Abstract
Coat components localize to specific membrane domains, where they sort selected transmembrane proteins. To study how clathrin coats are stabilized on such domains and to identify the protein networks involved, we combined proteomic screens and in vitro liposome-based assays that recapitulate the fidelity of protein sorting in vivo. Our study identifying ≈40 proteins on AP-1A-coated liposomes revealed that AP-1A coat assembly triggers the concomitant recruitment of Rac1, its effectors, and the Wave/Scar complex as well as that of Rab11 and Rab14. The coordinated recruitment of these different machineries requires a mosaic of membrane components comprising the GTPase ADP-ribosylation factor 1, sorting signals in selected transmembrane proteins, and phosphatidylinositol 4-phosphate. These results demonstrate that the combinatorial use of low-affinity binding sites present on the same membrane domain accounts not only for a selective coat assembly but also for the coordinated assembly of selected machineries required for actin polymerization and subsequent membrane fusion.
Keywords: membrane traffic, Rab GTPases, actin, sorting signals, phosphatidylinositol phosphate
Coat proteins are key molecular devices that contribute to maintaining the identity of organelles constituting the secretory and endocytic pathways of eukaryotic cells (1, 2). Clathrin coats are involved in cargo sorting between the plasma membrane, the Golgi apparatus, and endosomes. Clathrin associates with adaptor proteins (APs), which interact directly or indirectly with tyrosine- or dileucine-based sorting signals present in cytoplasmic domains of transmembrane proteins. This event leads to the segregation of cargo molecules into transport intermediates (3, 4). Mammals have four APs: whereas AP-2 functions in endocytosis, AP-1A and AP-3 are required for transport between the secretory pathway and the endosomal/lysosomal system (3, 5). The epithelial cell-specific AP-1B (6) and the ubiquitously expressed AP-4 (7) have been implicated in basolateral transport in polarized cells.
AP-1A and AP-1B cannot substitute for each other and distribute to distinct membrane domains of the trans-Golgi network (TGN) and endosomes (8, 9). Together with accessory proteins like Golgi-localized, γ-ear containing, ADP-ribosylation factor (ARF)-binding proteins (10) or phosphofurin acidic cluster-sorting proteins (11), AP-1A mediates the trafficking of transmembrane proteins shuttling between the TGN, endosomes, and the plasma membrane, such as the mannose 6-phosphate receptors (12) or the envelope glycoprotein I (gpI) of the varicella zoster virus (13). AP-3 is located on endosomes and the TGN (14, 15), where it sorts selected transmembrane proteins destined to lysosomes such as the lysosomal membrane glycoproteins Lamp-1, Lamp-2, and LimpII (16–18).
How APs are stabilized on different membrane domains, where they sort selected transmembrane proteins to maintain organelle identities, is not fully understood. ARF-1 and transmembrane proteins play a role in AP-1A and AP-3 coat assembly (16, 19–21). Phosphatidylinositol phosphates (PIPs), whose syntheses are restricted to specific membrane domains, have also emerged as key components regulating AP coat assembly and therefore protein sorting and membrane dynamics (22, 23). Thus, several membrane components can contribute to the stabilization of APs on selected membrane domains. Here we asked how many cytosolic proteins are involved in AP-1A coat assembly and how the combinatorial use of ARF-1, transmembrane proteins, and PIPs contributes to their high-affinity interactions with given membrane domains.
Results
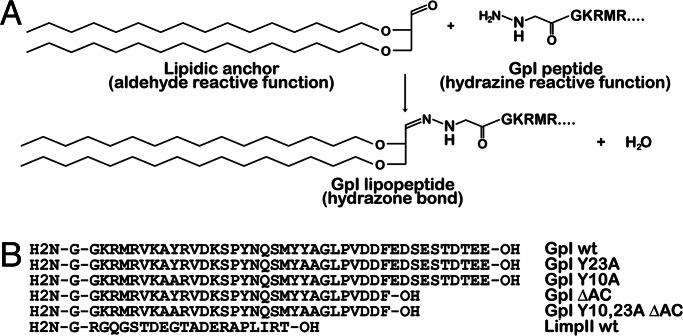
To introduce cytoplasmic domains of transmembrane proteins into liposomes, we took advantage of a lipid anchor with an aldehyde-derivatized head group, which reacts with a hydrazine group present at the N terminus of synthetic peptides. When these peptides were incubated with liposomes containing reactive anchors, a covalent hydrazone linkage was obtained (Fig. 1). In this manner, the peptide variants corresponding to cytoplasmic tails of the gpI envelope glycoprotein of the varicella zoster virus or LimpII could be covalently linked to the liposome surface with similar efficiencies. These liposomes were incubated with brain cytosol under various conditions, and the amount of bound ARF-1, AP-1A, AP-2, AP-3, COP-I, and clathrin was quantified.
Fig. 1.
Coupling of a lipid anchor to synthetic peptides. (A) Chemical reaction for coupling synthetic peptides to a lipid anchor incorporated into liposomes. (B) Amino acid sequences of cytoplasmic domains of transmembrane proteins used in this study. Note that the gpI tail referred here as wild type (wt) contains a truncation of the C terminus devoid of any trafficking signal (13).
AP-1A Binding Requires Selected Transmembrane Proteins and Their Sorting Signals.
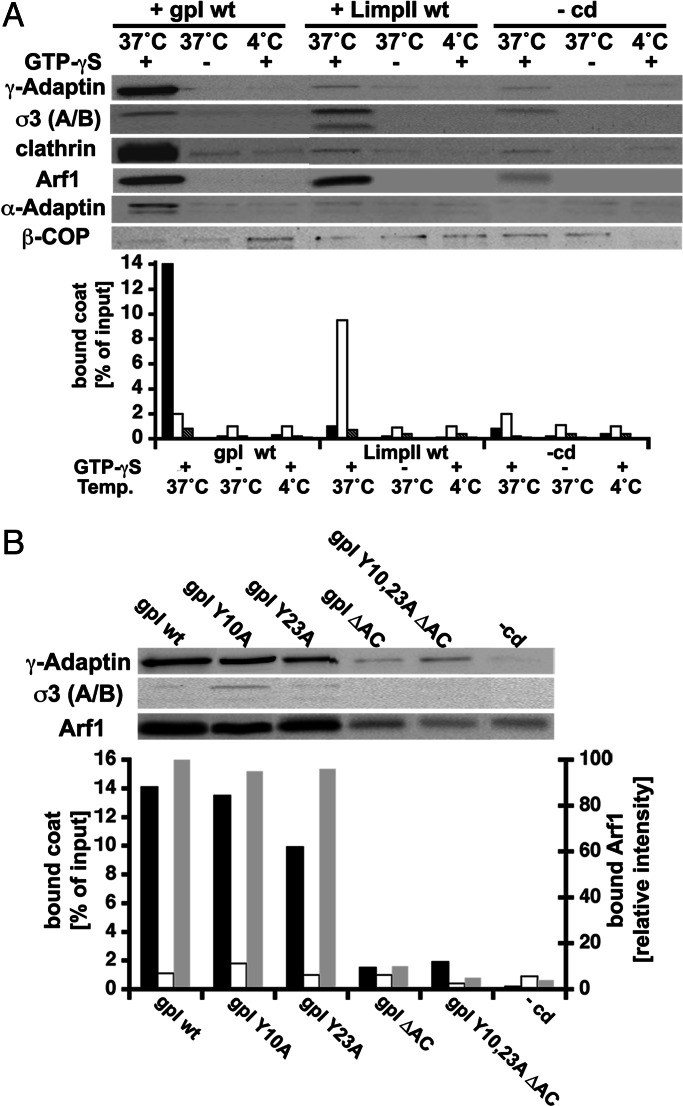
Fig. 2A shows that liposomes alone recruited ARF-1, AP-1A, AP-3, clathrin, and COP-I in the presence of guanosine 5′-[γ-thio]triphosphate (GTP-γS), which stabilizes ARF-1 in an active, membrane-bound conformation, as described (24–26). This recruitment was decreased when the incubation was performed in the presence of GTP or at 4°C, which reduces nucleotide exchange on ARF-1. However, when the same experiment was performed with liposomes exhibiting cytoplasmic domains of transmembrane proteins, AP-1A or AP-3 recruitment was more than 10 times increased and became specific. Thus, gpI-containing liposomes recruited AP-1A whereas little AP-3 binding was observed. In contrast, LimpII-containing liposomes recruited AP-3 but only little AP-1A. Interestingly, clathrin was efficiently recruited when AP-1A was bound onto gpI-containing liposomes but poorly recruited when AP-3 was bound onto LimpII-containing liposomes or on liposomes devoid of cytoplasmic domains. Under all conditions, AP-2 and COP-I binding remained negligible. Similar results were obtained when these experiments were done with ARF-depleted cytosol complemented with recombinant, myristoylated ARF-1 (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
AP-1A binding is cargo-specific and requires intact sorting signals. (A) Liposomes with or without gpI or LimpII wild-type tails were incubated with cytosol in the presence of GTP or GTP-γS at 37°C or at 4°C. After incubation, AP-1A, AP-2, AP-3, COP-I, clathrin, and ARF-1 bound to liposomes were detected after SDS/PAGE and Western blotting using specific antibodies. AP-1A (black bars), AP-3 (white bars), AP-2 (dark gray bars), and COP-I (light gray bars) were quantified. (B) Liposomes with wild-type or mutated (Y10A, Y23A, ΔAC, Y10,23A ΔAC) gpI cytoplasmic domains or with glycine (-cd) were incubated at 37°C with cytosol in the presence of GTP-γS. After incubation, AP-1A, AP-3, and ARF-1 bound to liposomes were detected after SDS/PAGE and Western blotting using specific antibodies. AP-1A (black bars), AP-3 (white bars), and ARF-1 (gray bars) were quantified. The charts represent the typical results obtained in four independent experiments.
gpI trafficking requires a dominant tyrosine-based sorting signal (Tyr-23), a weaker tyrosine-based sorting signal (Tyr-10), and an acidic cluster in its cytoplasmic tail (13). Fig. 2B shows that AP-1A was no longer recruited on liposomes when all sorting motifs of gpI were mutated or deleted. AP-1A recruitment onto liposomes with gpI tails mutated on a single tyrosine-based sorting motif at position Tyr-10 or Tyr-23 was only partially inhibited (10% and 40% reduction, respectively). However, when the acidic cluster was absent, AP-1A recruitment was reduced by 90%, giving similar values as those obtained with gpI tails devoid of trafficking signals. In contrast, serine phosphorylation of acidic clusters by recombinant casein kinase II was found to enhance AP-1A binding by 40% (Fig. 7, which is published as supporting information on the PNAS web site). These experiments demonstrate that an efficient binding of AP-1A on liposomes requires both ARF-1 and intact sorting signals present in selected transmembrane proteins like gpI.
AP-1A Binding Is Enhanced in the Presence of Phosphatidylinositol 4-Phosphate (PI-4P).
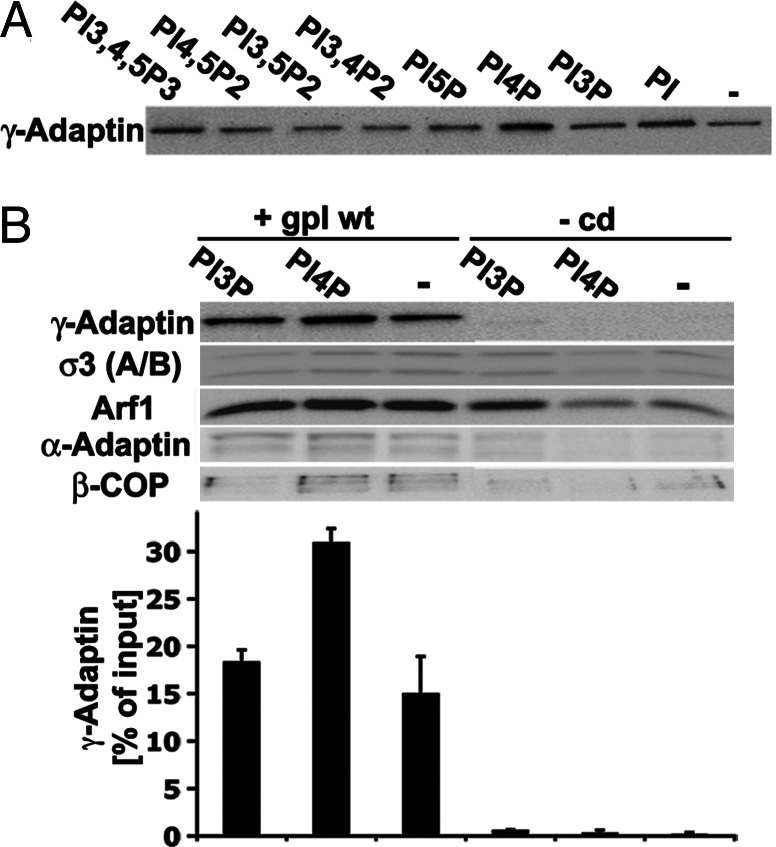
Liposomes containing gpI and different PIPs were then analyzed for their ability to recruit AP-1A. These liposomes were incubated with GTP-γS and sublimiting concentrations of cytosol (3 mg/ml) so that changes in AP-1A affinities could be evaluated by measuring an increase in AP-1A binding by Western blotting. Fig. 3 shows that AP-1A binding was two times increased when PI-4P was present in gpI-containing liposomes whereas the recruitment of AP-2, AP-3, and COP-I remained at background levels. No AP-1A binding was observed on PI-4P-rich liposomes devoid of gpI. The other PIPs tested had no significant effect. Under optimal conditions, ≈30% of cytosolic AP-1A was recruited onto gpI-containing liposomes. Because PI-4P does not induce AP-1A binding on liposomes devoid of gpI, we conclude that PI-4P strengthens the interactions of AP-1A with sorting signals present in gpI.
Fig. 3.
AP-1A binding and phosphoinositides. (A) gpI-liposomes containing various PIPs were incubated with cytosol (3 mg/ml) and GTP-γS. (B) Phosphatidylinositol 3-phosphate, PI-4P, or no PIP was added to liposomes with either gpI or glycine (-cd) and then incubated with cytosol (3 mg/ml) in the presence of GTP-γS. AP-1A, AP-2, AP-3, COP-I, and ARF-1 bound to liposomes were quantified after SDS/PAGE and Western blotting.
Proteomic Analysis of AP-1A-Coated Liposomes.
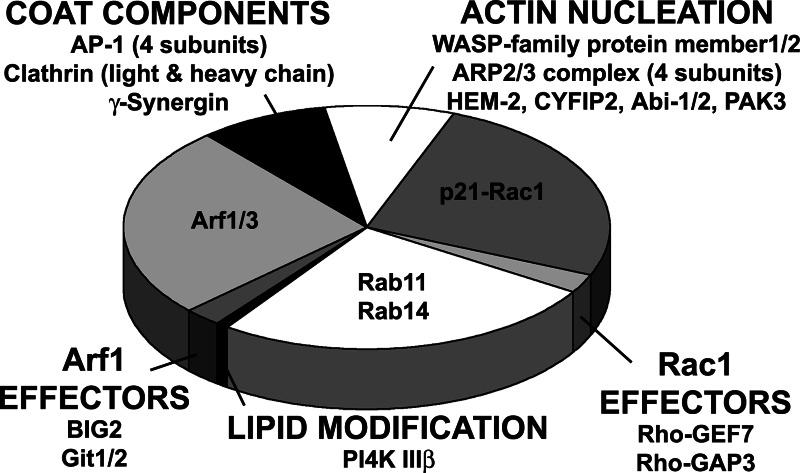
We examined AP-1A-coated liposomes by electron microscopy. These liposomes showed large clathrin lattices as well as still-attached clathrin-coated buds (Fig. 8, which is published as supporting information on the PNAS web site). No clathrin coat was detected on liposomes containing gpI devoid of sorting signals. We analyzed the protein content of AP-1A-coated liposomes by SDS/PAGE, which revealed that >20 major proteins, identified by MALDI-TOF/TOF MS, were specifically recruited (Fig. 8). In addition, the lanes were cut into 50 slices, which were analyzed by LC-MS/MS to detect minor proteins. Both analyses identified ≈40 proteins recruited along with AP-1A (Fig. 4 and Table 1, which is published as supporting information on the PNAS web site), and they were classified into three major groups. First, coat components, i.e., clathrin heavy and light chains, the AP-1A subunits, and γ-synergin, were identified, as well as ARF-1/ARF-3 (ARF-1 and ARF-3 could not be distinguished by MS), with its brefeldin A-inhibited exchange factor 2 (Big2) and its GTPase-activating proteins Git1 and Git2. Arfaptin 1 and 2, which equally bind to ARF-1 and Rac-1, were also present. Second, the MS analysis identified a Rac1-dependent actin nucleation module, i.e., the different subunits of the Wave/Scar complex, including CYFIP2, Nck-associated protein 1 (HEM-2), Abi-1 and 2, as well as members 1 and 3 of the Wasp family, which activate the ARP2/3 complex that was also detected. Rac1, as well as its exchange factor Rho-GEF 7 and its GTPase activating protein Rho-GAP3 (also called Wave-associated Rac-GAP), was identified. Third, two major Rab GTPases, Rab11 and Rab14, were detected. Finally, the phosphatidylinositol 4-kinase (PI-4-kinase) IIIβ and the p21-activativated serine/threonine kinase PAK3, known to interact with Rho-GEF7 or PIX (27), were recovered on gpI/PI-4P-containing liposomes. Coomassie blue staining indicates that one clathrin triskelion, one AP-1A, and one Wave/Scar complex were found together with approximately three ARF-1, approximately three Rac1, and approximately three Rab11/Rab14. The different GTPase effectors were found in substoichiometric amounts. Thus, machineries controlling AP-1A coat formation, actin nucleation, and membrane fusion are selectively recruited on gpI/PI-4P-containing liposomes. The recruitment of the different protein machineries was drastically reduced on liposomes devoid of gpI and PI-4P. Contamination by AP-2 was estimated to be <5% in mass when compared with AP-1A. The contamination by other coats was below this value.
Fig. 4.
Proteomics of AP-1A-coated liposomes. Proteins bound to liposomes were fractionated by SDS/PAGE and identified by MALDI-TOF and LC-MS/MS spectrometry. The surface areas on the diagram reflect an estimated stoichiometry between the different machineries (taking AP-1-γ and CYFIP2 as markers) and GTPases obtained after Coomassie blue staining.
AP-1A Coat Assembly Stabilizes Machineries Required for Actin Nucleation and Membrane Fusion.
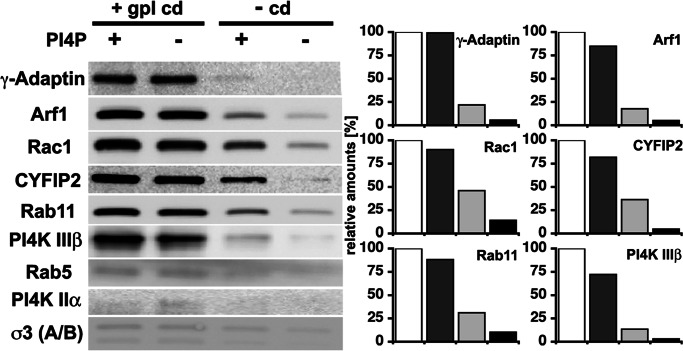
gpI cytoplasmic domains influence not only AP-1A recruitment but also that of ARF-1. ARF-1 recruitment was 20-fold increased in the presence of intact sorting signals in gpI, in particular acidic clusters (Figs. 2 and 5). This recruitment was saturable and maximal at ≈500 nM gpI (Fig. 9, which is published as supporting information on the PNAS web site). This effect was not observed when liposomes contained only PI-4P (Figs. 3 and 5). We then asked whether the recruitment of the different machineries identified in our proteomic screen is influenced by gpI and PI-4P. Fig. 5 shows that gpI also stabilizes the membrane association of Rac1, CYFIP2 taken as a marker of the Wave/Scar complex, Rab11, and PI-4-kinase IIIβ. Although PI-4P alone can stabilize to some degree the different components on membranes, the most efficient recruitment was observed in the presence of both gpI and PI-4P. In contrast, AP-3, Rab5, and PI-4-kinase IIα were not recruited. Thus, gpI not only stabilizes ARF-1 and AP-1A but also contributes to stabilizing machineries required for actin nucleation and membrane fusion.
Fig. 5.
AP-1A coat assembly stabilizes machineries required for actin nucleation and membrane fusion. Liposomes with or without gpI and with or without PI-4P were incubated with cytosol and GTP-γS. After incubation and purification by flotation gradients, they were analyzed by Western blotting using specific antibodies as indicated. The recruitment of these markers was quantified (white bar, gpI and PI-4P; dark gray bar, gpI no PI-4P; light gray bar, PI-4P and no gpI; black bar, no gpI and no PI-4P).
Discussion
AP-1A coat assembly on synthetic membranes triggers the concomitant recruitment of the Rac1-dependent Wave/Scar complex necessary for actin nucleation and Rab11/Rab14 required for membrane fusion. The coordinated recruitment of these different machineries requires the combinatorial use of low-affinity membrane components, minimally composed of ARF-1, selected transmembrane proteins providing specificity, and PI-4P strengthening affinities of AP-1A for selected transmembrane proteins. Our findings strongly suggest that these components, when present on the same membrane domain, provide parts of the molecular networks needed for the recruitment of interconnected cellular machineries. Such interaction networks may provide parts of the mechanisms required for establishing organelle identity and dynamics.
AP-1A Coat Assembly, Actin Nucleation, and Membrane Fusion.
Our proteomic screen allowed us to identify ≈40 components involved in early stages of AP-1A coat assembly stabilized by GTP-γS. They include not only the expected clathrin chains, AP-1A subunits, and ARF-1/ARF-3, but also γ-synergin, the ARF-1-GEF Big2, two ARF-GAPs (Git1 and Git2), and Arfaptin 1 and 2, two ARF-1-interacting proteins that also interact with Rac1 (28). Big2 localizes to the TGN and Rab11-positive recycling endosomes and regulates the structural integrity of recycling endosomes (29). Git1, which localizes to Rab11-positive membranes (30), and Git2 have a similar GAP activity for ARF-1 and ARF-6 in vitro (31). However, ARF-6 was not detected in our proteomic screen, and therefore we believe that the recovery of Git1 and Git2 on gpI/PI-4P-containing liposomes is ARF-1/ARF-3-mediated. Besides γ-synergin, we could not detect additional AP-1A-interacting proteins such as phosphofurin acidic cluster-sorting proteins, Golgi-localized, γ-ear containing, ARF-binding proteins, or EpsinR, probably because of the absence of appropriate transmembrane proteins or soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) on the used liposomes. Similarly, we did not detect dynamin, which could function during late stages of AP-1A-coated transport intermediate formation.
Our proteomic screen also revealed the presence of a Rac1-dependent actin nucleation system. Rac1 controls the recruitment of the Wave/Scar complex, which activates the ARP2/3 complex and therefore relays Rac1 signaling to actin nucleation (32). We identified the different subunits of the Wave/Scar and ARP2/3 complexes. Rac1, its GTPase-activating protein Rho-GAP3, a Wave-associated GAP, and its exchange factor Rho-GEF7, also known as p21-activated serine/threonine kinase PAK-interacting exchange factor, were also identified. Rho-GEF7 can interact with the ARF-1-GAP Git1 (33), also named APP-1 (34). Although Git1 has been linked to ARF-6, actin organization, and assembly of focal adhesions at the cell surface, Git1/AAP1 is detected mostly on Rab11-rich membranes (30). Therefore, such interactions between the Rac1-GEF and Git1 could couple AP-1A coat assembly and actin nucleation, a possibility that can now be tested. Actin dynamics has been linked with AP-2-dependent endocytosis. It is still unclear how actin polymerization contributes to AP-1A-dependent transport. It could provide forces required for clustering membrane-bound AP-1A in the same membrane domains or for budding AP-1A-coated transport intermediates or both. It will be important to address these issues, because actin depolymerization prevents the formation of mannose 6-phosphate receptor-containing, TGN-derived transport intermediates in vivo (35).
Our proteomic screen revealed that two major Rab GTPases, Rab11 and Rab14, are found on AP-1A-coated liposomes. Rab14 has been shown to control protein trafficking between the TGN and endosomes (36), and Rab11 localizes to the TGN and recycling endosomes, where it controls the dynamics of this latter compartment (37). Both AP-1A and AP-1B have been found on regions of the TGN and recycling endosomes (9). The reason why both Rab11 and Rab14 are equally recovered together with AP-1A is actually not clear. However, their presence provides clues on the transport pathways controlled by AP-1A and highlights the potential importance of recycling endosomes in connecting the secretory and the endocytic pathways. The molecular links between Rabs, AP-1A coats, and actin nucleation machineries are not yet identified. It is possible that GAP domain-containing proteins with unknown functions found in our screen (data not shown) provide the molecular links coupling these Rabs with the other machineries.
A Combination of Membrane Components Stabilizes AP-1A Coats and Associated Machineries.
The high-affinity interactions of AP-1A and its associated machineries require the combinatorial use of several low-affinity components, namely ARF-1, sorting signals in selected transmembrane proteins like gpI, and PI-4P. The production of PI-4P is restricted to specific membrane domains, whereas ARF-1 and gpI are found in several membrane domains. Thus, PI-4P contributes to restrict high-affinity interactions of AP-1A on specific domains containing both ARF-1 and gpI. PI-4P regulates protein sorting in the TGN, in particular AP-1A and EpsinR binding (38, 39). Our data suggest that the affinity of AP-1 for sorting signals in selected transmembrane proteins is increased in the presence of PI-4P. A similar mechanism has been found for AP-2 and phosphatidylinositol (4,5)-bisphosphate at the plasma membrane (40). The distribution of AP-1A also supports the notion that AP-1A can be found on PI-4P-rich membranes. The production of PI-4P on the TGN is controlled by ARF-1, which recruits the PI-4-kinase IIIβ (22). Because membrane association of ARF-1 is increased by gpI, it is possible that specific transmembrane proteins could indirectly favor the production of PI-4P, a notion consistent with the recovery of PI-4-kinase IIIβ on AP-1A-coated liposomes. Such an amplification loop could expand PI-4P- and ARF-1-rich membrane domains, thereby ensuring an efficient AP-1A coat assembly for subsequent rounds of protein sorting. This amplification loop could also indirectly favor an efficient actin nucleation and subsequent membrane fusion. It has been proposed that PI-4-kinase IIIβ is critical for the functional association of Rab11 (41), a major GTPase found together with Rab14 on AP-1A-coated liposomes. Such a GTPase-controlled amplification loop has been proposed in the context of Rab5, which stimulates phosphatidylinositol-3-phosphate synthesis on early endosomes, thereby enhancing its own stabilization on membranes by concomitant interactions with effectors interacting with both Rab5 and phosphatidylinositol-3-phosphate (23).
It has been reported that AP-1 binding on liposomes requires ARF-1, Lamp-1, phosphatidylinositol (4,5)-bisphosphate (42), and ARF-GAP1 (43), an ARF-GAP also involved in COP-I dynamics (44). Our study and others (38, 39) show that AP-1A binding requires PI-4P, probably produced by the PI-4-kinase IIIβ, and most likely the ARF-1-GAP Git1 and possibly Git2. It has been proposed that transmembrane proteins are captured by APs only after their ARF-1-dependent binding to membranes (24). Our findings showing that a mosaic of components is required for high-affinity AP-1A binding agree with our previous studies illustrating the importance of sorting signals in transmembrane proteins (16, 19). The simplest explanation for any apparent discrepancy is that AP-1A can be recruited onto membranes in an ARF-1-dependent and transmembrane protein- or PI-4P-independent process. Such interactions would be rather weak, and AP-1A coat assembly would be followed by a rapid disassembly. However, specific transmembrane proteins encountering proper PIP-rich domains could stabilize such ARF-1-dependent transient assemblies. Studies on AP-1A coat dynamics may provide some support to this hypothesis. Dynamic studies of AP-2- and clathrin-dependent endocytosis have proposed a model in which coated pits initiate randomly at the plasma membrane but collapse unless stabilized, perhaps by cargo capture (45).
An interesting point is that our study does not recapitulate AP-2 recruitment on liposomes with transmembrane proteins like gpI that are endocytosed as part of their recycling pathway (13). This observation could reflect a lower affinity of AP-2 for sorting signals or the absence of other key membrane components in our liposomes (GTPases, PIPs, or other lipids or proteins) that are critical for AP-2 binding. Thus, our study may highlight only a part of the components required for efficient AP coat assembly.
Trafficking Pathways Controlled by AP-1A.
The precise function of AP-1A is still unclear. Although AP-1A cooperates with Golgi-localized, γ-ear containing, ARF-binding proteins for TGN-to-endosome transport (46), knockout experiments have suggested that AP-1A functions in endosome-to-TGN transport (47, 48). PI-4P, produced on TGN membranes, enhances the interactions of AP-1A and associated machineries, in particular those of Rab11 and Rab14, with membranes containing ARF-1 and selected transmembrane proteins. Without excluding the potential importance of AP-1A in recycling pathways, our finding argues that AP-1A functions efficiently in TGN-to-endosome transport and may highlight Rab11-positive recycling endosomes in sorting events controlled by AP-1A. In the absence of PI-4P, the interactions would be weaker, allowing AP-1A dissociation, thereby leaving a partial access of transmembrane proteins to the cell surface, as expected for mannose 6-phosphate receptors and gpI, perhaps from Rab11-positive recycling endosomes. Thus, the binding properties of AP-1A, as shown here with synthetic membrane domains, could partly explain the trafficking pathways of the different transmembrane proteins it recognizes.
In vitro reconstitution systems have been instrumental to investigate key aspects of membrane traffic (49–51). We have now paved the way for approaches combining proteomic screens and liposome-based reconstitution systems. It is clear that some putative molecular interactions uncovered in our screen of AP-1A-coated liposomes need to be clarified. This approach provides a means to illuminate the precise roles and targets of coat proteins and to identify associated cellular machineries. It also allows dissecting the mechanisms by which the assembly of these supramolecular structures is coordinated, in particular to understand how the GTP cycles on ARF-1, Rac1, and Rab11/Rab14 are coordinated, presumably by interactions between their effectors. Such studies could contribute to understanding how organelle identities are maintained.
Materials and Methods
Chemicals and Antibodies.
Chemicals were purchased from Sigma, lipids were from Avanti Polar Lipids, and phosphoinositides were from Echelon Biosciences. The antibodies used were as follows: polyclonal antibodies against the AP-3 σ-subunit (16), Rab5 (gift of M. Zerial, Max Planck Institute of Molecular Cell Biology and Genetics), PI-4-kinase IIα and PI-4-kinase IIIβ (gifts of A. De Matteis, Consorzio Mario Negri Sud, Abruzzo, Italy), and Rab11 (United States Biological); monoclonal antibodies against ARF-1 (1D9, Dianova), COP-I β-subunit (maD, Sigma), AP-1 γ-subunit (100/3, Sigma), AP-2 α-subunit (100/2, Sigma), clathrin heavy chain (Transduction Laboratories), Rac1 (Cytoskeleton), and CYFIP2 (gift of A. Schenck, Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France).
Peptides, Lipid Anchor, and Liposomes.
Lipid anchors and hydrazino peptides were synthesized as described (52). Peptides were dissolved in buffer A (15.4 mM citric acid/69.2 mM dibasic sodium phosphate, pH 6.4). A mixture of phosphatidylcholine/phosphatidylethanolamine/phosphatidylserine/cholesterol/anchor (40:30:10:10:10 molar ratio) in chloroform/methanol (phosphoinositides in chloroform/methanol were added as 1% molar ratio when indicated) was evaporated to dryness and resuspended in buffer A. Unilamellar liposomes were formed and sized to 400 nm. Peptides were coupled to liposomes, and unbound peptides were removed by gel filtration in buffer B (25 mM Hepes-KOH, pH 7.2/125 mM potassium acetate/2.5 mM magnesium acetate/1 mM DTT). Glycine was used as a control. In standard assays, 10 μl of liposomes was used, corresponding to ≈10 nmol of lipid and 0.5 nmol of peptide per reaction.
Coat Recruitment Assay.
Pig brain cytosol was prepared as described (19). Assays were performed in a total volume of 200 μl in buffer B containing cytosol (10 mg/ml), liposomes (0.5 nmol of peptide per reaction), GTP (1 mM), or GTP-γS (0.1 mM). Binding reactions were initiated by transfer to 37°C. After 20 min, membranes were recovered by centrifugation and washed with buffer B. Pellets were analyzed by SDS/PAGE followed by Western blotting using relevant antibodies. For MS analyses, gpI/PI-4P-containing lipososomes were incubated with mouse brain cytosol and GTP-γS, purified by flotation on sucrose density gradients, and concentrated by centrifugation. Bound proteins were separated by SDS/PAGE.
Protein Identification by MS.
Sample preparation, in-gel digestion, peptide extraction, and spectra acquisition were performed as described (53). In brief, MALDI-MS measurements were performed by using an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) in reflectron mode by using α-cyano-4-hydroxycinnamic acid as a matrix. Spectra were processed by using flexanalysis software. LC-MS/MS experiments were performed on a quadrupole orthogonal acceleration time-of-flight mass spectrometer Q-TOF Ultima (Micromass, Manchester, U.K.) equipped with a Z-spray nanoelectrospray source. Peptides were separated as described (53). Protein identification, by both peptide mass fingerprinting and fragment ion analysis, was performed by using mascot (Matrix Science). Search criteria were as follows: taxonomy, mouse; mass accuracy, 50 ppm for peptide mass fingerprinting and 0.5 Da for fragment analysis; modifications, carbamidomethylation and methionine oxidation; maximum of one missed cleavage site. The National Center for Biotechnology Information nonredundant protein database (version 20050623, 2,564,994 sequences) and SwissProt (version 170205, 172,233 sequences) were searched.
Electron Microscopy.
AP-1A-coated liposomes were recovered by centrifugation. Membrane pellets were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) and then postfixed with 1% osmium tetroxide. Membranes were then embedded in Epon. Thin sections were contrasted with uranyl acetate and lead citrate.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for helpful discussion and M. Zerial for critical reading of the manuscript. We are grateful to A. Blanpain for synthesizing the hydrazino peptides; M. Wilsch-Breuninger for performing the EM analysis; and A. De Matteis, A. Schenck, and J.-L. Mandel (Institute of Genetics and Molecular and Cellular Biology) for their gifts of antibodies. This work was supported in part by grants from Dresden University of Technology (HWP-1207), Sächsisches Ministerium für Wissenschaft und Kunst–Europäischer Fond für Regionale Entwicklung 1203, and Deutsche Forschungsgemeinschaft (Transregio 6031).
Abbreviations
- TGN
trans-Golgi network
- AP
adaptor protein
- PIP
phosphatidylinositol phosphate
- ARF
ADP-ribosylation factor
- gpI
glycoprotein I
- GTP-γS
guanosine 5′-[γ-thio]triphosphate
- PI-4P
phosphatidylinositol 4-phosphate
- PI-4-kinase
phosphatidylinositol 4-kinase
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kirchhausen T. Nat. Rev. Mol. Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 2.Antonny B., Schekman R. Curr. Opin. Cell Biol. 2001;13:438–443. doi: 10.1016/s0955-0674(00)00234-9. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino J. S., Traub L. M. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 4.Owen D. J., Collins B. M., Evans P. R. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 5.Robinson M. S. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Folsch H., Ohno H., Bonifacino J. S., Mellman I. Cell. 1999;99:189–198. doi: 10.1016/s0092-8674(00)81650-5. [DOI] [PubMed] [Google Scholar]
- 7.Simmen T., Honing S., Icking A., Tikkanen R., Hunziker W. Nat. Cell Biol. 2002;4:154–159. doi: 10.1038/ncb745. [DOI] [PubMed] [Google Scholar]
- 8.Ang A. L., Taguchi T., Francis S., Folsch H., Murrells L. J., Pypaert M., Warren G., Mellman I. J. Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folsch H., Pypaert M., Maday S., Pelletier L., Mellman I. J. Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonifacino J. S. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 11.Wan L., Molloy S. S., Thomas L., Liu G., Xiang Y., Rybak S. L., Thomas G. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh P., Dahms N. M., Kornfeld S. Nat. Rev. Mol. Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 13.Alconada A., Bauer U., Hoflack B. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 14.Dell’Angelica E. C., Ohno H., Ooi C. E., Rabinovich E., Roche K. W., Bonifacino J. S. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Borgne R., Alconada A., Bauer U., Hoflack B. J. Biol. Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- 17.Dell’Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S. Mol. Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 18.Feng L., Seymour A. B., Jiang S., To A., Peden A. A., Novak E. K., Zhen L., Rusiniak M. E., Eicher E. M., Robinson M. S., et al. Hum. Mol. Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- 19.Le Borgne R., Griffiths G., Hoflack B. J. Biol. Chem. 1996;271:2162–2170. doi: 10.1074/jbc.271.4.2162. [DOI] [PubMed] [Google Scholar]
- 20.Stamnes M. A., Rothman J. E. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- 21.Ooi C. E., Dell’Angelica E. C., Bonifacino J. S. J. Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Matteis M. A., Godi A. Nat. Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 23.Zerial M., McBride H. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y., Traub L. M., Kornfeld S. Mol. Biol. Cell. 1999;10:537–549. doi: 10.1091/mbc.10.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake M. T., Zhu Y., Kornfeld S. Mol. Biol. Cell. 2000;11:3723–3736. doi: 10.1091/mbc.11.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spang A., Matsuoka K., Hamamoto S., Schekman R., Orci L. Proc. Natl. Acad. Sci. USA. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manser E., Leung T., Lim L. Methods Mol. Biol. 1998;84:295–305. doi: 10.1385/0-89603-488-7:295. [DOI] [PubMed] [Google Scholar]
- 28.Tarricone C., Xiao B., Justin N., Walker P. A., Rittinger K., Gamblin S. J., Smerdon S. J. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- 29.Shin H. W., Morinaga N., Noda M., Nakayama K. Mol. Biol. Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matafora V., Paris S., Dariozzi S., de Curtis I. J. Cell Sci. 2001;114:4509–4520. doi: 10.1242/jcs.114.24.4509. [DOI] [PubMed] [Google Scholar]
- 31.Vitale N., Patton W. A., Moss J., Vaughan M., Lefkowitz R. J., Premont R. T. J. Biol. Chem. 2000;275:13901–13906. doi: 10.1074/jbc.275.18.13901. [DOI] [PubMed] [Google Scholar]
- 32.Stradal T. E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Z. S., Manser E., Loo T. H., Lim L. Mol. Cell. Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Cesare A., Paris S., Albertinazzi C., Dariozzi S., Andersen J., Mann M., Longhi R., de Curtis I. Nat. Cell Biol. 2000;2:521–530. doi: 10.1038/35019561. [DOI] [PubMed] [Google Scholar]
- 35.Waguri S., Dewitte F., Le Borgne R., Rouille Y., Uchiyama Y., Dubremetz J. F., Hoflack B. Mol. Biol. Cell. 2003;14:142–155. doi: 10.1091/mbc.E02-06-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junutula J. R., De Maziere A. M., Peden A. A., Ervin K. E., Advani R. J., van Dijk S. M., Klumperman J., Scheller R. H. Mol. Biol. Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R. G. J. Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 39.Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., McMahon H. T. J. Cell Biol. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 41.de Graaf P., Zwart W. T., van Dijken R. A., Deneka M., Schulz T. K., Geijsen N., Coffer P. J., Gadella B. M., Verkleij A. J., van der Sluijs P., van Bergen en Henegouwen P. M. Mol. Biol. Cell. 2004;15:2038–2047. doi: 10.1091/mbc.E03-12-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crottet P., Meyer D. M., Rohrer J., Spiess M. Mol. Biol. Cell. 2002;13:3672–3682. doi: 10.1091/mbc.E02-05-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer D. M., Crottet P., Maco B., Degtyar E., Cassel D., Spiess M. Mol. Biol. Cell. 2005;16:4745–4754. doi: 10.1091/mbc.E05-06-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W., Duden R., Phair R. D., Lippincott-Schwartz J. J. Cell Biol. 2005;168:1053–1063. doi: 10.1083/jcb.200410142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M. L., Kirchhausen T. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 47.Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 48.Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C. A., Sollner T. H., Rothman J. E., Wieland F. T. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 50.Matsuoka K., Orci L., Amherdt M., Bednarek S. Y., Hamamoto S., Schekman R., Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y., Traub L. M., Kornfeld S. Mol. Biol. Cell. 1998;9:1323–1337. doi: 10.1091/mbc.9.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bourel-Bonnet L., Pecheur E. I., Grandjean C., Blanpain A., Baust T., Melnyk O., Hoflack B., Gras-Masse H. Bioconjug. Chem. 2005;16:450–457. doi: 10.1021/bc049908v. [DOI] [PubMed] [Google Scholar]
- 53.Czupalla C., Mansukoski H., Pursche T., Krause E., Hoflack B. Proteomics. 2005;5:3868–3875. doi: 10.1002/pmic.200402059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.