Abstract
In budding yeast, the MLH1-PMS1 heterodimer is the major MutL homolog complex that acts to repair mismatches arising during DNA replication. Using a highly sensitive mutator assay, we observed that Saccharomyces cerevisiae strains bearing the S288c-strain-derived MLH1 gene and the SK1-strain-derived PMS1 gene displayed elevated mutation rates that conferred a long-term fitness cost. Dissection of this negative epistatic interaction using S288c-SK1 chimeras revealed that a single amino acid polymorphism in each gene accounts for this mismatch repair defect. Were these strains to cross in natural populations, segregation of alleles would generate a mutator phenotype that, although potentially transiently adaptive, would ultimately be selected against because of the accumulation of deleterious mutations. Such fitness “incompatibilities” could potentially contribute to reproductive isolation among geographically dispersed yeast. This same segregational mutator phenotype suggests a mechanism to explain some cases of a human cancer susceptibility syndrome known as hereditary nonpolyposis colorectal cancer, as well as some sporadic cancers.
Keywords: colorectal cancer, incompatibility
The highly conserved mismatch repair (MMR) system contributes to genome stability by repairing errors that occur during DNA replication (1). In Escherichia coli, MMR is initiated by the binding of MutS protein to DNA mismatches. MutL interacts with the MutS-mismatch complex and activates downstream repair factors. Multiple MutS homologs (MSH) and MutL homologs (MLH) have evolved in eukaryotes that form heterodimers with specialized functions in DNA repair and recombination (2, 3). In Saccharomyces cerevisiae, MSH2-MSH3 and MSH2-MSH6 function in mismatch recognition, and MLH1-PMS1 is the primary MLH heterodimer in postreplicative MMR. Mutations in MSH and MLH genes that act in MMR elevate mutation rate, as measured in reversion and forward mutation assays, and reduce spore viability of diploid cells due to the accumulation of recessive lethal mutations (4–6). In addition, MMR proteins act to prevent recombination between divergent DNA sequences. This activity has been shown to prevent chromosomal rearrangements (7, 8) and to enforce reproductive barriers between species (9, 10).
Previously we created 60 alleles of the S. cerevisiae MLH1 gene from the S288c strain (cMLH1) in which clusters of charged residues were simultaneously changed to Ala (11). These alleles were tested for defects in MMR in the S288c (12) and SK1 (13) strains. More than one-third of the mutation set conferred a more severe MMR defect in SK1 strains than in S288c strains. Two mutations, cmlh1-29 and cmlh1-56, conferred wild-type-like phenotypes in S288c but null-like phenotypes in SK1. Introduction of the S288c PMS1 gene into the SK1 strain suppressed the mutator phenotype of these mutants, suggesting that the MMR phenotype was due to incompatibility, or negative epistasis, between MLH components (11).
The influences of epistatic interactions on a wide variety of traits and processes have garnered increasing attention (14–17). Few examples, however, have been characterized in molecular detail. Here we show that the strain-dependent MMR phenotypes observed previously for site-specific mlh1 mutants were due in part to an underlying defect between wild-type MMR genes from S288c and SK1. We identified the specific amino acid polymorphisms in MLH1 and PMS1, whose combined effect in hybrid strains leads to an elevation in mutation rate and a generalized reduction in long-term fitness. As described below, the generation of mutators may influence the evolution of natural yeast populations in several ways. Also, the negative epistatic interaction observed between yeast MMR gene variants suggests a mechanism to explain the genetic basis of some hereditary nonpolyposis colorectal cancer (HNPCC)-like cancers displaying atypical inheritance (18).
Results
Strain-Dependent MMR Defects Are Due to MLH1 and PMS1 Polymorphisms.
Previously we identified S288c-derived mlh1 alleles (cmlh1-29 and cmlh1-56) that appeared to confer a wild-type MMR phenotype in the S288c background of S. cerevisiae but severe MMR defects in SK1 strains (11). To test whether polymorphisms within MLH1 and PMS1 were responsible for the strain-dependent phenotypes, we crossed a wild-type SK1 strain to S288c strains bearing the mlh1 alleles. Spore progeny from these crosses were genotyped and analyzed for a mutator phenotype.
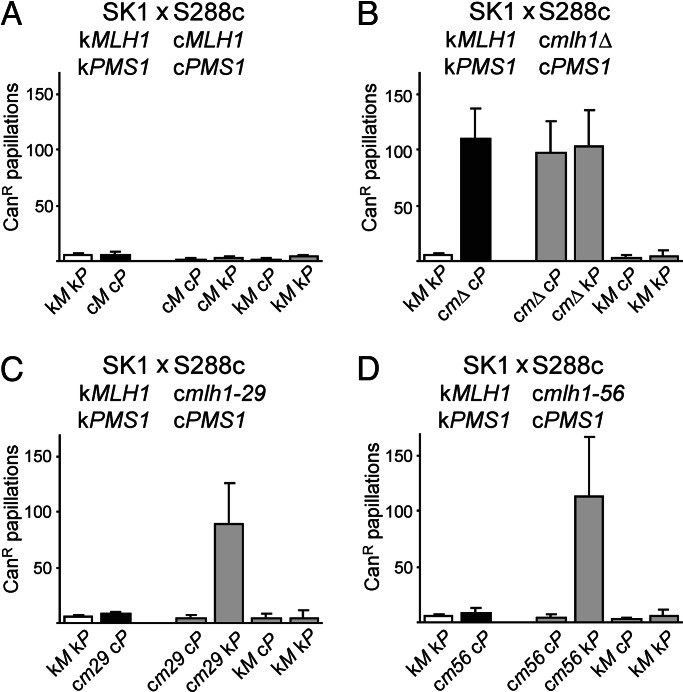
In a control cross, SK1 × S288c hybrids bearing only wild-type MLH1 and PMS1 genes yielded spore progeny that displayed no MMR defect as measured in a semiquantitative canavanine resistance assay (Fig. 1A) (11). In a second control cross in which wild-type SK1 was crossed to an S288c strain bearing a deletion in MLH1 (mlh1Δ), only progeny bearing the mlh1Δ mutation displayed a detectable defect in MMR (Fig. 1B). When wild-type SK1 was crossed to S288c strains bearing the mlh1 site-specific mutations, progeny containing SK1 PMS1 (kPMS1) and the cmlh1-29 or cmlh1-56 alleles displayed a MMR defect, whereas all others appeared wild-type (Fig. 1 C and D). This result indicates that the MMR defect results from negative epistasis between the mlh1 and PMS1 alleles from the different parental strains. Within the limits of the canavanine assay, no other differences in strain background contributed to the MMR phenotype. Although these results do not exclude the potential contributions of loci linked to MLH1 or PMS1, additional experiments in which cmlh1-29 and cmlh1-56 were each expressed with cPMS1 or kPMS1 within S288c or SK1 confirmed that these two genes are the major determinants of the observed defect (data not shown).
Fig. 1.
MMR-proficient strains can give rise to MMR-defective progeny. A wild-type SK1 strain (EAY1080) was crossed to S288c strains containing PMS1 marked with HIS3 and either MLH1 (EAY1093) (A), mlh1Δ (EAY1091) (B), mlh1-29 (EAY1095) (C), or mlh1-56 (EAY1096) (D) marked with KanMX. For each cross, canavanine resistance (CanR) frequency was determined for spore clones from at least seven complete tetrads. The median number of resistant colonies per spore clone (±SD) is presented. c, S288c; k, SK1. Unshaded bars represent the SK1 parent, black bars represent the S288c parent, and gray bars represent the spore progeny.
The mlh1-56 mutation (E680A, D681A, and E682A) maps to the putative PMS1 interaction domain, whereas mlh1-29 (K393A and R394A) lies within a region of MLH1 for which no function has been identified (11). Two-hybrid interactions for all combinations of SK1 and S288c MLH1 and PMS1 alleles were measured by using a GAL10 UAS-LacZ reporter. No difference in β-galactosidase level was observed in mlh1-PMS1 combinations that yielded functional or defective MMR, indicating that the observed genetic incompatibility was unlikely to be due to a simple defect in MLH1–PMS1 interactions (Fig. 5, which is published as supporting information on the PNAS web site) (11).
The Wild-Type S288c MLH1 and SK1 PMS1 Gene Combination Confers an Elevated Mutation Rate.
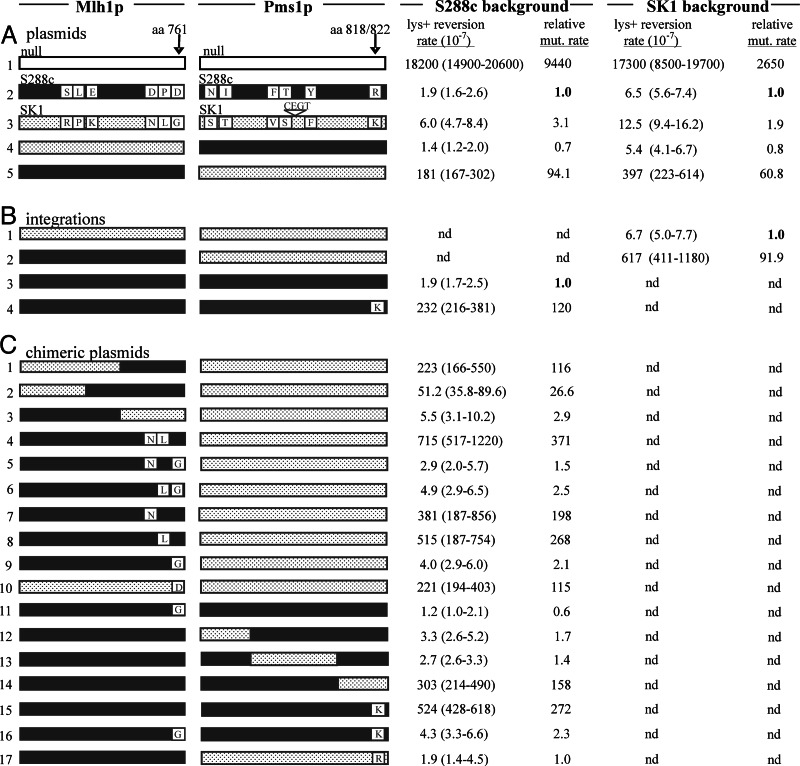
The interactions observed among mutant mlh1 and PMS1 alleles raised the question of whether the wild-type cMLH1 and kPMS1 alleles are fully compatible. We did not observe a defect for any combination of the wild-type genes in the canavanine assay or by two-hybrid analysis (Figs. 1 and 5 and data not shown). However, the difference in mutation rate between wild-type and MMR-defective strains in the canavanine assay is modest (10- to 20-fold), relative to that seen in assays that measure the reversion of specific frameshift mutations (4, 19–22). For example, MMR-defective strains bearing the lys2::insE-A14 frameshift allele revert to Lys+ at a 3,000- to 10,000-fold higher rate than wild-type (Fig. 2) (19).
Fig. 2.
S288c MLH1 and SK1 PMS1 confer a mutator phenotype. The MLH1 and PMS1 combinations indicated were tested in the lys2::insE-A14 mutator assay. The amino acid differences between the S288c (solid red) and SK1 (stippled blue) sequences are shown (11). (A) Lys+ reversion rates of S288c and SK1 mlh1Δ pms1Δ strains (EAY1365 and EAY1362) bearing the indicated S288c and SK1 MLH1 and PMS1 genes expressed from ARS CEN plasmids. (B) SK1 strains bearing S288c MLH1 (EAY1363) or SK1 MLH1 (EAY1364), both marked with KanMX4, and S288c strains bearing S288c PMS1 (EAY1369) or S288c PMS1R818K (EAY1370), both marked with HIS3, were tested for reversion to Lys+. (C) Lys+ reversion of S288c mlh1Δ pms1Δ strains (EAY1365) bearing S288c–SK1 MLH1 and S288c–SK1 PMS1 chimeras expressed on ARS CEN vectors. Rates are relative to the cMLH1–cPMS1 combination (construct A2) in the S288c background (1.9 × 10−7).
We tested all combinations of S288c and SK1 MLH1 and PMS1 genes in the lys2-A14 reversion assay. Three of the four combinations displayed low mutation rates similar to the S288c wild-type strain (Fig. 2, constructs A2, A3, and A4). The cMLH1–kPMS1 pairing, however, increased the mutation rate by ≈100-fold (Fig. 2, construct A5). This increase was observed regardless of whether the two genes were expressed in S288c or SK1 strains. Furthermore, chromosomal integrations of cMLH1 in an SK1 strain (Fig. 2, construct B2) and cPMS1R818K in an S288c strain (Fig. 2, construct B4) resulted in similar elevations in mutation rate.
To test whether the cMLH1–kPMS1 defect could be observed in hybrid strains, we carried out an experiment similar to that shown in Fig. 1, with strains bearing the lys2::insE-A14 allele. SK1 (EAY1364) and S288c (EAY1369) strains were crossed, and haploid progeny (36 spore clones from nine tetrads) were analyzed for reversion to Lys+ in a patch assay (data not shown). Fourteen independent colonies derived from each spore clone were patched onto media lacking Lys. Spore clones containing the cMLH1–cPMS1 or kMLH1–cPMS1 combinations displayed a low median number of revertants (n = 0–10), similar to that obtained when these combinations were tested within S288c or SK1 (Fig. 2, constructs A2 and A4). Seven of ten kMLH1–kPMS1 spore clones displayed a similar phenotype; however, three clones showed a slightly higher (n = 18–33) median number of revertants. For the cMLH1–kPMS1 genotype, six of eight spore clones displayed a median number of revertants (n = 120–750) that was similar to that observed for the mutator combinations shown in Fig. 2. Two spore clones displayed lower median numbers (n = 21 and 24). These results indicate that no S288c–SK1 incompatibilities other than cMLH1–kPMS1 confer a major effect on mutation rate. The variability suggests that other gene combinations may modify the cMLH1–kPMS1 defect. We also genotyped the spore progeny for S288c and SK1 MSH2, MSH3, and MSH6 MMR genes and found no correlation between strain origin of the MSH genes and mutator phenotype (Supporting Appendix 1, which is published as supporting information on the PNAS web site, and data not shown).
The cMLH1–kPMS1 MMR Defect Is Due to a Single Polymorphism in Each Gene.
Both MLH1 and PMS1 from S288c and SK1 contain multiple amino acid and silent polymorphisms (Supporting Appendix 1) (11). The two strains display 0.7% nucleotide sequence divergence in a 32-kb region that includes the 2.6-kb PMS1 ORF, and the MLH1 and PMS1 genes display 0.5% and 1.3% divergence, respectively (23). We made S288c–SK1 chimeras of both genes to determine whether specific polymorphisms could be identified that confer the cMLH1–kPMS1 defect. Substituting the 5′ end of kMLH1 into cMLH1 retained the defect with kPMS1 (Fig. 2, construct C1), whereas making the reciprocal chimera abolished it (Fig. 2, construct C3). These results suggested that residue(s) in the C terminus of cMLH1 are required for the defective interaction. Fine structure mapping with additional constructs allowed us to determine that a single amino acid in cMLH1, the Asp residue at amino acid 761, is sufficient for creating a mutator phenotype in the presence of kPMS1. Most of the MLH1 constructs encoding D761 conferred an ≈100- to 370-fold elevation in mutation rate when combined with kPMS1; constructs encoding a Gly residue at this position, as found in kMLH1, were fully functional for MMR (Fig. 2C). One MLH1 construct encoding D761 (Fig. 2, construct C2) displayed only a 27-fold elevation of Lys+ reversion rate when combined with kPMS1, indicating that other polymorphic sites can influence the MLH1–PMS1 compatibility. However, it is clear that the D761G polymorphism is the major determinant within MLH1 of the defect with kPMS1. Making the single G761D change within kMLH1 recreated this defect (Fig. 2, construct C10).
We found that the C terminus of kPMS1, containing a single amino acid polymorphism, was sufficient for the mutator phenotype observed with cMLH1 (Fig. 2, construct C14). Lys-822 in the kPMS1 polypeptide corresponds to Arg-818 in cPMS1. Because the subcloned region encoding the Lys residue also had two silent polymorphisms that could influence gene expression, we made the single R818K change in the cPMS1 gene (Fig. 2, construct C15). This construct displayed the same elevated mutation rate when paired with cMLH1, a result that was confirmed by integrating the chimera into an S288c strain (Fig. 2, construct B4). Finally, expressing the S288c versions of MLH1 and PMS1 with single SK1 polymorphisms at both loci (cMLH1D761G and cPMS1R818K) restored MMR to the same level as kMLH1 and kPMS1 (Fig. 2, construct C16). Thus the segregation of a single polymorphism in each of the two genes generates the cMLH1–kPMS1 defect in MMR.
Chimera analysis indicated that the cmlh1-29–kPMS1 mutator phenotype shown in Fig. 1C was due primarily to Lys-822 in the kPMS1 protein (data not shown). In the canavanine assay, cmlh1-29 displayed as severe a defect with cPMS1R818K as with the wild-type kPMS1. These observations suggest that the S288c mlh1 site-specific mutations exacerbated the defect observed in the cMLH1–kPMS1 combination.
Is There a Fitness Cost Associated with the cMLH1–kPMS1 MMR Defect?
Complete loss of MMR activity in S. cerevisiae leads to reductions in fitness due to the accumulation of deleterious mutations (4–6, 24, 25). We observed an ≈100-fold increase in mutation rate for the cMLH1–kPMS1 combination in the lys2-A14 reversion assay. We suspected that even this moderate increase would yield a fitness cost because most mutations with a phenotypic effect are deleterious. Genomic mutation rates tend to vary by less than an order of magnitude in organisms and viruses as diverse as λ, M13, E. coli, Neurospora crassa, and S. cerevisiae (24, 26), suggesting that deviations from the typical rates for S. cerevisiae would usually be selected against. Moreover, a high proportion of yeast genes contain microsatellite sequences that are particularly prone to mutation in the absence of MMR (19). For example, 143 ORFs, including 22 that are essential, contain A or T homopolymer runs of 10 or more nucleotides.
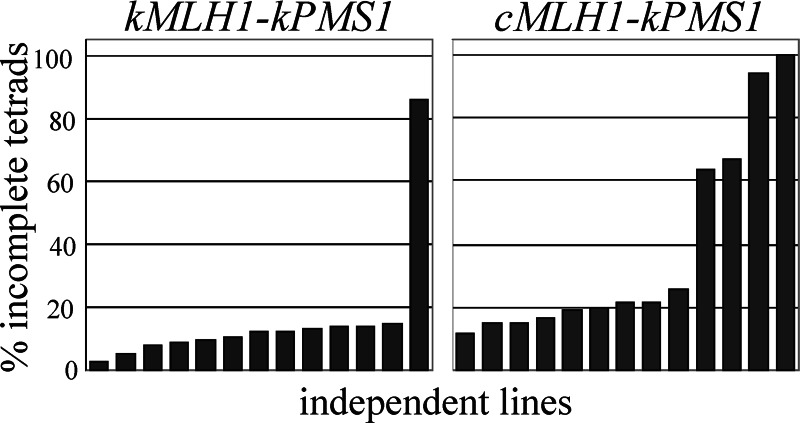
To test whether the cMLH1–kPMS1 defect affects long-term fitness through the accumulation of deleterious mutations, we assessed spore viability of homozygous diploid SK1 strains grown for ≈120 generations (Fig. 3). We examined 13 replicate lines each of a wild-type strain (kMLH1–kPMS1) and a strain in which the kMLH1 allele was replaced with cMLH1 (cMLH1–kPMS1) as well as five lines of an mlh1Δ strain. The initial spore viability of the mlh1Δ strain was 70.6%, with 50.6% of the tetrads being incomplete, i.e., containing less than four viable spores (data not shown). Much of this initial reduction in spore viability is attributable to the meiotic crossover defect associated with the mlh1Δ mutation (27). The wild-type and cMLH1–kPMS1 strains displayed initial spore viabilities of 94.3% and 91.1%, respectively, with 15.5% and 17.5% incomplete tetrads, respectively. These values reflect spore viability before mutation accumulation and are expected to be similar. The cMLH1–kPMS1 genotype was previously shown not to have a meiotic phenotype (11).
Fig. 3.
The percentage of tetrads that form four viable spores can be used as an indicator of fitness. Thirteen replicate lines each of a wild-type strain (kMLH1::KanMX4-kPMS1; EAY1459 × EAY1460) (A) and the MMR-defective cMLH1::KanMX4-kPMS1 strain (EAY1455 × EAY1456) (B) were analyzed for spore viability after 120 generations of growth. At least 65 tetrads were examined for each line. The percentage of tetrads containing less than four viable spores is indicated.
After 80 generations, overall spore viability of the five replicate mlh1Δ lines dropped to 2.5–29%, and no four-spore-viable tetrads were observed in any of the lines. In addition, the surviving spore clones varied greatly in size. One wild-type line displayed a large decrease in spore viability after 120 generations, dropping to 45.2%, with 85.9% incomplete tetrads. This observation of a low frequency drop in fitness in wild-type lines is not unprecedented, because it was observed in a previous mutation accumulation experiment in which one of 30 nonpetite wild-type lines displayed a large reduction in fitness (24). Spore viability for the other 12 wild-type lines remained high through 120 generations (Fig. 3). In contrast, many of the cMLH1–kPMS1 lines, which displayed the ≈100-fold increase in mutation rate in the lys2-A14 reversion assay, displayed a decrease in spore viability (Fig. 3), and surviving spore clones more often showed poor growth. The stochastic nature of the occurrence of mutations means that not all replicates would be expected to incur mutations that confer fitness effects (24). Although the difference in total spore viability between the wild-type and cMLH1–kPMS1 data sets was not significant (P = 0.072), the two data sets differed significantly with respect to the percentage of incomplete (less than four viable) tetrads (Fig. 3) (P = 0.002, Wilcoxon–Mann–Whitney test). Thus, mutations affecting fitness accumulated at a greater rate in the SK1 cMLH1–kPMS1 diploids compared with the SK1 wild-type diploids.
Evolution of the MMR Phenotype.
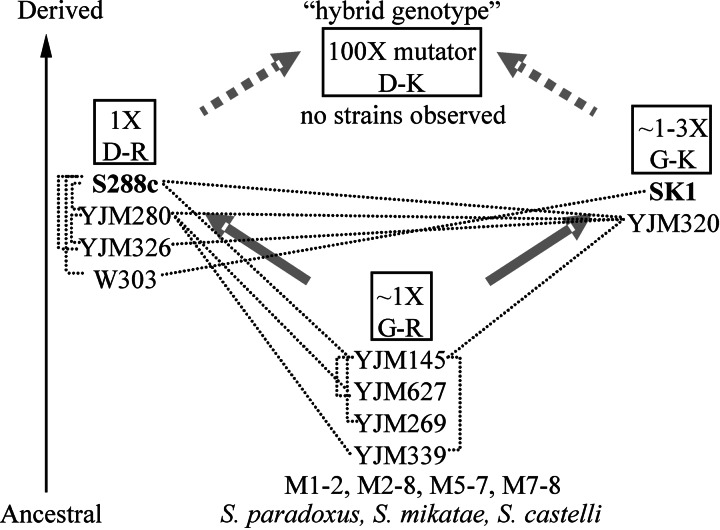
We were interested in determining whether negative epistatic interactions observed between MMR genes in laboratory strains could arise in natural yeast populations. Such interactions would yield a mutator phenotype that could be important for adaptive evolution (28). We examined MLH1 and PMS1 sequences from 12 other S. cerevisiae strains, including laboratory, clinical, and wild isolates (23), and compared them with those of several other Saccharomyces species (Supporting Appendix 1). We classified the genes according to residues 761 of MLH1 (Asp in S288c; Gly in SK1) and 818/822 of PMS1 (Arg in S288c; Lys in SK1), which confer the MMR defect described in this study, and the strains fell into three groups (Fig. 4). Both alleles causing the incompatibility between S288c and SK1 are found in natural strains but not in the same strain. Eight S. cerevisiae strains as well as Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces castelli contain the Gly–Arg combination; we infer that this is the ancestral combination. This finding is consistent with the kMLH1–cPMS1 (Gly–Arg) pairing conferring a low mutation rate (Fig. 2, construct A4). Four S. cerevisiae strains, including S288c, contain the Asp–Arg combination. Besides SK1, only the S. cerevisiae strain YJM320, a clinical isolate, contains the Gly–Lys combination.
Fig. 4.
Likely reconstruction of events resulting in the observed defect in MMR in the contemporary S. cerevisiae gene pool. Fourteen S. cerevisiae strains and three closely related Saccharomyces species are schematically grouped according to their amino acid residues: 761 (Gly or Asp) in MLH1 and 818/822 (Arg or Lys) in PMS1. Bold arrows indicate transitions between genotypes resulting from single mutational events. The relative mutation rates of genotypes, based on Lys+ reversion experiments using S288c and SK1 strains (1X, wild-type S288c), are shown. Thin dotted lines denote predicted recombination events (P < 0.05) between 10 of these strains based on a sequenced 32-kb region (23). In this reconstruction, the S288c [Asp–Arg (D–R)] and SK1 [Gly–Lys (G–K)] strains diverged from an ancestral [Gly–Arg (G–R)] population. Genetic exchange between Asp–Arg and Gly–Lys strains would generate a mutator combination (Asp–Lys) at a 25% frequency (indicated by thick dotted arrows).
The inferred ancestral combination, Gly–Arg, conferred the lowest mutation rate in the lys2-A14 assay (Fig. 2). The Asp–Arg combination seen in S288c was slightly higher, with Gly–Lys from SK1 slightly higher still, for an ≈4-fold range among these three genotypes (within a >10,000-fold range overall). These data suggest the possibility that the two derived alleles are each slightly deleterious. However, because optimal mutation rates likely reflect a balance between genome stability and adaptability, we cannot predict whether these slight differences in MMR function are neutral, deleterious, or even beneficial.
To determine whether YJM320 MMR genes would also confer a mutator phenotype in combination with MMR genes from other strain backgrounds, we cloned MLH1 and PMS1 from YJM320 and tested them in combination with the S288c and SK1 alleles in a qualitative mutator assay. YJM320 MLH1 displays 0.1% and 0.3% divergence from the SK1 and S288c alleles, respectively, whereas YJM320 PMS1 displays 0.6% and 1.4% divergence from the SK1 and S288c alleles, respectively. A defect was observed only for the cMLH1–YJM320 PMS1 (Asp–Lys) combination, with an elevated mutation frequency similar to that observed for cMLH1–kPMS1 (data not shown). All other combinations displayed low mutation frequencies. These results are also consistent with the residues at 761 of MLH1 and 818/822 of PMS1 controlling the mutator phenotype.
As shown in Fig. 4, the identified Asp–Arg (S288c, YJM280, YJM326, and W303) and Gly–Lys (SK1, YJM320) combinations could be generated from the predicted ancestral state by single amino acid changes in MLH1 and PMS1. Both of these combinations were functional for MMR (Fig. 2). However, no strains were identified in this small data set that displayed the Asp–Lys mutator combination. We found the absence of this combination interesting because analysis of the distribution of sequence polymorphisms among 10 S. cerevisiae strains for which a 32-kb region was sequenced indicated recent recombination among Gly–Lys and Asp–Arg strains (Fig. 4). In particular, YJM320 displayed extensive evidence of recombination with the Asp–Arg group. We speculate that the Asp–Lys mutator genotype has been generated through interstrain crosses but was not observed due to the fitness cost associated with the MMR defect (Fig. 3).
Discussion
Strains bearing the S288c MLH1–SK1 PMS1 combination displayed an ≈100-fold increased reversion rate to Lys+ in a lys2-A14 reversion assay that correlated with a long-term decrease in spore viability, consistent with a genome-wide increase in the rate of mutation accumulation. The spore viability assay was conducted only through 120 generations, or ≈2.5 weeks under laboratory conditions; over many thousands of generations, the MMR defect would likely have a more dramatic impact on fitness. Mutational analysis revealed that a single polymorphism in each gene was responsible for this mutator phenotype. The simple, bilocus interaction underlying the observed MMR defect may have relevance to two distinct phenomena, the inheritance of the cancer susceptibility syndrome HNPCC and the evolution of yeast populations.
Implications for HNPCC Inheritance.
MMR defects have been implicated in a dominantly inherited cancer susceptibility syndrome known as HNPCC (18, 29). Of the ≈500 MMR gene variants listed in the HNPCC database, many are missense mutations whose associations with cancer predisposition are unclear (18, 29). In almost half of families with HNPCC, a germline mutation in a MMR gene has not been identified, and some familial cancers resemble HNPCC but do not display the typical, dominant inheritance pattern (18). For example, Kariola et al. (30) identified two independent HNPCC families in which the affected individuals had one mutation in hMSH2 and another in hMSH6. Inheritance of HNPCC in these families appeared to be recessive. The authors speculated that a MMR defect and consequently HNPCC arose only when both mutations were present in the same individual. Based on our studies in yeast, we hypothesize that human MMR gene variants that confer defects only in particular genetic backgrounds and/or environmental conditions could account for a portion of HNPCC and HNPCC-like cases, particularly those displaying atypical inheritance patterns.
The inheritance of a MMR defect in haploid yeast strains, as shown in Fig. 1, can easily be extended to diploid organisms. The following example uses the yeast nomenclature presented in this study: Phenotypically wild-type parents of genotype kMLH1/kMLH1 kPMS1/kPMS1 and cmlh1-29/kMLH1 cPMS1/kPMS1 would have one in four offspring with predisposition to HNPCC. For the offspring of genotype cmlh1-29/kMLH1 kPMS1/kPMS1, loss of heterozygosity at one locus (kMLH1) would confer a MMR defect; i.e., cancer risk for these individuals is the same as in typical HNPCC families. Interactions among polymorphic loci have been identified in other cancers but have not yet been studied extensively for HNPCC (31). Our observations provide an additional incentive to investigate whether such a mechanism underlies some cases of familial colorectal cancer.
Negative Epistasis Between MMR Gene Variants Could Contribute to Adaptive Evolution.
What are the consequences of an epistatic interaction of MMR genes for natural yeast populations? The answer depends largely on the distribution of alleles within and among populations. Although we have little information regarding the populations from which S288c and SK1 were originally isolated (12, 13), we have shown that amino acid residues that cause the incompatibility between the strains are found in wild strains. Moreover, there is evidence for recombination among these strains, suggesting that the mutator combination of MLH1 and PMS1 alleles has been generated in the wild (Fig. 4).
The effects of yeast mutators might be comparable with what has been observed in natural and laboratory populations of bacteria. In E. coli populations, spontaneous loss of MMR gives rise to mutators at low frequency, and such mutators may contribute to the adaptive evolution of a population in several ways (28). When adapting to a new environment, mutators may have an advantage due to their increased probability of acquiring the first adaptive mutation within a population. The mutation conferring mutator status can hitchhike to fixation with the beneficial mutation. Over time, however, the accumulation of deleterious mutations will outweigh the advantages of the beneficial mutation. Phylogenetic studies of E. coli strains suggest that mutators can reacquire MMR function through horizontal gene transfer (32). It is possible that segregation of various MLH1 and PMS1 alleles in yeast results in similar loss and reacquisition of MMR function. Loss or reduction of MMR function may result in a burst of divergence; interestingly, reacquisition of fully functional MMR genes may sustain that divergence, because of their role in suppressing recombination between diverged sequences. MMR genes have been implicated in enforcing species barriers in bacteria (10) and in yeast (9). In yeast, suppression of meiotic recombination can result in chromosome missegregation and spore death. Hunter et al. (9) demonstrated that functional MSH2 contributes to the sterility of hybrids of S. cerevisiae and S. paradoxus, which display 30% sequence divergence.
Reductions in MMR function, as described here, may result from hybridization of different yeast strains followed by segregation of gene variants within progeny (Fig. 4). This mechanism for generating MMR defects is similar to the epistatic interactions that are thought to underlie hybrid incompatibility between established (33–38) or incipient species (14). Dobzhansky and Muller (39–42) first proposed a model to explain how hybrid incompatibilities can arise without also causing defects within the parental strains or species. In their model, two geographically isolated populations arising from a common ancestor sustain mutations that are neutral or beneficial within the population in which they originate but potentially deleterious within the genetic background of the other population. These incompatibilities will only be observed in hybrids upon secondary contact of the two populations. The evolution of the cMLH1–kPMS1 MMR incompatibility (Fig. 4) correlates to this model, although the cMLH1 and kPMS1 alleles each confer slightly elevated mutation rates and may therefore be deleterious. Postzygotic reproductive barriers appear to be present between the SK1 and S288c strains, which display ≈1% nucleotide sequence divergence. SK1–S288c interstrain crosses yielded 73% spore viability, compared with 97% for S288c–S288c and 93% for SK1–SK1 intrastrain crosses (Fig. 6, which is published as supporting information on the PNAS web site). The incompatibility between MLH1 and PMS1 alleles may cause further fitness reductions in later generations of hybrids. Although our work offers a speculative scenario for the contribution of MMR genes to hybrid incompatibility, we have established that the relevant MLH1 and PMS1 alleles are segregating in natural populations. It is important for future work to obtain sufficient population sample and allele frequency information to determine whether this interaction represents a Dobzhansky–Muller-type incompatibility.
Materials and Methods
Strains and Plasmids.
The yeast strains used in this study are described in Supporting Appendix 2, which is published as supporting information on the PNAS web site. Integrations and disruptions were constructed by single-step gene replacement; the plasmids used in these constructs are available from the authors upon request. The W303–2B strain was obtained from Lorraine Symington (Columbia University, New York). YJM145, YJM269, YJM280, JM320, YJM326, YJM339, and YJM627 were obtained from John McCusker (Duke University, Durham, NC) and M1-2, M2-8, M5-7, and M7-8 wild yeast isolates from Montalcino, Italy were obtained from Jeffrey Townsend (University of California, Berkeley). Yeast strains were grown in yeast extract/peptone/dextrose (YPD), minimal complete, or minimal selective media (43). When required, canavanine (Sigma) was included in minimal selective media at 60 mg/l and geneticin (GIBCO) was included in YPD at 200 mg/l (43, 44). Sporulation plates and procedures and yeast two-hybrid analysis (Fig. 5) were performed as described in refs. 11 and 21.
Plasmids pEAA213 and pEAA214 contain the S288c and SK1 MLH1 genes, respectively, inserted into the polylinker of pRS415 (ARSH4 CEN6, LEU2) (45). The expression of both genes is driven through the S288c MLH1 promoter. All of the chimeric MLH1 constructs are derivatives of pEAA213 and pEAA214. pEAA238 and pEAA239 contain the S288c and SK1 PMS1 genes, respectively, inserted into the polylinker of pRS413 (ARSH4 CEN6, HIS3) (45). Both genes are expressed from the S288c PMS1 promoter. All of the chimeric PMS1 constructs are derivatives of these two plasmids. pEAA248 and pEAA249 contain the S288c and SK1 PMS1 genes, respectively, inserted into the polylinker of pRS416 (ARSH4 CEN6, URA3). The pEAA238/239 and pEAA248/249 plasmids were used to monitor complementation in the S288c and SK1 strains, respectively. Details on the subcloning required to make the chimeras are available on request.
Other Methods.
The semiquantitative canavanine-resistance assay was performed as described in ref. 11. Rates of lys2::insE-A14 reversion were calculated as μ = f/ln(N·μ), where f is reversion frequency and N is the total number of revertants in the culture (19). For all mutator assays, the median mutation rate was obtained from at least 11 independent cultures. For the spore viability analysis, initial spore viabilities were determined by using the zero growth mating protocol (4, 22). Briefly, haploid strains were mated for 4 h on minimal media, after which they were plated onto sporulation media. Tetrads were dissected on minimal complete plates (43). Stable diploid lines were propagated by serial transfer of randomly chosen colonies on YPD plates. Colonies were frozen every two transfers (≈40 generations). To determine spore viability at 80 or 120 generations, cells were patched from frozen stocks onto YPD plates, grown overnight, and then plated on sporulation media. To infer gene exchange as shown in Fig. 4, geneconv, version 1.81, was used to survey for recombination events among 10 strains in a 32-kb region (23).
Supplementary Material
Acknowledgments
We thank Daniel Barbash, Andrew Clark, Dmitry Gordenin, Martha Hamblin, Allen Orr, Tom Petes, Jeffrey Townsend, Amy Lyndaker, and Jennifer Surtees for helpful discussions; John Schmidt for contributions to this work; and Jeffrey Townsend and John McCusker for strains. This work was supported by National Institutes of Health Grants GM53085 (to E.A.) and GM36431 (to C.F.A.), a Coordenação de Aperfeiçaomento de Pessoal de Nível Superior Fellowship awarded by the Brazilian government (to J.L.A.), Department of Education and National Institutes of Health training grants (to A.B. and J.A.H.), and a Cornell Undergraduate fellowship (to Z.G.). R.G.R. was supported by a Cornell Biotechnology Institute grant (to C.F.A. and Mariana Wolfner).
Abbreviations
- MMR
mismatch repair
- HNPCC
hereditary nonpolyposis colorectal cancer
- MSH
MutS homolog
- MLH
MutL homolog
Footnotes
References
- 1.Schofield M. J., Hsieh P. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner R. D., Marsischky G. T. Curr. Opin. Genet. Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 3.Harfe B. D., Jinks-Robertson S. Annu. Rev. Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 4.Reenan R. A., Kolodner R. D. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prolla T. A., Christie D. M., Liskay R. M. Mol. Cell. Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson M. S., Game J. C., Fogel S. Genetics. 1985;110:609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petit M. A., Dimpfl J., Radman M., Echols H. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myung K., Datta A., Chen C., Kolodner R. D. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 9.Hunter N., Chambers S. R., Louis E. J., Borts R. H. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 10.Rayssiguier C., Thaler D. S., Radman M. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 11.Argueso J. L., Kijas A. W., Sarin S., Heck J. A., Waase M., Alani E. Mol. Cell. Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortimer R. K., Johnston J. R. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane S. M., Roth R. J. Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawson P. D., Burton R. S. Proc. Natl. Acad. Sci. USA. 2002;99:12955–12958. doi: 10.1073/pnas.202335899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brem R. B., Storey J. D., Whittle J., Kruglyak L. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caicedo A. L., Stinchcombe J. R., Olsen K. M., Schmitt J., Purugganan M. D. Proc. Natl. Acad. Sci. USA. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroymann J., Mitchell-Olds T. Nature. 2005;435:95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- 18.Chung D. C., Rustgi A. K. Ann. Intern. Med. 2003;138:560–570. doi: 10.7326/0003-4819-138-7-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 19.Tran H. T., Keen J. D., Kricker M., Resnick M. A., Gordenin D. A. Mol. Cell. Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni T. T., Marsischky G. T., Kolodner R. D. Mol. Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 21.Pang Q., Prolla T. A., Liskay R. M. Mol. Cell. Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Argueso J. L., Smith D., Yi J., Waase M., Sarin S., Alani E. Genetics. 2002;160:909–921. doi: 10.1093/genetics/160.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., McCusker J. H., Davis R. W. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 24.Zeyl C., DeVisser J. A. Genetics. 2001;157:53–61. doi: 10.1093/genetics/157.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wloch D. M., Szafraniec K., Borts R. H., Korona R. Genetics. 2001;159:441–452. doi: 10.1093/genetics/159.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drake J. W., Charlesworth B., Charlesworth D., Crow J. F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter N., Borts R. H. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 28.Giraud A., Radman M., Matic I., Taddei F. Curr. Opin. Microbiol. 2001;4:582–585. doi: 10.1016/s1369-5274(00)00254-x. [DOI] [PubMed] [Google Scholar]
- 29.Lynch H. T., de la Chapelle A. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 30.Kariola R., Otway R., Lonnqvist K. E., Raevaara T. E., Macrae F., Vos Y. J., Kohonen-Corish M., Hofstra R. M., Nystrom-Lahti M. Hum. Genet. 2003;112:105–109. doi: 10.1007/s00439-002-0866-4. [DOI] [PubMed] [Google Scholar]
- 31.Popanda O., Schattenberg T., Phong C. T., Butkiewicz D., Risch A., Edler L., Kayser K., Dienemann H., Schulz V., Drings P., et al. Carcinogenesis. 2004;25:2433–2441. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- 32.Denamur E., Lecointre G., Darlu P., Tenaillon O., Acquaviva C., Sayada C., Sunjevaric I., Rothstein R., Elion J., Taddei F., et al. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 33.Wu C. I., Ting C. T. Nat. Rev. Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 34.Coyne J. A., Orr H. A. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 35.Ting C. T., Tsaur S. C., Wu M. L., Wu C. I. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 36.Barbash D. A., Siino D. F., Tarone A. M., Roote J. Proc. Natl. Acad. Sci. USA. 2003;100:5302–5307. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 38.Wittbrodt J., Adam D., Malitschek B., Maueler W., Raulf F., Telling A., Robertson S. M., Schartl M. Nature. 1989;341:415–421. doi: 10.1038/341415a0. [DOI] [PubMed] [Google Scholar]
- 39.Muller H. J., Pontecorvo G. Nature. 1940;146:114–122. [Google Scholar]
- 40.Muller H. J. Biol. Rev. Cambridge Philos. Soc. 1939;14:261–280. [Google Scholar]
- 41.Dobzhansky T. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orr H. A. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. Woodbury, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 44.Wach A., Brachat A., Pohlmann R., Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski R. S., Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.