Abstract
Engaging mammalian Toll-like receptors (TLRs) activate both the NF-κB and mitogen-activated protein kinase signaling pathways. Here we establish that mitogen-activated protein 3 kinase Tpl2, levels of which are markedly reduced in nfkb1−/− cells, is required for extracellular signal-regulated kinase (ERK) activation in bone marrow-derived macrophages and B cells stimulated with diverse TLR ligands. Despite rescuing TLR-dependent ERK activation in nfkb1−/− bone marrow-derived macrophages by using an estrogen receptor-regulated version of the mitogen-activated protein 3 kinase, c-Raf (Raf:ER), CpG or LPS induction of IL-10 was only partially restored in nfkb1−/− cells expressing Raf:ER, a finding consistent with NF-κB1 regulating IL-10 by a combination of ERK-independent and -dependent mechanisms. Collectively, our findings indicate that the Tpl2/MEK/ERK signaling module is a master regulator of ERK-dependent gene expression downstream of TLRs in different hemopoietic cells.
Keywords: mitogen-activated protein kinase, Rel/NF-kB
Toll-like receptors (TLRs) comprise a family of transmembrane proteins that recognize and bind different microbial components termed pathogen-associated molecular patterns (1). A universal outcome of engaging all TLRs is the activation of the Rel/NF-κB and the mitogen-activated protein kinase (MAPK) signaling pathways (2). Collectively, MAPKs comprise the extracellular signal regulated kinases (ERKs) and stress-activated protein kinases, the latter including c-Jun-N-terminal kinases (JNKs) and p38. Among the substrates phosphorylated by MAPKs in response to TLR signals are transcription factors required for the expression of genes encoding proinflammatory cytokines and immune mediators. Importantly, in the context of TLR-induced transcription, certain genes depend on the interplay of NF-κB and MAPK-regulated transcription factors (3–6).
A key step in TLR-dependent NF-κB and MAPK activation involves engaging mitogen-activated protein 3-kinases (MAP3Ks). One MAP3K, TAK1, is required for the activation of NF-κB and MAPKs (7–9). NF-κB transcription factors are retained as latent complexes in the cytoplasm by IκB proteins. In response to TLR engagement, TAK1 activation of the IκB kinase complex results in IκB phosphorylation, targeting it for ubiquitin-dependent proteasome-mediated degradation (7, 8). This event liberates NF-κB dimers for translocation to the nucleus (10). TAK1 activates JNKs and p38 via the kinase intermediates MKK3 and MKK6 (2, 8). In addition to TAK1, TLR4 signals have been shown to engage another MAP3K, Tpl2 or Cot (11). Tpl2 was first identified as a target for proviral integration in retrovirus-induced T cell lymphomas and mammary carcinomas (12–14). Enforced overexpression of Tpl2 was subsequently shown to activate ERK, JNK, and p38 and the transcription factors NF-AT and NF-κB (15–17). However, recent physiologically relevant studies using tpl2−/− mice have established that in LPS-stimulated macrophages, Tpl2 is required for ERK, but not JNK or p38 activation (3, 11).
In certain cell lines, Tpl2 was shown to exist in a cytoplasmic complex with p105, the precursor of the p50 NF-κB1 transcription factor (18). This interaction, mediated through the C termini of both proteins, is essential for stabilizing Tpl2 (18, 19). LPS stimulation of wild-type macrophages, which results in Tpl2 activation and dissociation from p105, leads to MEK1-dependent ERK activation (19). Because of rapid turnover of Tpl2 in the absence of p105 NF-κB1, nfkb1−/− macrophages like those from tpl2−/− mice fail to activate ERK in response to LPS and, as a consequence, are deficient in the production of proinflammatory mediators such as TNF-α and Cox2 that depend on ERK-regulated transcription factors for their expression (3, 11, 19).
The inability of nfkb1−/− macrophages to activate ERK when stimulated with the TLR4 ligand LPS (19) prompted us to determine whether diverse TLRs use the p105/Tpl2 complex for ERK activation or it is specific to TLR4. Here we report that ERK is activated via the Tpl2/MEK1 pathway in bone marrow-derived macrophages (BMDMs) in response to engaging TLR2, TLR3, TLR7, or TLR9 but fails to be phosphorylated in nfkb1−/− BMDMs stimulated with ligands for these TLRs. Furthermore, B cells, but not keratinocytes from nfkb1−/− mice, were also defective in ERK activation when stimulated with LPS or CpG. Coupled with the finding that Tpl2 expression was absent in normal keratinocytes indicated the requirement for TLR-dependent ERK activation may be cell type-specific. Consistent with TLR-mediated IL-10 expression being dependent on ERK-regulated transcription factors (20), TLR activated nfkb1−/− BMDMs make significantly less IL-10. Although a conditionally active version of the oncogenic MAP3K c-Raf could bypass the TLR-dependent ERK activation defect in nfkb1−/− macrophages, IL-10 expression was not restored to wild-type levels, revealing a hitherto unknown ERK-independent role for NF-κB1 in IL-10 expression. Collectively, our findings indicate that in macrophages and B cells, Tpl2 is an essential mediator of ERK activation in response to diverse TLR signals.
Results
ERK Activation in nfkb1−/− BMDMs Is Defective in Response to TLR2, TLR3, TLR7, and TLR9 Stimuli.
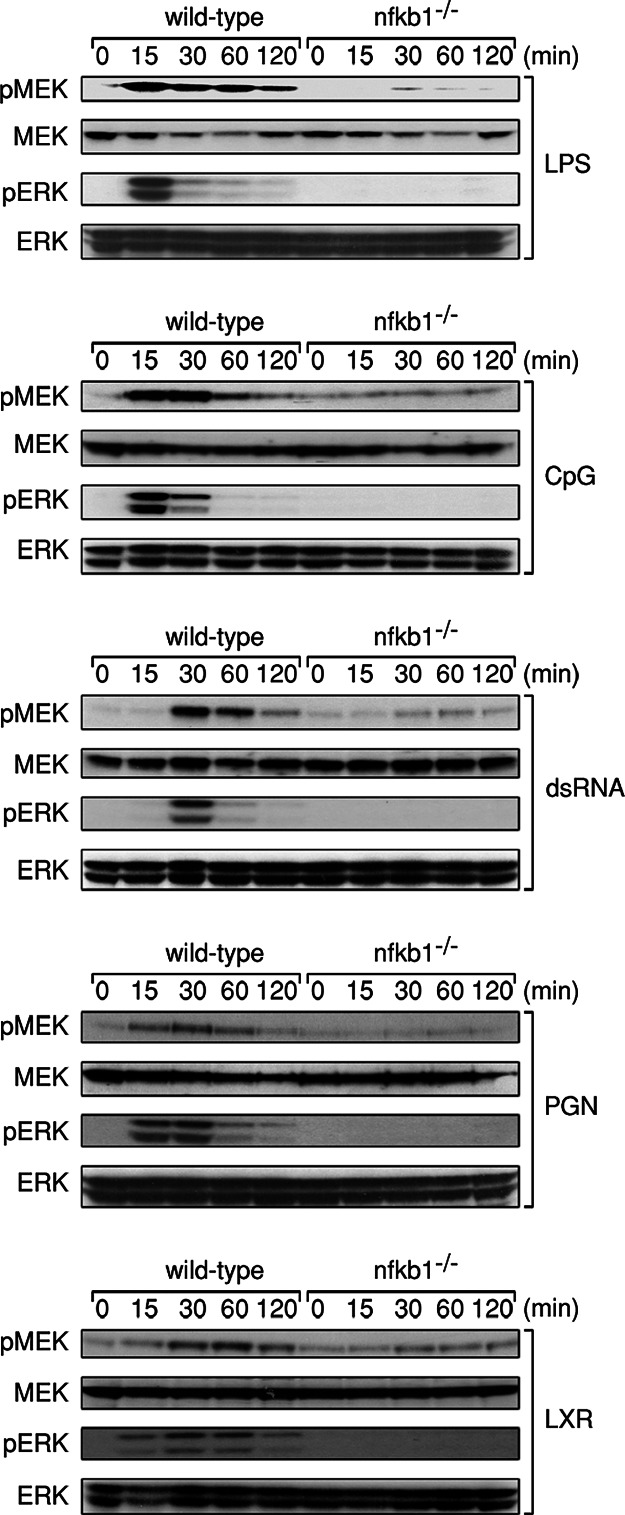
The ERK activation defect in LPS-treated nfkb1−/− BMDMs, which coincides with markedly reduced levels of Tpl2, prompted us to determine whether ERK phosphorylation normally induced in wild-type BMDMs by other TLR ligands was also defective in these cells. Wild-type and nfkb1−/− BMDMs were treated with peptidoglycan (PGN), double-stranded RNA (dsRNA), LPS, loxoribine (LXR), and CpG DNA, ligands for TLR2, TLR3, TLR4, TLR7, and TLR9, respectively. Cells of both genotypes were stimulated for various times over a 2-h period, lysed, and immunoblotting performed with phospho-ERK-specific antibodies (Fig. 1). Consistent with previous reports, all TLR ligands activated ERK in wild-type macrophages, albeit with different activation kinetics and signal duration. In sharp contrast to normal BMDMs and as shown previously for the TLR4 ligand LPS (19), none of the other TLR ligands examined were able to induce detectable levels of phospho-ERK in nfkb1−/− BMDMs (Fig. 1).
Fig. 1.
ERK activation is defective in nfkb1−/− BMDMs stimulated with diverse TLR ligands. Wild-type and nfkb1−/− BMDMs were treated with LPS, CpG DNA, dsRNA, PGN, or loxoribine (LXR) for the indicated times, and total cell lysates were subjected to immunoblot analysis of phospho-MEK, phospho-ERK, MEK, and ERK. This data are representative of three independent experiments.
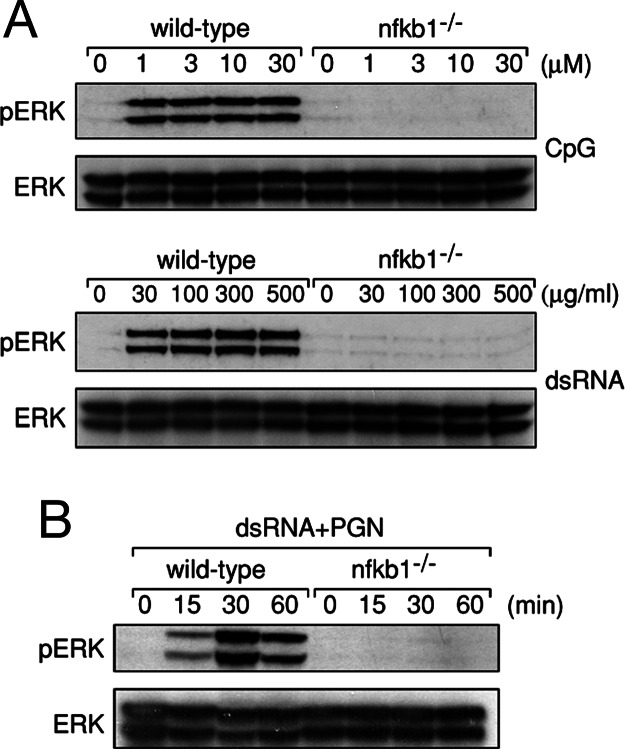
The ERK activation defect afflicting nfkb1−/− BMDMs appeared to be a direct consequence of the failure to activate MEK1, the MAP2K directly upstream of ERK. Whereas MEK1 phosphorylation kinetics in wild-type BMDMs coincided with the induction of phospho-ERK, phosphorylation of MEK1 in response to the TLR ligands examined was absent in nfkb1−/− BMDMs (Fig. 1). To ensure the inability of TLR signals to activate ERK in nfkb1−/− BMDMs did not simply reflect hyporesponsiveness of these cells to the concentrations of TLR ligands used in our initial experiments, wild-type and nfkb1−/− BMDMs were stimulated over a range of CpG and dsRNA concentrations. Although ERK was activated to the same extent in wild-type cells at all concentrations of CpG and dsRNA tested, even the highest ligand concentrations (30 μM and 500 μg/ml for CpG and dsRNA, respectively), which were 5–10 fold greater than those reported to effectively activate ERK, were unable to invoke ERK phosphorylation in nfkb1−/− BMDMs (Fig. 2A). As reported for TLR4, JNK1/2 and p38 were still activated in response to CpG and dsRNA in the absence of p105 NF-κB1 (Fig. 7, which is published as supporting information on the PNAS web site). Collectively, these findings indicate that ERK, but not JNK or p38 activation in BMDMs by a range of different TLR stimuli, is impaired in the absence of p105 NF-κB1.
Fig. 2.
The ERK activation defect in nfkb1−/− BMDMs is not overcome by multiple TLR signals. (A) Wild-type and nfkb1−/− BMDMs were stimulated with a range of CpG and dsRNA concentrations for 30 min. (B) Cells of both genotypes were costimulated with PGN and dsRNA for indicated times. TLR stimulated wild-type and nfkb1−/− BMDMs were lysed and subjected to immunoblotting for phospho-ERK and ERK. This data are representative of two independent experiments.
In addition to engaging the ubiquitous intracellular adaptor MyD88, TLRs use a number of adaptors that modulate the specificity, intensity, and duration of downstream signaling pathway activation (21–24). With the ERK pathway activated by TLR signals in both a MyD88-dependent and -independent manner (24, 25), coupled with delayed ERK phosphorylation in myd88−/− macrophages (25), multiple adaptors appear to be required for optimal ERK activation. These observations raised the possibility that there may be a signal threshold for TLR-mediated ERK activation that depends on distinct signaling intermediates regulated via different adaptors. To determine whether the TLR-dependent ERK activation defect exhibited by nfkb1−/− BMDMs represents an inability to breach a signal threshold delivered via a particular adaptor, wild-type and nfkb1−/− BMDMs were costimulated with PGN and dsRNA. In addition to using Myd88, these TLR2 and TLR3 ligands also signal through the adaptors TIRAP and TRIF, respectively. As shown in Fig. 2B, nfkb1−/− BMDMs fail to activate ERK, even when costimulated through two TLRs that can use different adaptors to engage downstream signaling pathways.
Tpl2 MEK Kinase Activity Is Induced in BMDMs by Diverse TLR Ligands.
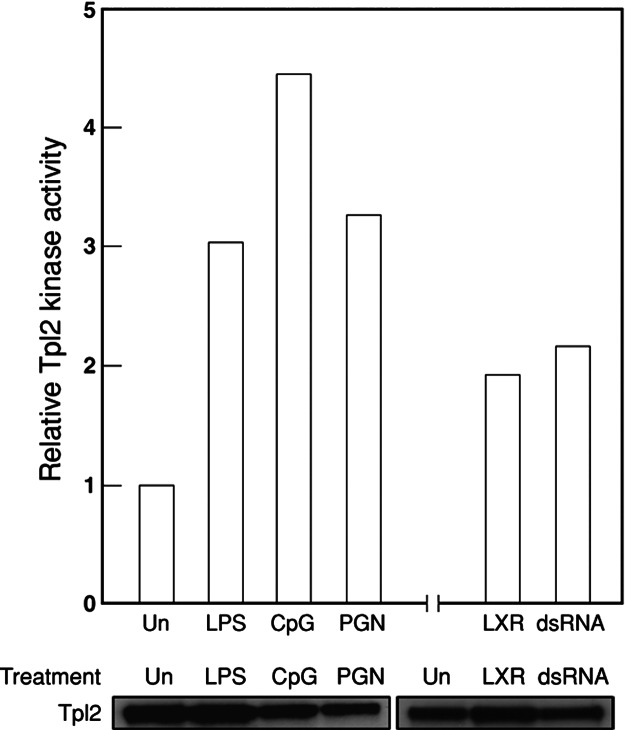
LPS stimulation of BMDMs leads to activation of the MAP3K Tpl2, which mediates ERK activation through MEK1 (19). This finding, coupled with the inability of diverse TLR ligands to activate MEK1 and ERK in nfkb1−/− BMDMs, prompted us to establish whether Tpl2 is normally activated by different TLR ligands. After a ≈30-min treatment of wild-type BMDMs with different TLR ligands, Tpl2 was immunoprecipitated from cell lysates by using a Tpl2-specific antiserum and the Tpl2/antibody conjugates tested for the capacity to promote the phosphorylation of an ERK substrate in a coupled in vitro kinase assay. Kinase assays were not performed on nfkb1−/− BMDMs, because Tpl2 kinase levels were previously shown to be undetectable in these cells because of Tpl2 instability in the absence of p105 (19). As reported for LPS, stimulation with other TLR ligands (CpG, dsRNA, PGN, and loxoribine) led to a modest, yet reproducible, increase in activated Tpl2 levels in wild-type BMDMs (Fig. 3), indicating that engaging each of the TLRs examined in this study leads to the activation of the MAP3K Tpl2.
Fig. 3.
Tpl2 is activated in BMDMs by diverse TLR ligands. Wild-type BMDMs were treated with TLR ligands for 30 min, lysed, and IP kinase assays were performed. Each bar represents the fold increase in Tpl2 activity versus normalized levels in untreated cells. Total amount of Tpl2 immunoprecipitated after each treatment was analyzed by Western blotting and used to normalize the kinase assay. This data are representative of independent experiments.
Expression of an Inducible Form of Raf Can Overcome the ERK Activation Defect in nfkb1−/− Macrophages.
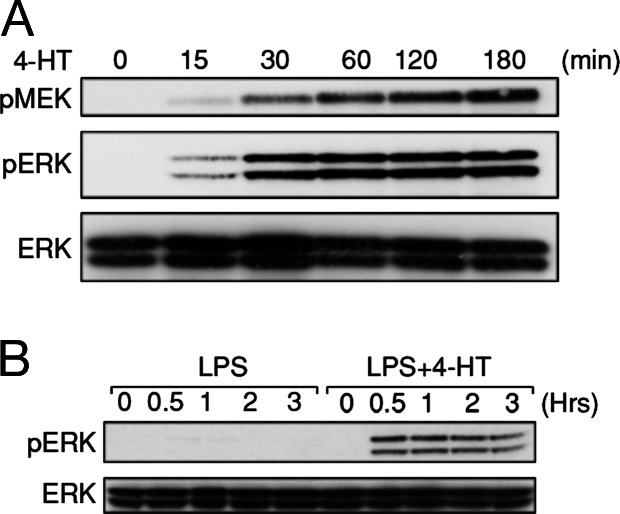
Given Tpl2 appeared to be essential for initiating ERK activation in BMDMs stimulated with different TLR ligands, we investigated whether ERK phosphorylation in LPS-stimulated nfkb1−/− BMDMs could be restored by an alternate MAP3K. To test this possibility an inducible form of the MAP3K Raf, comprising the catalytic domain of c-Raf fused to a version of the estrogen receptor hormone binding domain that specifically responds to the estrogen analogue 4-hydroxy-tamoxifen (4-HT) (26) was used. Normally, endogenous c-Raf is not activated in LPS-stimulated BMDMs (19), reinforcing the notion that Tpl2 is the ERK-specific MAP3K engaged by TLRs in these cells. Treatment of nfkb1−/− BMDMs expressing the Raf:ER fusion protein (hereafter referred to as nfkb1−/−-Raf:ER) with 4-HT led to a dose-dependent induction of ERK1/2 phosphorylation (Fig. 8, which is published as supporting information on the PNAS web site). Furthermore, treatment of nfkb1−/−-Raf:ER cells with an optimal dose (50 nM) of 4-HT led to rapid activation of MEK1 and ERK1/2 with kinetics similar to that exhibited by TLR ligand-treated wild-type cells (Fig. 4A). However, unlike the transient activation of ERK in response to LPS, 4-HT-induced ERK activation in cells expressing Raf:ER is sustained (26) due, in part, to a failure of negative feedback regulation by MAPK phosphatases to operate effectively. Importantly, in the context of the impaired TLR-mediated ERK activation observed in nfkb1−/− BMDMs, whereas treatment of nfkb1−/−-Raf:ER BMDMs with LPS alone failed to activate ERK, a combined LPS plus 4-HT treatment led to robust and sustained ERK activation (Fig. 4B). These data indicate that the impaired phosphorylation of ERK in nfkb1−/− BMDMs during LPS stimulation can be overcome by using a conditionally active version of Raf.
Fig. 4.
Expression of an inducible raf:ER fusion protein overcomes the ERK activation defect in nfkb1−/− BMDMs. nfkb1−/− BMDMs expressing the Raf:ER fusion protein were treated with 50 nM 4-HT (A) or LPS in the absence or presence of 4-HT (B) for indicated times. Cells were then lysed, and the presence of phospho-MEK, phospho-ERK, or ERK in lysates were analyzed by Western blotting. This data are representative of two independent experiments.
Induction of IL-10 After Treatment with LPS or CpG Requires ERK Activation.
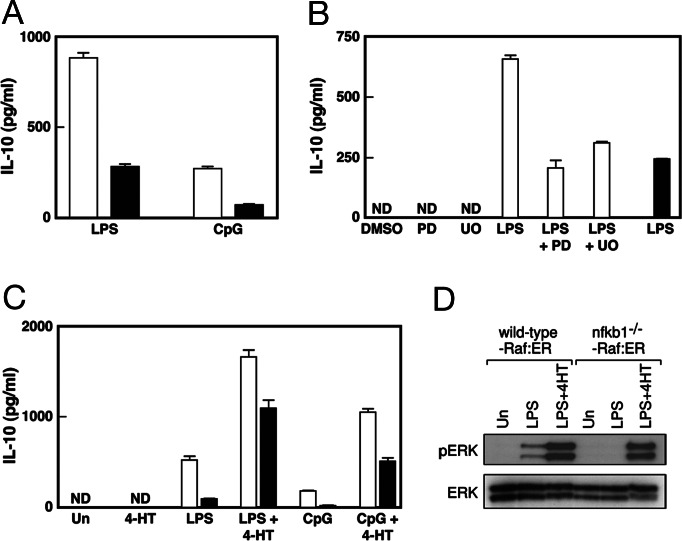
Stimulation of cells with TLR ligands leads to the expression of genes encoding pro- and antiinflammatory cytokines, the balance of which determines the outcome of an immune response (27, 28). IL-10 is one such cytokine, the transcription of which is ERK-dependent in TLR-stimulated dendritic cells and macrophages (20, 29). The TLR-dependent ERK activation defect exhibited by nfkb1−/− BMDMs prompted an assessment of IL-10 expression in these cells after TLR stimulation. Wild-type and nfkb1−/− BMDMs were stimulated with LPS or CpG for 6 h, and IL-10 levels in culture supernatants were determined by using an IL-10 specific ELISA. The data summarized in Fig. 5A shows that nfkb1−/− BMDMs produce significantly less IL-10 than wild-type cells in response to LPS or CpG. Moreover, when wild-type BMDMs were treated with LPS in the presence of the MEK inhibitors PD98059 or UO126, these cells now expressed IL-10 at reduced levels, similar to that of LPS-treated nfkb1−/− BMDMs (Fig. 5B). Although these data suggested that the compromised IL-10 expression in TLR-activated nfkb1−/− BMDMs was likely to be a direct consequence of the Tpl2-dependent ERK activation defect, examination of IL-10 expression in nfkb1−/−-Raf:ER BMDMs indicated that the relationship between TLR-mediated ERK activation and IL-10 induction was complex. Data summarized in Fig. 5C shows that the levels of IL-10 made by nfkb1−/−-Raf:ER BMDMs in response to 4-HT plus LPS or CpG were as expected, higher than these cells treated with TLR stimuli alone. Nevertheless, a comparison of IL-10 expression by wild-type and nfkb1−/− BMDMs expressing Raf:ER when treated with LPS or CpG in the presence of 4-HT revealed that nfkb1−/− BMDMs still made significantly less IL-10 than wild-type cells. The difference in IL-10 expression by wild-type and nfkb1−/− BMDMs expressing Raf:ER was not simply due to a differential level of ERK activation in response to the combined stimuli, because wild-type and nfkb1−/− BMDMs expressing Raf:ER when treated with LPS and 4-HT displayed similar levels of phospho-ERK (Fig. 5D). It also did not reflect a difference in the survival of wild-type and nfkb1−/− BMDMs (C. White, unpublished results). Rather, these results, coupled with the finding that 4-HT activation of ERK in the absence of LPS was unable to induce IL-10 expression in wild-type or nfkb1−/− BMDMs expressing Raf:ER (Fig. 5C), indicated that IL-10 expression is also reliant on TLR-induced ERK-independent signals. Although NF-κB had not been thought to directly regulate IL-10 transcription, a recent report identified a novel κB site in the murine IL-10 promoter that in macrophages was required for phorbol ester-dependent IL-10 transcription and bound RelA in response to LPS stimulation (30). Although the nuclear induction of RelA is not affected by the absence of NF-κB1 (A.B., unpublished results), it was plausible that RelA binding to the IL-10 κB element depended on NF-κB1 and, as a consequence, was impaired in LPS activated nfkb1−/− BMDMs. Whereas electrophoretic mobility shift assays demonstrated the rapid up-regulation of a nuclear complex that bound the IL-10 κB element in LPS-stimulated wild-type BMDMs, this complex was not induced in nfkb1−/− cells (Fig. 9A, which is published as supporting information on the PNAS web site). Supershift analysis (Fig. 9B) confirmed that the induced complex mainly comprised NF-κB1/RelA, with NF-κB/c-Rel heterodimers also bound to this site. These findings indicate that NF-κB1 appears to promote TLR-induced IL-10 transcription through a combination of direct NF-κB-dependent and indirect ERK-dependent regulation.
Fig. 5.
ERK activation is required for the TLR ligand-mediated induction of IL-10 in BMDMs. (A and B) Wild-type (open bars) and nfkb1−/− (filled bars) BMDMs were treated with LPS or CpG (A) or LPS in the presence or absence of PD98059 (PD) or U0126 (UO) (B) for 6 h. (C) BMDMs from both genotypes (identification codes as above) expressing the Raf:ER fusion protein were treated with LPS or CpG in the presence or absence of 4-HT for 6 h. After treatments, culture supernatants were harvested and analyzed for IL-10 by ELISA. These assays were performed in triplicate ± SD for each experiment. (D) Wild-type and nfkb1−/− BMDMs expressing the Raf:ER fusion protein were treated with LPS in the presence or absence of 4-HT for 30 min, lysed, and the presence of phospho-ERK or ERK was determined by Western blotting. Un and ND refer to untreated and undetectable, respectively. This data are representative of three independent experiments.
p105 NF-κB1 Is Required for ERK Activation in B Cells but Not Keratinocytes Treated with TLR Stimuli.
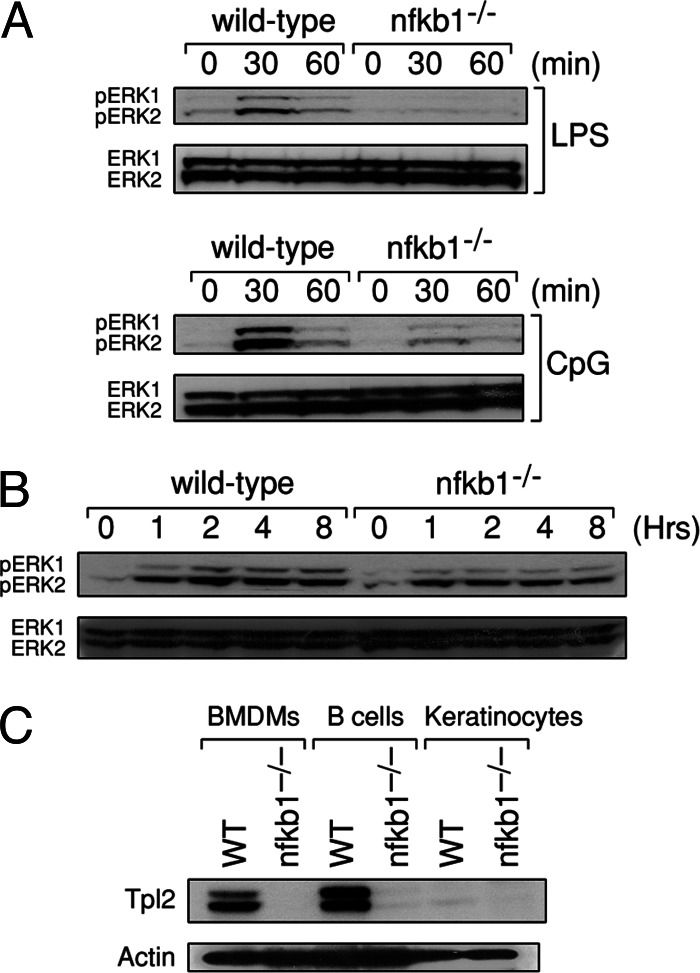
B cells and keratinocytes are examples of cell types other than macrophages that express TLRs (31, 32). Consequently, we investigated whether ERK activation in B cells and keratinocytes stimulated with ligands for TLRs expressed by these cells also depended on the p105 NF-κB1/Tpl2 signaling pathway. These findings are summarized in Fig. 6. In wild-type B cells, ERK is strongly, but transiently, activated in response to LPS or CpG. nfkb1−/− B cells, however, only display weak activation of ERK in response to either TLR ligand, indicating that like BMDMs, p105 NF-κB1 appears to be required for ERK activation downstream of TLR4 and TLR9 in B cells. In contrast, ERK phosphorylation in LPS-treated nfkb1−/− keratinocytes was only marginally diminished compared to wild-type cells (Fig. 6B), and unlike macrophages or B cells, was sustained in keratinocytes of both genotypes exposed to LPS. The finding that in the absence of p105 NF-κB1, ERK activation was impaired in LPS-activated macrophages and B cells, but not keratinocytes, prompted an examination of Tpl2 protein expression in these different cell types (Fig. 6C). Although Tpl2L and Tpl2S were expressed in wild-type macrophages and B cells, neither isoform was detectable in keratinocytes. Moreover, consistent with Tpl2 being stabilized by p105 NF-κB1 in BMDMs, Tpl2 was also undetectable in nfkb1−/− B cells. These findings indicate that distinct MAP3Ks are used for TLR4-mediated ERK activation in a cell type-dependent manner controlled, in part, by the expression pattern of MAP3Ks.
Fig. 6.
ERK activation is impaired in TLR-stimulated B cells but not keratinocytes lacking NF-κB1. (A and B) B cells (A) or keratinocytes (B) from wild-type and nfkb1−/− mice were treated with LPS or CpG for indicated times, followed by lysis and immunoblotting for phospho-ERK or ERK. (C) Total cell lysates from wild-type and nfkb1−/− BMDMs, B cells, and keratinocytes were subjected to immunoblotting for Tpl2 and actin. Data are representative of two independent experiments.
Discussion
Despite considerable advances having been made into understanding how TLRs engage key intracellular signaling pathways crucial in controlling innate and adaptive immune responses, the detailed mechanisms governing TLR-dependent transcription has received less attention. Multiple signaling pathways are activated in response to engaging TLRs, of which the IRF, NF-κB, and MAP kinase pathways feature prominently. Of particular interest is how TLR-activated signaling pathways are co-coordinated to regulate transcription. Previous studies have shown TLR4 activation of ERK is impaired in nfkb1−/− BMDMs due to an indirect effect the loss of p105 NF-κB1 has on the stability of the MAP3K Tpl2 (19). We have exploited this Tpl2 deficiency in nfkb1−/− mice to examine the role of Tpl2 in ERK activation induced by diverse TLR signals in different cell types and to start examining how NF-κB and ERK activation might coordinate gene expression.
Here we establish that ERK is not activated in nfkb1−/− BMDMs treated with ligands for TLR2, TLR3, TLR7, and TLR9, even though these cells maintain normal activation profiles for JNK and p38 in response to these stimuli. Two independent lines of evidence indicate the inability of nfkb1−/− BMDMs to activate ERK via diverse TLR signals does not simply reflect a failure to optimize TLR-dependent activation. First, TLR ligands used over a wide range of concentrations do not induce ERK phosphorylation in nfkb1−/− BMDMs. Second, costimulation of nfkb1−/− BMDMs with TLR2 and TLR3 ligands, which engage TIRAP and TRIF, respectively, also fail to rescue the ERK activation defect. This latter finding, which exploits the use of distinct TLR adaptors to activate signaling pathways, also indicates that this ERK activation defect appears to be distal to the TLR/adaptor complex. Instead, the LPS-dependent ERK activation defect in nfkb1−/− BMDMs, originally described for LPS that is associated with a marked reduction of Tpl2 in these cells, also applies to other TLRs and indicates that most, if not all, TLR-mediated ERK activation in BMDMs is Tpl2-dependent. Consistent with this conclusion is the finding that Tpl2 is activated in wild-type BMDMs stimulated with pathogen-associated molecular patterns for TLR2, TLR3, TLR4, TLR7, and TLR9.
The apparent role of Tpl2 in TLR-dependent ERK activation was not restricted to BMDMs, with nfkb1−/− B cells unable to activate ERK normally in response to LPS or CpG DNA. Despite the clear link between p105 NF-κB1, Tpl2, and TLR-dependent ERK activation in B cells and BMDMs, ERK activation was found to be relatively normal in LPS-stimulated nfkb1−/− keratinocytes, a cell type that we show normally does not express Tpl2. Together, these findings indicate that TLR-dependent ERK activation is not exclusively mediated by Tpl2. Coupled with a recent report showing Tpl2 was dispensable for ERK activation downstream of TLR9 in thioglycollate elicited peritoneal macrophages (TEPMs) (33), the relationship between Tpl2 and TLR-mediated ERK activation in different cell types appears to be complex. In particular, the finding that TLR9-mediated ERK activation appears to use different MAP3Ks in two types of macrophages is intriguing. These observations may indicate that the activation status of a particular cell type, which is different for BMDMs and TEPMs, or prior encounter with microbial stimuli can also determine which MAP3K a cell will use to activate ERK in response to a TLR signal.
Although the reduced levels of Tpl2 in nfkb1−/− BMDMs coincided with the TLR-dependent ERK activation defect, it remained to be shown formally that the ERK defect was indeed due to impaired MAP3K activity. This issue was examined by determining whether a distinct MAP3K normally not activated in BMDMs by TLR ligands could overcome the ERK defect in TLR-stimulated nfkb1−/− cells. Expression of a conditionally active Raf:ER fusion protein was found to rescue the ERK defect in LPS-treated nfkb1−/− BMDMs. Coupled with TLR activation of the JNK and p38, pathways remaining intact in nfkb1−/− macrophages despite these MAPK pathways using the same TLR adaptors as the ERK pathway (25) strongly supports the notion that the ERK activation defect in certain nfkb1−/− cells responding to diverse TLR signals indeed lies at the level of Tpl2.
Rescue of the ERK defect in nfkb1−/− BMDMs by 4-HT activation of a Raf:ER fusion protein permitted an investigation of ERK-dependent gene expression in nfkb1−/− BMDMs induced by TLR signals. Consistent with studies showing IL-10 transcription in dendritic cells and macrophage cell lines was ERK-dependent (20, 29), IL-10 induction by TLR ligands was found to be defective in nfkb1−/− BMDMs. A requirement for ERK signals as the explanation for defective TLR induced IL-10 synthesis by nfkb1−/− BMDMs was supported by the finding that the MEK inhibitors PD98059 and U0126 reduced IL-10 levels in LPS-stimulated wild-type BMDMs to that observed for TLR4-activated nfkb1−/− cells. However, 4-HT activation of Raf:ER, despite being able to induce ERK phosphorylation in both wild-type and nfkb1−/− BMDMs, was unable to induce detectable levels of IL-10. Therefore, IL-10 production by BMDMs, in response to LPS or CpG, appears to be under both ERK-dependent and -independent transcriptional control. This notion was supported by the finding that a combination of LPS and 4-HT induced higher levels of IL-10 in nfkb1−/− BMDMs expressing Raf:ER than LPS treatment alone. Although NF-κB1 had not previously been shown to regulate IL-10 transcription, a comparison of IL-10 production by wild-type and nfkb1−/− BMDMs expressing Raf:ER revealed wild-type cells still made 30–50% more IL-10 when costimulated with 4-HT and LPS or CpG. This finding suggests that p50/p105 itself also contributes to TLR-mediated IL-10 production in an ERK-independent manner. This conclusion is supported by a recent report showing RelA can directly regulate IL-10 transcription via a κB element in its promoter (30), which we show binds RelA/NF-κB1 and c-Rel/NF-κB1 dimers in LPS-activated wild-type macrophages. Notably, the absence of p50/NF-κB1 abolishes the binding of transactivating RelA and c-Rel dimer partners to the IL-10 κB site, a finding that supports a direct role for NF-κB1 in TLR-induced IL-10 transcription.
In summary, the results presented here show that Tpl2 is the MAP3K essential for ERK activation in TLR ligand-stimulated BMDMs and B cells but not keratinocytes. This finding, coupled with the expression pattern of Tpl2, raises the interesting possibility that Tpl2 is the primary regulator of ERK-mediated gene expression downstream of TLRs in the hemopoietic compartment. Finally, the finding that p50 NF-κB1 and its precursor, p105 NF-κB1, contribute to TLR-induced IL-10 expression in BMDMs in a direct and indirect ERK-dependent manner, respectively, highlights the utility of the model described here to study the joint contribution of the NF-κB and ERK pathways in controlling TLR-mediated gene expression.
Materials and Methods
Reagents and Mice.
Phospho-specific antibodies were from Cell Signaling Technology (Beverly, MA) and antibodies recognizing ERK, JNK, and p38 were from Santa Cruz Biotechnology. Polyclonal Tpl2 antiserum was a kind gift from S. Ley (National Institute for Medical Research, London). TLR ligands were obtained from the following sources: Escherichia coli LPS (Sigma), PGN (Fluka), dsRNA (Amersham Pharmacia Biosciences), CpG 1668 (Geneworks, Adelaide, Australia) and loxoribine (Invivogen, San Diego). PD98059 and U0126 were purchased from Calbiochem. All C57BL/6 and nfkb1−/− mice (34) (backcrossed to C57BL/6 for nine generations) used in this study were 6–10 weeks of age.
Retroviral Construct, Hemopoietic Stem Cell Infections, and Recipient Engraftment.
The retroviral construct expressing a truncated version of c-Raf under the control of the estrogen receptor (pMy-IRES-GFP-Raf:ER) was made by subcloning the Raf:ER coding sequence from pMXi-EGFP:dRaf1:ER* (26) into the pMY-IRES-GFP retroviral vector (35).
Mice were injected with 5-Fluoro-Uracil (188 μg/g of body weight) and 4 days later, bone marrow cells were harvested and cultured in DMEM containing 15% FCS, 10 ng/ml mouse stem cell factor, 500 ng/ml Flk ligand, 50 ng/ml mouse Thrombopoetin, and 50 ng/ml IL-6. On days 1, 2, and 3 of culture, cells were infected by using centrifugation (90 min at 1,200 × g at 32°C) with polybrene (4 μg/ml) supplemented supernatant containing pMy-IRES-GFP-Raf:ER retrovirus. On day 5 of culture, bone marrow cells were harvested and ≈106 cells injected into the tail vein of irradiated (2 × 550 Rad) C57BL/6 Ly5.1 mice. Engrafted recipients were maintained on water containing neomycin sulfate in a pathogen-free environment for 8–12 weeks.
Cell Culture and Cell Sorting.
BMDMs were prepared as described in ref. 36. Splenic B cells were prepared as described in ref. 37 with purity routinely of the order of ≈98%. Viral-infected Ly5.2 GFP+ bone marrow cells were isolated by flow cytometry from Ly5.1 recipients by first staining total bone marrow with a biotinylated anti-Ly5.1 antibody, followed by phycoerythrin-conjugated streptavidin to exclude residual recipient cells. These cells were then cultured to generate macrophages as outlined above.
Isolation of Mouse Keratinocytes.
Mouse keratinocytes were isolated from adult tail skin as described in ref. 38. Keratinocytes were cultured in keratinocyte serum-free media supplemented with 0.07 mg/ml bovine pituitary extract/0.02 μg/ml EGF/0.5 μg/ml hydrocortisone/5 μg/ml insulin/8 × 10−13 M triiodothyronine/5 μg/ml transferrin/0.1 μg/ml cholera toxin/6 μg/ml gentamycin at a density of 1 × 106 cells in 3.5-cm plates precoated with collagen IV (20 μg/ml). At ≈70% confluency, cells were washed in PBS and incubated for a further 18 h in supplement-free keratinocyte media before stimulating with LPS.
Stimulation of Cells.
BMDMs were starved in mouse toxicity-RPMI medium 1640 and 0.5% FCS for 18 h before treatment with 1 μg/ml LPS/3 μM CpG/100 μg/ml dsRNA/10 μg/ml PGN or 100 μM loxoribine. Keratinocytes were stimulated with 10 μg/ml LPS. BMDMs and keratinocytes were lysed as described in ref. 39. BMDMs expressing Raf:ER were stimulated with TLR ligands in the absence or presence of 4-HT before lysis. B cells were treated with 50 μg/ml LPS or 5 μM CpG, then lysed in 20 mM Tris, pH 7.4/135 mM NaCl/1.5 mM MgCl2/1 mM EDTA/1% Triton X-100/10% glycerol/1 mM sodium vanadate/1 mM sodium molybdate/1 mM β-glycerophosphate/1 mM sodium pyrophosphate/10 mM sodium fluoride/protease inhibitors (Roche Diagnostics, Mannheim, Germany). Lysates were stored at −80°C before use.
Immunoblotting and Tpl2 MEK Kinase Assay.
Equal amounts of total protein were loaded on 10% Novex gels (Invitrogen), subjected to electrophoresis and Western blotting performed as described in ref. 37. For Tpl2 immunoprecipitation and MEK kinase assay, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
ELISA.
Wild-type and nfkb1−/− BMDMs, including cells expressing Raf:ER, were treated with LPS or CpG in the absence or presence of 4-HT or the MEK inhibitors PD98059 (40 μM) or UO126 (10 μM) for 6 h, following which supernatants were collected and analyzed for IL-10 by ELISA according to the manufacturer's instructions (BD Biosciences).
Electrophoretic Mobility Shift Assays.
A 32P-dATP end-labeled probe corresponding to the κB site (5’-TTA CAC AAA GGG GAA TTC CAC ATT GGC TG-3’) from the murine IL-10 promoter (30) was incubated with 1–2 μg of nuclear extracts, prepared from wild-type or nfkb1−/− BMDMs stimulated with LPS (37). For supershift analysis, antibodies that specifically recognize NF-κB1, RelA, or c-Rel (Santa Cruz Biotechnology) were used.
Supplementary Material
Acknowledgments
We thank Dr. Baltimore for the generous gift of nfkb1−/− mice, R. Grumont and C. White for help and advice, and K. Brown and J. Merryfull for animal husbandry. This work was supported by a program grant from National Health and Medical Research Council of Australia.
Abbreviations
- BMDM
bone marrow-derived macrophage
- ERK
extracellular signal regulated kinase
- 4-HT
4-hydroxy-tamoxifen
- JNK
c-Jun-N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MAP3K
mitogen-activated protein 3-kinase
- PGN
peptidoglycan
- TLR
Toll-like receptor
- Tpl2
tumor progression locus 2
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Barton G. M., Medzhitov R. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 3.Eliopoulos A. G., Dumitru C. D., Wang C. C., Cho J., Tsichlis P. N. EMBO J. 2002;21:4831–4840. doi: 10.1093/emboj/cdf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guha M., Mackman N. Cell. Signalling. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 5.Guha M., O’Connell M. A., Pawlinski R., Hollis A., McGovern P., Yan S. F., Stern D., Mackman N. Blood. 2001;98:1429–1439. doi: 10.1182/blood.v98.5.1429. [DOI] [PubMed] [Google Scholar]
- 6.Saccani S., Pantano S., Natoli G. Nat. Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 7.Irie T., Muta T., Takeshige K. FEBS Lett. 2000;467:160–164. doi: 10.1016/s0014-5793(00)01146-7. [DOI] [PubMed] [Google Scholar]
- 8.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 10.Karin M., Ben-Neriah Y. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 11.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 12.Ceci J. D., Patriotis C. P., Tsatsanis C., Makris A. M., Kovatch R., Swing D. A., Jenkins N. A., Tsichlis P. N., Copeland N. G. Genes Dev. 1997;11:688–700. doi: 10.1101/gad.11.6.688. [DOI] [PubMed] [Google Scholar]
- 13.Erny K. M., Peli J., Lambert J. F., Muller V., Diggelmann H. Oncogene. 1996;13:2015–2020. [PubMed] [Google Scholar]
- 14.Patriotis C., Makris A., Bear S. E., Tsichlis P. N. Proc. Natl. Acad. Sci. USA. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiariello M., Marinissen M. J., Gutkind J. S. Mol. Cell. Biol. 2000;20:1747–1758. doi: 10.1128/mcb.20.5.1747-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmeron A., Ahmad T. B., Carlile G. W., Pappin D., Narsimhan R. P., Ley S. C. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 17.Tsatsanis C., Patriotis C., Bear S. E., Tsichlis P. N. Proc. Natl. Acad. Sci. USA. 1998;95:3827–3832. doi: 10.1073/pnas.95.7.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belich M. P., Salmeron A., Johnston L. H., Ley S. C. Nature. 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 19.Waterfield M. R., Zhang M., Norman L. P., Sun S. C. Mol. Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 20.Yi A. K., Yoon J. G., Yeo S. J., Hong S. C., English B. K., Krieg A. M. J. Immunol. 2002;168:4711–4720. doi: 10.4049/jimmunol.168.9.4711. [DOI] [PubMed] [Google Scholar]
- 21.O’Neill L. A. Trends Immunol. 2002;23:296–300. doi: 10.1016/s1471-4906(02)02222-6. [DOI] [PubMed] [Google Scholar]
- 22.Horng T., Barton G. M., Medzhitov R. Nat. Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 23.Horng T., Barton G. M., Flavell R. A., Medzhitov R. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 26.McMahon M. Methods Enzymol. 2001;332:401–417. doi: 10.1016/s0076-6879(01)32218-8. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal S., Agrawal A., Doughty B., Gerwitz A., Blenis J., Van Dyke T., Pulendran B. J. Immunol. 2003;171:4984–4989. doi: 10.4049/jimmunol.171.10.4984. [DOI] [PubMed] [Google Scholar]
- 28.Pulendran B. J. Immunol. 2005;174:2457–2465. doi: 10.4049/jimmunol.174.5.2457. [DOI] [PubMed] [Google Scholar]
- 29.Dillon S., Agrawal A., Van Dyke T., Landreth G., McCauley L., Koh A., Maliszewski C., Akira S., Pulendran B. J. Immunol. 2004;172:4733–4743. doi: 10.4049/jimmunol.172.8.4733. [DOI] [PubMed] [Google Scholar]
- 30.Saraiva M., Christensen J. R., Tsytsykova A. V., Goldfeld A. E., Ley S. C., Kioussis D., O’Garra A. J. Immunol. 2005;175:1041–1046. doi: 10.4049/jimmunol.175.2.1041. [DOI] [PubMed] [Google Scholar]
- 31.Peng S. L. Curr. Opin. Immunol. 2005;17:230–236. doi: 10.1016/j.coi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Pivarcsi A., Bodai L., Rethi B., Kenderessy-Szabo A., Koreck A., Szell M., Beer Z., Bata-Csorgoo Z., Magocsi M., Rajnavolgyi E., Dobozy A., Kemeny L. Int. Immunol. 2003;15:721–730. doi: 10.1093/intimm/dxg068. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto K., Ohata M., Miyoshi J., Ishizaki H., Tsuboi N., Masuda A., Yoshikai Y., Takamoto M., Sugane K., Matsuo S., et al. J. Clin. Invest. 2004;114:857–866. doi: 10.1172/JCI20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sha W. C., Liou H. C., Tuomanen E. I., Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 35.Onai N., Zhang Y., Yoneyama H., Kitamura T., Ishikawa S., Matsushima K. Blood. 2000;96:2074–2080. [PubMed] [Google Scholar]
- 36.Sester D. P., Beasley S. J., Sweet M. J., Fowles L. F., Cronau S. L., Stacey K. J., Hume D. A. J. Immunol. 1999;163:6541–6550. [PubMed] [Google Scholar]
- 37.Grumont R. J., Strasser A., Gerondakis S. Mol. Cell. 2002;10:1283–1294. doi: 10.1016/s1097-2765(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 38.Gugasyan R., Voss A., Varigos G., Thomas T., Grumont R. J., Kaur P., Grigoriadis G., Gerondakis S. Mol. Cell. Biol. 2004;24:5733–5745. doi: 10.1128/MCB.24.13.5733-5745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweet M. J., Campbell C. C., Sester D. P., Xu D., McDonald R. C., Stacey K. J., Hume D. A., Liew F. Y. J. Immunol. 2002;168:392–399. doi: 10.4049/jimmunol.168.1.392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.