units, which mimics a weak agonist function (44), we postulate that CARM1 and CBP/p300 recruitment in this case accounts for this quantitative difference in agonist versus antagonist activation (Fig. 3A). The implication of these data is that different ligands cause allosteric modification of recruited coactivators, and, hence, composition of coactivator complexes accounts for ligand-specific differences in transcriptional outcome. These findings are of particular interest because of the findings that many nuclear receptors contain multiple LXXLL helical motifs (6, 11–12, 15–17). Thus, we suggest that this finding potentially provides a “rule” for allosteric regulation: By binding distinct LXXLL motifs, conformational changes in these platform proteins change the ability to recruit specific cofactors, thereby setting the fine-tuned transcriptional response. Together, these findings provide mechanistic insights into SARM actions in prostate cancer, revealing the unique coactivator complex recruitment in response to different ligands as a basic consequence of ligand-dependent effects on AR C-terminal conformation (Fig. 4E).
Experimental Procedures
Materials and Reagents.
The following antibodies were obtained from Santa Cruz Biotechnology: anti-AR, anti-HDAC1, anti-HDAC2, anti-HDAC3, anti-MBD3, anti-MEKK1, anti-MEK1, anti-CBP, anti-p300, anti-p/CAF, anti-SRC1, anti-p/CIP, anti-GRIP1, anti-PBP, anti-BRG1, and anti-mSin3A/B. The following commercially available antibodies were used: anti-Tip60, anti-CARM1, anti-acetylated histone H3, anti-acetylated histone H4, anti-dimethyl-histone H3(R17), anti-dimethyl-histone H3(R26), and anti-dimethyl-histone H4(R3) (Upstate Biotechnology, Lake Placid, NY), anti-RNA polymerase II antibodies (Berkeley Antibody, Richmond, CA). Fluorogenic histone deacetylase substrate [Boc-Lys(Ac)-7-amino-4-methylcoumarin] and human IL-1β were from Calbiochem. DHT and CPA were purchased from Sigma.
siRNA.
Sequences for siRNA were as follows: for N-CoR (5′-GCACCGAAGUAUUGUCCAA-3′) and for CARM1 (5′-CUUCUGGUACCAGCCAUCU-3′). siRNA pool for HDAC1, HDAC3, and nonspecific control (SMARTpool HDAC1 and HDAC3) were purchased from Dharmacon Research (Lafayette, CO).
Single-Cell Nuclear Microinjection Assays.
Microinjection assays were carried out as described in refs. 46 and 47. Each experiment was performed on three independent coverslips consisting of 1,000 cells, with >300 cells injected. Where no experimental antibody was used, preimmune rabbit or guinea pig IgG was coinjected, allowing the unambiguous identification of injected cells in addition to serving as a preimmune control. Antibodies to HDACs are as described in ref. 47.
ChIP Assays.
The ChIP assay was conducted as described in refs. 44–46, with average size of sheared fragments ≈300–500 bp. For PCR, 1 μl from 50 μl of DNA extract and 25–30 cycles of amplification were used.
Acknowledgments
We thank M. Stallcup for kindly providing CMV-CARM1 and HMT mutant CARM1 plasmids and J. Hightower for figure preparation. M.G.R. and C.L.S. are investigators for Howard Hughes Medical Institute. This work was supported by the Ministry of Health and Welfare National Research and Design program for cancer control, the Ministry of Education Brain Korea 21 research fellowship, the Ministry of Science and Technology (MOST) and Korea Science and Engineering Foundation (KOSEF) Science Research Center/Engineering Research Center (SRC/ERC) program (to S.H.B.), and grants from the National Institutes of Health/National Cancer Institute (to D.W.R., and M.G.R.) and from the Prostate Cancer Research program (to M.G.R.).
Abbreviations
- AF
activation function
- AR
androgen receptor
- ARE
androgen-response element
- CARM1
coactivator-associated arginine methyltransferase 1
- CBP
cAMP response element-binding protein
- ChIP
chromatin immunoprecipitation
- CPA
cyproterone acetate
- DHT
5α-dihydrotestosterone
- HAT
histone acetyltransferase
- HDAC
histone deacetylases
- LXDs
LXXLL helices
- N-CoR
nuclear receptor–corepressor
- PSA
prostate-specific antigen
- SARMs
selective androgen receptor modulators
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Feldman B. J., Feldman D. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Onate S. A., Boonyaratanakomkit V., Spencer T. E., Tsai S. Y., Tsai M. J., Edwards D. P., O’Malley B. W. J. Biol. Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 3.Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. Mol. Cell. Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He B., Lee L. W., Minges J. T., Wilson E. M. J. Biol. Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 5.Douarin B. L., Zechel C., Garnier J. M., Lutz Y., Tora L., Pierrat P., Heery D., Gronemeyer H., Chambon P., Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 7.Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 8.Hong H., Kohli K., Garabedian M. J., Stallcup M. R. Mol. Cell. Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 10.Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P J., Baxter J. D., Fletterick R. J., Yamamoto K. R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renaud J. P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 14.Wagner R. L., Apriletti J. W., McGrath M. E., West B. L., Baxter J. D., Fletterick R. J. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 15.Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engstrom O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 16.Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 17.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 18.Moras D., Gronemeyer H. Curr. Opin. Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 19.Berry M., Metzger D., Chambon P. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debes J. D., Tindall D. J. Cancer Lett. (Shannon, Irel.) 2002;187:1–7. doi: 10.1016/s0304-3835(02)00413-5. [DOI] [PubMed] [Google Scholar]
- 21.Perlmann T., Evans R. M. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 22.Hassig C. A., Schreiber S. L. Curr. Opin. Chem. Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 23.Glass C. K., Rosenfeld M. G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 24.McKenna N. J., O’Malley B. W. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 26.Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K., Rosenfeld M. G. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen J. D., Evans R. M. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 28.Sande S., Privalsky M. L. Mol. Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 29.Jackson T. A., Richer J. K., Bain D. L., Takimoto G. S., Tung L., Horwitz K. B. Mol. Endocrinol. 1997;11:693–705. doi: 10.1210/mend.11.6.0004. [DOI] [PubMed] [Google Scholar]
- 30.Smith C. L., Nawaz Z., O’Malley B. W. Mol. Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 31.Lavinsky R. M., Jepsen K., Heinzel T., Torchia J., Mullen T. M., Schiff R., Del-Rio A. L., Ricote M., Ngo S., Gemsch J., et al. Proc. Natl. Acad. Sci. USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. Genes Dev. 2000;4:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen Y.-D., Perissi V., Staszewski L. M., Yang W.-M., Krones A., Glass C. K., Rosenfeld M. G., Seto E. Proc. Natl. Acad. Sci. USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Kalkum M., Chait B. T., Roeder R. G. Mol. Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 36.Baek S. H., Ohgi K. A., Rose D. W., Koo E. H., Glass C. K., Rosenfeld M. G. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 37.Heinzel T., Lavinsky R. M., Mullen T. M., Soderstrom M., Laherty C. D., Torchia J., Yang W. M., Brard G., Ngo S. D., Davie J. R., et al. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 38.Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. R. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alland L., Muhle R., Hou H., Jr, Potes J., Chin L., Schreiber-Agus N., DePinho R. A. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 40.Hu X., Lazar M. A. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 41.Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 43.McDonnell D. P., Norris J. D. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 44.Chakravarti D., LaMorte V. J., Nelson M. C., Nakajima T., Schulman I. G., Juguilon H., Montminy M., Evans R. M. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 45.Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 46.Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 47.Jepsen K., Hermanson O., Onami T. M., Gleiberman A. S., Lunyak V., McEvilly R. J., Kurokawa R., Kumar V., Liu F., Seto E., et al. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]