Abstract
Lutein, a dihydroxy derivative of α-carotene (β,ε-carotene), is the most abundant carotenoid in photosynthetic plant tissues where it plays important roles in light-harvesting complex-II structure and function. The synthesis of lutein from lycopene requires at least four distinct enzymatic reactions: β- and ε-ring cyclizations and hydroxylation of each ring at the C-3 position. Three carotenoid hydroxylases have already been identified in Arabidopsis, two nonheme diiron β-ring monooxygenases (the B1 and B2 loci) that primarily catalyze hydroxylation of the β-ring of β,β-carotenoids and one heme-containing monooxygenase (CYP97C1, the LUT1 locus) that catalyzes hydroxylation of the ε-ring of β,ε-carotenoids. In this study, we demonstrate that Arabidopsis CYP97A3 (the LUT5 locus) encodes a fourth carotenoid hydroxylase with major in vivo activity toward the β-ring of α-carotene (β,ε-carotene) and minor activity on the β-rings of β-carotene (β,β-carotene). A cyp97a3-null allele, lut5-1, causes an accumulation of α-carotene at a level equivalent to β-carotene in wild type, which is stably incorporated into photosystems, and a 35% reduction in β-carotene-derived xanthophylls. That lut5-1 still produces 80% of wild-type lutein levels, indicating at least one of the other carotene hydroxylases, can partially compensate for the loss of CYP97A3 activity. From these data, we propose a model for the preferred pathway for lutein synthesis in plants: ring cyclizations to form α-carotene, β-ring hydroxylation of α-carotene by CYP97A3 to produce zeinoxanthin, followed by ε-ring hydroxylation of zeinoxanthin by CYP97C1 to produce lutein.
Keywords: α-carotene, carotenoid hydroxylase, cytochrome P450, xanthophyl, substrate channeling
Carotenoids are a group of >600 red, yellow, and orange pigments (most commonly C40) that contain extended conjugated double-bond systems (1, 2). This tremendous structural diversity has presumably evolved in relation to the many functions of carotenoids, which include acting as structural components of membranes and photosystems, accessory light-harvesting pigments, components for photoprotection and substrates for hormone syntheses (3, 4). Carotenoids are derived from isoprenoid precursors and are generally divided into two groups, the carotenes (acyclic or cyclic hydrocarbons) and the xanthophylls (oxygenated derivatives of carotenes). The carotenes of bacteria, plants, and protists are chemically and structurally similar, indicating that the initial steps of carotene synthesis are similar among organisms (3). Much of the enormous structural diversity of carotenoids occurs in the later steps of synthesis, including various ring cyclization and oxygenation reactions.
Cyclization of lycopene is a key step in generating carotenoid diversity because it marks a branch point to two major cyclic carotenoid groups (Fig. 1): the β,β- and β,ε-carotenoids. β,β-Carotenoids contain two identical β-rings formed by the symmetrical action of the β-ring cyclase (β-cyclase), whereas β,ε-carotenoids contain two different ring structures (β and ε) formed by the action of the β-cyclase and ε-ring cyclase (ε-cyclase) (encoded by the Arabidopsis LUT2 locus). β-Rings contain a double bond in conjugation with the polyene chain, which results in a rigid ring structure with only one conformation. In contrast, the ε-ring double bond is not in conjugation, and ε-rings have relatively free rotation around the C6′–C7′ carbon. Unlike the ubiquitous β-ring, which is found in organisms as diverse as archaeobacteria and plants, ε-rings occur exclusively in green plants (5–7), red algae (8), and one extant prochlorophyte (9, 10). Therefore, ε-ring formation and modifications should postdate that of β-rings in evolutionary time and would be expected to have evolved only in this subgroup of ε-ring carotenoid-containing organisms.
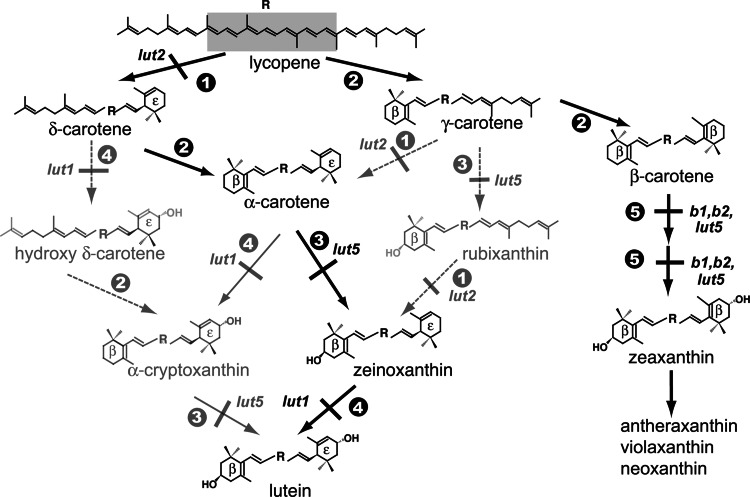
Fig. 1.
Pathway showing all possible routes to xanthophyll synthesis in Arabidopsis. Enzymatic reactions are indicated by numbers:  , ε-cyclization;
, ε-cyclization;  , β-cyclization;
, β-cyclization;  , β-ring hydroxylation of β,ε- and β,ψ-carotenoids;
, β-ring hydroxylation of β,ε- and β,ψ-carotenoids;  , ε-ring hydroxylation;,
, ε-ring hydroxylation;,  , β-ring hydroxylation of β,β-carotenoids. Reactions blocked by mutation of the indicated loci are shown: lut1 (ε-ring hydroxylase), lut2 (ε-cyclase), lut5, b1, and b2 (three β-ring hydroxylases). Solid gray arrows indicate a reaction sequence that is supported by mutant phenotypes and/or enzyme activity assays in E. coli, whereas dashed gray arrows are not. Solid black arrows, compounds, and mutant loci indicate major biosynthetic routes.
, β-ring hydroxylation of β,β-carotenoids. Reactions blocked by mutation of the indicated loci are shown: lut1 (ε-ring hydroxylase), lut2 (ε-cyclase), lut5, b1, and b2 (three β-ring hydroxylases). Solid gray arrows indicate a reaction sequence that is supported by mutant phenotypes and/or enzyme activity assays in E. coli, whereas dashed gray arrows are not. Solid black arrows, compounds, and mutant loci indicate major biosynthetic routes.
Lutein (3R,3′R-β,ε-carotene-3,3′-diol) is a dihydroxy β,ε-carotene and the most abundant carotenoid in plant photosynthetic tissues (e.g., accounting for >50% of the carotenoids in an Arabidopsis leaf). Lutein synthesis requires hydroxylation of C-3 of both the β- and ε-rings by the action of β-ring and ε-ring hydroxylases. Numerous studies have shown that a class of nonheme diiron monooxygenases (nonheme) that are present in most carotenoid-containing organisms (11) can efficiently hydroxylate the β-rings of several carotenoid substrates (12, 13). Arabidopsis contains two genes encoding nonheme β-ring hydroxylases (the B1 and B2 loci). Their primary in planta substrates are β-rings of β,β-carotenoids (Fig. 1) because a b1b2 double-null mutant reduced β,β-carotene-derived xanthophyll (β,β-xanthophyll) levels 80%, but the level of lutein (a β,ε-xanthophyll) was slightly increased relative to the WT (14). These data suggest that β,β- and β,ε-xanthophyll synthesis operate relatively independently of each other, which presumably reflects the independent evolution of the two pathway branches in plants.
Recently, two heme-containing cytochrome P450 monooxygenases (P450) have been identified that define two new classes of carotenoid hydroxylases. The archetypal members are the Arabidopsis ε-ring hydroxylase (CYP97C1) encoded by the LUT1 locus (15) and the β-carotene hydroxylase (CYP175A1) of the eubacteria, Thermus thermophilus (16). Expression of T. thermophilus CYP175A1 in Escherichia coli engineered to produce β-carotene resulted in the production of zeaxanthin, a dihydroxy β,β-carotene, and is a clear example of convergent evolution in β-ring hydroxylases (the nonheme and P450 classes). In Arabidopsis, a T-DNA insertion mutant in the LUT1 gene accumulates zeinoxanthin (β,ε-carotene with a hydroxylated β-ring) in place of lutein, consistent with LUT1 being the primary enzyme responsible for ε-ring hydroxylation in Arabidopsis. In the triple carotenoid hydroxylase mutant, b1b2lut1, β,β-xanthophylls are reduced 80% relative to WT, but zeinoxanthin is still accumulated to high levels suggesting at least one additional carotene hydroxylase must exist in Arabidopsis with activity toward the β-rings of β,ε- and β,β-carotenoids (14). In this article, we report that a second member of the Arabidopsis CYP97 P450 family, CYP97A3, encodes a β-ring hydroxylase with major activity toward the β-ring of α-carotene (β,ε-carotene) and, to a lesser extent, the β-rings of β-carotene.
Results
Mutation of Arabidopsis CYP97A3 Causes Accumulation of High Levels of α-Carotene.
Our initial search for a fourth Arabidopsis carotenoid hydroxylase focused on members of the CYP97 clade because it already contains one carotenoid hydroxylase (CYP97C1, the LUT1 locus). Although all three Arabidopsis CYP97 clade members are predicted to be targeted to the chloroplast by chlorop 1.1 (17), CYP97A3 (At1g31800) was selected because it has the highest (52%) amino acid identity with CYP97C1. To determine whether loss of CYP97A3 activity affected the carotenoid composition in Arabidopsis leaf tissue, 5-week-old leaves of two independent cyp97a3 mutant alleles were analyzed. lut5-1 contains a T-DNA insertion in the third exon of CYP97A3, whereas lut5-2 contains a single amino acid change (E283K).
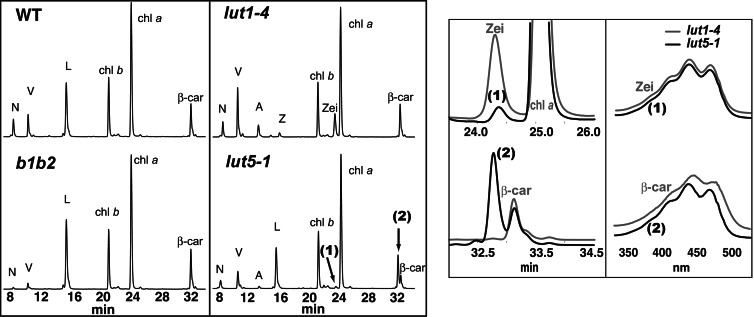
Fig. 2 shows the HPLC profiles of leaf extracts from WT, b1b2, lut1-4, and lut5-1. All lines are in the Columbia-0 (Col-0) background except b1b2 (disrupted in both nonheme β-ring hydroxylases), which is Wassilewskija. Individual and total carotenoid levels in Col-0 and Wassilewskija were not significantly different (data not shown), and only Col-0 is shown for WT. WT accumulates four major carotenoids: three xanthophylls (neoxanthin, violaxanthin, and lutein) and one carotene (β-carotene). Although the total carotenoid level is not significantly different between mutants and their respective WT, the carotenoid composition of each mutant genotype dramatically differs from WT (Table 1). As reported in ref. 14, the b1b2 mutant has lower levels of β,β-xanthophylls and increased β-carotene and lutein. lut1-4 (a null lut1 allele) virtually lacks lutein and accumulates a high level of zeinoxanthin, β,ε-carotene with a hydroxylated β-ring, and elevated levels of β,β-xanthophylls (zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin) (15, 18).
Fig. 2.
Identification and characterization of two unknown peaks in lut5-1. (Left) HPLC analyses (440-nm absorption) of leaf extracts of the indicated genotypes. N, neoxanthin; V, violaxanthin; A, antheraxanthin; L, lutein; Z, zeaxanthin; chl a, chlorophyll a; chl b, chlorophyll b; zei, zeinoxanthin; β-car, β-carotene. (Right) An overlay of sections of the lut5-1 (black) and lut1-4 (gray) HPLC chromatograms containing unknown peaks 1 and 2 and UV-visible absorption spectra of zeinoxanthin, α-carotene, and unknown peaks 1 and 2.
Table 1.
Leaf tissue carotenoid composition of the indicated genotypes
| Lutein | α-Cryptoxanthin | Zeinoxanthin | α-Carotene | β-Carotene | VAZ | Neoxanthin | tot.car | Hβ | β,ε/β,β | |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 127.0 ± 9.1 (51) | 0.7 ± 0.3† (0.3) | 0.7 ± 0.3† (0.3) | 1.4 ± 0.5 (0.6) | 55.4 ± 4.0 (22) | 32.1 ± 5.1 (13) | 32.5 ± 3.8 (13) | 249 ± 21 | 257 ± 26 | 1.1 |
| b1b2 | 157.2 ± 0.8* (65) | 1.0 ± 0.3† (0.4) | 1.0 ± 0.3† (0.4) | 1.4 ± 0.2 (0.6) | 68.4 ± 0.8* (28) | 11.9 ± 1.2* (5) | 3.9 ± 1.1* (1.6) | 244 ± 2 | 190 ± 3* | 1.9 |
| lut1-4 | 0.6 ± 0.1* (0.3) | ND | 60.5 ± 5.8* (26) | 1.4 ± 0.3 (0.6) | 57.1 ± 4.7 (25) | 87.1 ± 7.4* (37) | 26.3 ± 1.8* (11) | 233 ± 17 | 288 ± 22 | 0.4 |
| b1b2lut1-3 | 0.7 ± 0.3* (0.3) | ND | 100.6 ± 4.4* (45) | 1.5 ± 0.2 (0.7) | 97.4 ± 3.3* (43) | 22.4 ± 3.6* (10) | 1.9 ± 0.9* (0.8) | 225 ± 4 | 150 ± 5* | 0.8 |
| lut5-1 | 103.9 ± 7.4* (46) | 5.3 ± 0.7* (2.3) | ND | 49.6 ± 0.7* (22) | 27.2 ± 2.8* (12) | 23.2 ± 3.2* (10) | 18.7 ± 2.7* (8) | 228 ± 10 | 193 ± 8* | 2.3 |
| lut5-2 | 110.7 ± 1.8* (49) | 3.1 ± 0.7* (1.3) | ND | 34.0 ± 4.8* (15) | 32.5 ± 4.6* (14) | 22.4 ± 1.8* (10) | 24.7 ± 1.4* (11) | 228 ± 4 | 208 ± 2* | 1.9 |
| lut5-1lut1-4 | 0.4 ± 0.1* (0.2) | ND | 52.8 ± 5.2* (23) | 53.9 ± 3.1* (23) | 37.9 ± 3.5* (16) | 69.6 ± 5.6* (30) | 19.9 ± 2.4* (9) | 235 ± 17 | 232 ± 20 | 0.8 |
Carotenoids are expressed as millimoles of pigment per moles of chlorophyll a + b, with the relative molar percentage of each carotenoid given in parentheses. Each value is the mean result of four experiments ± SD. Student’s t test for two samples. VAZ, the sum of violaxanthin, antheraxanthin, and zeaxanthin; tot.car, total carotenoids; Hβ, the moles of hydroxylated β-rings; β,ε/β,β, the molar ratio of total β,ε-carotenoids to total β,β-carotenoids. ND, not detectable.
*, P < 0.05.
†Indicates a monohydroxy α-carotene derivative that could not be identified because of the low levels present.
lut5-1 has an 18% reduction in lutein and contains two novel peaks (minor and major) relative to WT, with retention times (24.3 and 32.7 min), mass, and absorption spectra that are consistent with those of monohydroxy α-carotene derivatives (zeinoxanthin and/or α-cryptoxanthin; see Fig. 1) and α-carotene, respectively (Fig. 2). Zeinoxanthin and α-cryptoxanthin cannot be distinguished on the basis of HPLC retention time or spectra. Nevertheless, carotenoids that have an allylic hydroxyl group, for example at C-3′ of an ε-ring, readily eliminate water when positively ionized, whereas a nonallylic hydroxyl group, such as at the C-3 of a β-ring, does not (18). The major ion of the 24.3-min unknown peak was [MH+-H2O] (see Fig. 5A, which is published as supporting information on the PNAS web site); based on this comparatively easy loss of water, we can conclude its single hydroxyl group is on the ε-ring of α-carotene, and hence, it is α-cryptoxanthin.
Accumulation of high and low levels of α-carotene and α-cryptoxanthin, respectively, occurs in both lut5 alleles and is accompanied by a reduction in total β,β-carotenoids and an increase in total β,ε-carotenoids (Table 1). The lut5-1 phenotype is generally more severe than lut5-2, consistent with lut5-2 being a weaker allele. The lut5 and lut1 mutations are additive: the lut5-1lut1-4 double mutant accumulates α-carotene and zeinoxanthin at levels nearly identical to lut5-1 and lut1-4, respectively.
α-Carotene Is Incorporated into Photosystems in lut5-1.
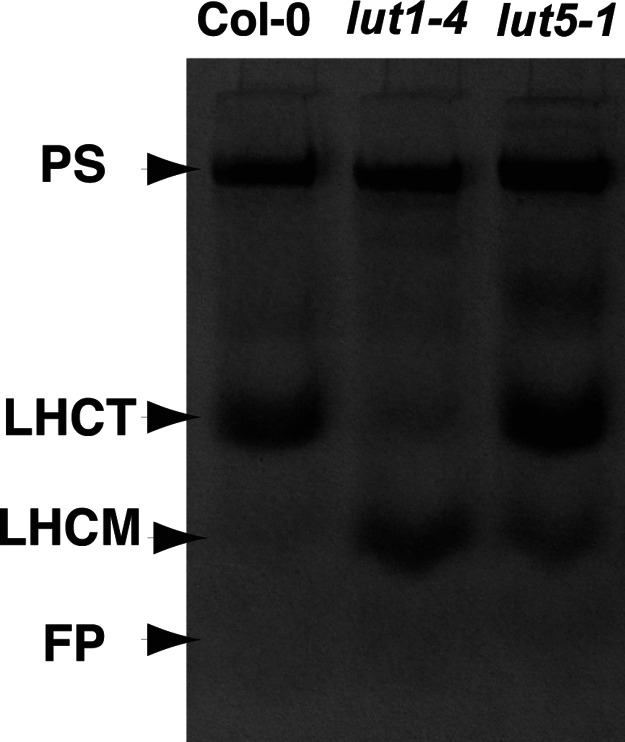
To determine the location of α-carotene in lut5, we purified thylakoid membranes from WT, lut1-4, and lut5-1, separated the pigment–protein complexes by nondenaturing gel electrophoresis (Fig. 3), and analyzed their pigment compositions (Table 2). Fig. 3 shows that the photosystem I holocomplex and photosystem II core complex comigrated and are well separated from light-harvesting complex (LHC) monomers and trimers in all genotypes (19, 20). The photosystems are the most likely place for α-carotene incorporation because they contain β-carotene, a structural isomer of α-carotene (19, 21–23). Like β-carotene in WT, α-carotene in lut5-1 was localized almost exclusively in photosystems, accounting for 39% of photosystem carotenoids (Table 2). In terms of carotenoid stoichiometry in lut5-1 photosystems, the increase in α-carotene was mirrored by a corresponding decrease in β-carotene. Fig. 3 also shows lut1-4 had a higher level of LHC monomers and lower level of LHC trimers, consistent with a report for this mutant showing a key role for lutein in LHC trimerization (20). lut5-1 also shows an increase in LHC monomers, suggesting the 18% reduction in lutein in the mutant also affects LHC trimer stability. The faint bands migrating between the photosystem and LHC trimer bands in lut1 and lut5 were variable in occurrence, and levels between experiments presumably reflect reduced photosystem stability in the mutants.
Fig. 3.
Nondenaturing gel electrophoretic separation of pigment–protein complexes from thylakoid membranes of the indicated genotypes. PS, photosystem I holocomplex and photosystem II core complex; LHCT, trimeric form of LHC; LHCM, monomeric form of LHC; FP, free-pigment zone.
Table 2.
Carotenoid composition in photosystems (PS I holocomplex and PS II core complex) of the indicated genotypes
| Lutein | α-Cryptoxanthin | Zeinoxanthin | α-Carotene | β-Carotene | VAZ | Neoxanthin | |
|---|---|---|---|---|---|---|---|
| WT | 54.0 ± 8.1 (28) | ND | ND | 1.9 ± 0.3 (1) | 112.6 ± 7.7 (59) | 26.1 ± 3.1 (12) | 0.8 ± 0.4 (0.6) |
| lut1-4 | 1.3 ± 1.6* (1.7) | ND | 14.6 ± 0.9* (12) | 1.3 ± 0.5 (1) | 86.1 ± 2.9* (68) | 21.6 ± 1.2 (17) | 0.5 ± 1.1 (<0.5) |
| lut5-1 | 51.4 ± 3.3 (26) | 1.0 ± 1.1 (0.5) | ND | 77.3 ± 5.8* (39) | 47.7 ± 1.7* (24) | 17.9 ± 1.2 (9.1) | 0.6 ± 1.1 (<0.5) |
The amount of carotenoid is expressed as millimoles of pigment per moles of chlorophyll a. Each value is the mean result from three experiments ± SD, with the relative molar percentage of each carotenoid given in parentheses. Student’s t test for two samples; ND, not detectable; VAZ, the sum of violaxanthin, antheraxanthin, and zeaxanthin.
*, P < 0.05.
Effect of Mutating P450-Type Carotenoid Hydroxylase on the Expression of Other Carotenoid Biosynthetic Enzymes.
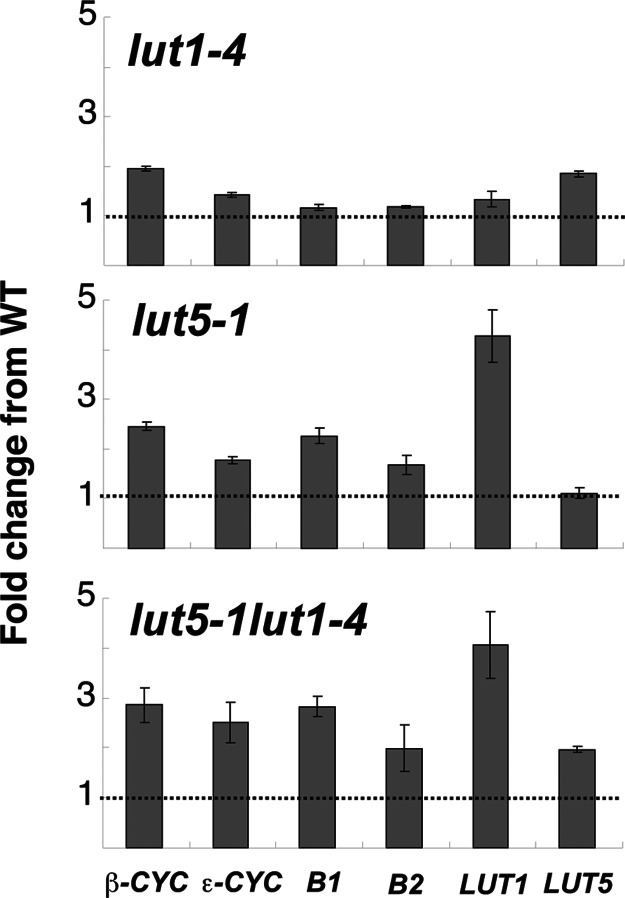
As shown in Table 1, the ratio of total β,ε-carotenoids to β,β-carotenoids in lut5-1 was over twice that of WT, suggesting that β- and/or ε-cyclase activities might be affected in the mutant. Because it is not possible to directly assay carotenoid cyclase or hydroxylase activities in Arabidopsis leaf extracts, we determined the steady-state transcript levels of these genes to indirectly assess the effect of the lut5 and lut1 mutations on the pathway. As shown in Fig. 4, both cyclase mRNAs are modestly increased in lut1-4 and lut5-1 relative to WT, the β-cyclase more so than the ε-cyclase, but this increase appears unrelated to changes in the β,ε- to β,β-carotenoid ratios in the mutants. We also quantified mRNAs for the four known carotenoid hydroxylase genes (B1, B2, LUT1, and LUT5) to determine whether altered gene expression plays a role in compensating for the absence of P450-type carotenoid hydroxylases in the mutants. The detection of each P450 gene transcript in the corresponding gene-knockout mutant was possible because the real-time probe is positioned upstream of each T-DNA insertion site. With the exception of a slight increase in LUT5 mRNA levels, the expression of other carotene hydroxylases is not affected in the lut1-4 mutant. In contrast, B1, B2, and LUT1 mRNAs were all up-regulated in lut5-1 and lut5-1lut1-4, most notably LUT1 mRNA, which was >4-fold higher than WT.
Fig. 4.
Expression of carotenoid biosynthetic genes in lut1-4, lut5-1, and lut5-1lut1-4 relative to WT. The dotted line refers to the expression level of each gene in WT. Transcripts were quantified by real-time PCR by using elongation factor-1α as a reference control.
Discussion
The current study and prior work (12, 13, 18) have defined a minimum of four carotenoid hydroxylase genes involved in xanthophyll biosynthesis in Arabidopsis: two nonheme β-ring hydroxylases (encoded by the B1 and B2 loci), a P450-type ε-ring hydroxylase (CYP97C1, encoded by the LUT1 locus), and now a P450-type β-ring hydroxylase (CYP97A3, encoded by the LUT5 locus). Analyses of mutants defective in one or more carotenoid hydroxylase activities (Table 1) (14) provide insight into the specific and overlapping activities of each enzyme in vivo. All mutant genotypes exhibit specific alterations in xanthophyll and carotene compositions (Table 1) (14), indicating the loss of any single activity cannot be fully compensated by the remaining activities. The degree of compensation observed in a given mutant genotype reflects the preferential activity of the missing enzyme(s) and the regulation and substrate specificities of the remaining active enzymes in the mutant. For example, LUT1 is the primary ε-ring hydroxylase activity in Arabidopsis because lutein synthesis is nearly completely blocked in the null lut1-3 (15) and lut1-4 (Fig. 2) alleles and the presumed LUT1 monohydroxy substrate, zeinoxanthin, accumulates. Similarly, disruption of both nonheme β-ring hydroxylases in the b1b2 double-null mutant reduces β,β-xanthophylls 76% without affecting β-ring hydroxylation in lutein synthesis, suggesting that the β-rings of β,β-carotenoids are the preferred in planta substrates for B1 and B2.
LUT5 is a P450 β-ring hydroxylase with activity toward the β-rings of both α-carotene and β-carotene in vivo. The major effect of the null lut5-1 mutation is an accumulation of α-carotene at a level equivalent to β-carotene in WT, consistent with the β-ring of α-carotene being a preferred LUT5 substrate in planta. Because lutein is only reduced 18% in lut5-1 relative to WT, at least one of the other carotene hydroxylases must also be able to catalyze hydroxylation of the β-ring of α-carotene, although less efficiently than LUT5. B1 and B2 are clearly able to hydroxylate the β-ring of α-carotene as in the lut5-1lut1-4 double mutant zeinoxanthin is still present at levels similar to lut1-4. LUT1 may also have some level of β-ring hydroxylase activity in vivo because LUT1 expression is strongly up-regulated in the lut5-1 background, as one might expect for a compensating enzyme (Fig. 4), and the level of total hydroxylated β-rings is further reduced in the b1b2lut1-3 mutant relative to the b1b2 mutant (Table 1) (15).
LUT5 also appears to have a minor in vivo activity toward the β-rings of β-carotene as β,β-xanthophylls (primarily violaxanthin and neoxanthin) are reduced 35% in lut5-1 versus a 76% reduction in the b1b2 genotype (Table 1). Although LUT5 is likely to be responsible for synthesis of at least a portion of the β,β-xanthophylls present in b1b2, contributions from LUT1, an additional uncharacterized hydroxylase activity, or indirect affects resulting from the elevated levels of α-carotene produced in lut5-1 can also not be excluded. Unfortunately, attempts to assay LUT1 and LUT5 in vitro by heterologous expression in Saccharomyces cerevisiae and E. coli and in vivo in E. coli engineered to accumulate carotenoid substrates have not been successful (data not shown), and therefore, we cannot directly assay potential substrates for the two enzymes. The generation of triple mutant genotypes in which only one of the four carotenoid hydroxylase activities remains would be informative in this regard.
A Model for Lutein Biosynthesis and Regulation in Plants.
As shown in Fig. 1, there are several potential biosynthetic routes leading from lycopene to lutein that have only partially been delineated by prior genetic studies in Arabidopsis (18, 24, 25). The reactions leading to lutein must be highly efficient or tightly associated because many of the potential pathway intermediates in WT Arabidopsis leaf tissue are near or below the 1-ng carotenoid HPLC detection limit (Table 1). The activities of Arabidopsis β- and ε-cyclases expressed in lycopene producing E. coli suggested that the preferential substrate for the ε-cyclase is lycopene rather than γ-carotene (24). Consistent with this, mutation of the Arabidopsis LUT2 locus (encoding the ε-cyclase) eliminates lutein synthesis without causing accumulation of the pathway intermediates rubixanthin or γ-carotene (18). These data indicate that ε-ring cyclization of lycopene to produce δ-carotene is the first step in lutein synthesis. δ-Carotene is undetectable in WT Arabidopsis leaf tissue, but α-carotene is detectable suggesting that the δ-carotene produced by LUT2 is efficiently used by the β-cyclase. The isolation and characterization of mutations in the LUT5 and LUT1 loci now allow us to infer the primary reaction sequence for lutein synthesis in plant tissues from among the remaining possible steps in Fig. 1.
The pathway shown in Fig. 1 has two possible routes leading to lutein from α-carotene: β-ring hydroxylation to zeinoxanthin followed by ε-ring hydroxylation to lutein or ε-ring hydroxylation to α-cryptoxanthin followed by β-ring hydroxylation to lutein. Whether one pathway is favored or both occur depends on the substrate specificities and regulation of the hydroxylases involved, LUT5 (CYP97A3) and LUT1 (CYP97C1). Accumulation of zeinoxanthin in lut1 while the α-carotene level remains identical to that in WT is consistent with zeinoxanthin, rather than α-carotene, being a preferred substrate for ε-ring hydroxylation by LUT1. If α-carotene were a preferred LUT1 substrate, the lut5 mutant (which still has LUT1 activity) would be expected to accumulate significant amounts of α-cryptoxanthin, which it does not. lut5 instead accumulates an α-carotene level 10 times that of α-cryptoxanthin, which is consistent with α-carotene being a preferred substrate for β-ring hydroxylation by LUT5 and LUT1 having, at best, a minor activity toward the ε-ring of α-carotene. Biochemical regulation (e.g., feedback inhibition by α-cryptoxanthin) may also contribute to the lut5-1 phenotype, but this hypothesis seems less likely because the lut5-1lut1-4 double mutant, which cannot synthesize α-cryptoxanthin, has a phenotype that is essentially the combination of the lut1 and lut5 single mutants. If α-cryptoxanthin played a regulatory role, one would expect a different phenotype when the compound was removed in the double mutant. Together, the data presented are consistent with the preferred pathway for lutein synthesis being two ring cyclizations to yield α-carotene and then proceeding to zeinoxanthin and lutein by the sequential action of LUT5 and LUT1.
The general reaction sequence for lutein synthesis, ring cyclizations followed by hydroxylations, is the same as that of its closest structural isomer, zeaxanthin, a dihydroxy β-carotene derivative. However, zeaxanthin is produced primarily by the action of the nonheme B1 and B2 β-ring hydroxylases, whereas lutein is produced primarily by the action of P450-mediated α-carotene ring hydroxylases, and the regulatory mechanisms of these two classes of enzymes appear to differ significantly. In lutein synthesis the bicyclic intermediate α-carotene is barely detectable in WT, whereas the corresponding intermediate in zeaxanthin synthesis, β-carotene, is 22% of total WT leaf carotenoids. Accumulation of high levels of α-carotene and zeinoxanthin in the lut5 and lut1 mutants excludes the possibility that these intermediates do not accumulate in WT simply because they are unstable in vivo. The high level of α-carotene in lut5 is especially intriguing because it suggests that the presence of LUT5 is specifically required for efficient lutein synthesis and that the two nonheme β-ring hydroxylases, B1 and B2, cannot substitute for LUT5 in this regard.
Why Would Two Members of the P450 Class of Carotenoid Hydroxylases Be Specifically Required for Efficient Lutein Biosynthesis in Vivo?
We propose that in WT the reaction sequence from lycopene to lutein is catalyzed by a protein complex composed of two lycopene cyclases (the β and ε-cyclases) and two P450 class hydroxylases (CYP97A3 and CYP97C1) that allows channeling of substrates between reactions. There is precedent for this finding because cytochrome P450 enzymes are known to form dimers (26, 27), functionally interact with other cytochrome P450 enzymes (28–30), or act as anchors for soluble and membrane biosynthetic complexes (31) in other systems. Given the clear importance of lutein in LHC structure and photosystem function (20, 32, 33), it is relatively easy to rationalize a strong selective pressure for the evolution of ε-ring cyclization and hydroxylation activities for lutein synthesis. However, the forces driving the evolution of different biochemical machinery (P450 and nonheme carotenoid hydroxylases) for hydroxylation of α-carotene and β-carotene are less obvious. Perhaps the answer lies in the need to efficiently synthesize lutein for LHC structure and function while simultaneously tightly controlling α-carotene production, due to the potential negative consequences of producing α-carotene-containing photosystems.
Plants produce β-carotene-containing photosystems almost exclusively under most environmental conditions (e.g., high light). α-Carotene-containing photosystems are also produced in many plant genera but generally only in shade-grown or low-light-adapted plants, where α-carotene can be present in excess of β-carotene (34–36). Presumably α-carotene-containing photosystems provide a competitive advantage under low-light conditions, but at higher light levels, such photosystems show increased photooxidation, α-carotene levels decrease, and β-carotene-containing photosystems predominate (37). These data suggest tight control of the α/β-carotene ratio is an important adaptive response to low and high light (38, 39). Consistent with this hypothesis, the constitutive production of α-carotene in lut5 renders the mutant much more sensitive than WT to high-light exposure (see Fig. 6, which is published as supporting information on the PNAS web site). LUT5 clearly plays a key role in carotenoid synthesis by allowing efficient lutein production while limiting α-carotene accumulation and provides a straightforward biochemical mechanism for producing α-carotene and lutein when both are needed under low-light conditions; this mechanism is by regulation of CYP97A3.
When taken together, the current genetic data in Arabidopsis are consistent with in vivo synthesis of the two major groups of xanthophylls in plants being preferentially catalyzed by two different classes of carotene hydroxylases: P450-type enzymes for synthesis of lutein and nonheme enzymes for synthesis of β,β-xanthophylls. The evolution of what at first appears to be an unnecessarily complex system can be understood when considering the contrasting needs of plants in response to changing light conditions. In high (normal) light, it is competitively advantageous to produce lutein without α-carotene, whereas under low-light conditions, it is advantageous to produce both lutein and α-carotene. β-Carotene and β,β-xanthophylls are needed at different levels and ratios under these conditions. Thus, the P450 carotenoid hydroxylases may be best suited to fulfilling these contrasting demands by allowing formation of a separate biosynthetic complex for efficient metabolic channeling to lutein while providing a regulatory mechanism for α-carotene production that is independent of the synthesis and regulation of β-carotene and β,β-xanthophylls. Such independent regulation of the two branches of the carotenoid pathway allows plants to respond efficiently, effectively, and adaptively to ever-changing light conditions.
Materials and Methods
Plant Material.
Plants were grown under a 12-h photoperiod (100–120 μmol·m−2·s−1; 22°C) and 18°C at night. lut1-4 and lut5-1 are T-DNA-knockout mutant alleles of CYP97C1 (At3g53130) and CYP97A3 (At1g31800), respectively. lut5-2 was identified by HPLC screening of nine missense mutant alleles isolated by targeting induced local lesions in genomes (TILLING) screening (40). The lut5-1lut1-4 double mutant was selected by PCR screening of F2 progeny from a cross of lut5-1 and lut1-4. HPLC separation, identification, and quantification by spectra and retention time were performed as described in ref. 12, except that quantification of monohydroxy α-carotenes was performed at 475 nm.
TaqMan Real-Time PCR Assay.
The transcript levels of six different carotenoid biosynthetic genes (β-cyclase, ε-cyclase, B1, B2, LUT1, and LUT5) were quantified by TaqMan real-time PCR by using elongation factor-1α mRNA levels for normalization. The LUT5 primers and TaqMan probe are as follows: 5′-GTTTGATTGGACTGGTTCTGACC-3′ (forward primer), 5′-TTCCGGACCGCCTGAAT-3′ (reverse primer), and 5′-ACCCCAAGGTTCCTGAGGCTAAAGGCT-3′ (TaqMan probe). Primers and probes for other genes are described in ref. 14. The relative quantity of the transcripts was calculated by using the comparative threshold cycle (CT) method (41).
Isolation of Thylakoid Membranes and Nondenaturing PAGE.
Thylakoid membranes from Col-0, lut1-4, and lut5-1 were isolated, and the photosystems and peripheral LHCs were separated by nondenaturing PAGE as described in ref. 20. Individual pigment-containing bands were excised and homogenized in gel running buffer (19), and the pigments were extracted and analyzed as described in ref. 12.
Determination of Unknown Monohydroxy α-Carotene.
TLC separation on Si250F·PA silica plates (Mallinckrodt Baker, Phillipsburg, NJ) with hexane/isopropanol solvent (9:1) was used to enrich the unknown monohydroxy α-carotene from lut5-1 saponified leaf extracts (18). A band showing the same Rf value as that of zeinoxanthin was isolated and subjected to further mass analysis. The elutant from HPLC was chemically ionized by atmospheric pressure chemical ionization and subsequently analyzed by MS. Lutein was used as a control to show water loss from a hydroxylated ε-ring when ionized, whereas zeaxanthin and zeinoxanthin were used to show no water loss from hydroxylated β-rings when ionized (18).
Supplementary Material
Acknowledgments
We thank Dr. Daniel Jones of the Michigan State University Mass Spectroscopy facility for assistance with MS, Dr. Li Tian for invaluable discussions, and Maria Magallanes-Lundback for technical assistance with RNA isolation. This work was supported by grants from HarvestPlus and National Science Foundation Grant IBN 0131253 (to D.D.).
Abbreviations
- β-cyclase
β-ring cyclase
- ε-cyclase
ε-ring cyclase
- β,β-xanthophyll
β,β-carotene-derived xanthophylls
- P450
cytochrome P450 monooxygenase
- LHC
light-harvesting complex
- Col-0
Columbia-0.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. NM_102914).
References
- 1.Straub O. In: Key to Carotenoids. Pfander H., Gerspacher M., Rychener M., Schwabe R., editors. Basel: Birkhäuser; 1987. pp. 11–218. [Google Scholar]
- 2.Kull D., Pfander H. In: Carotenoids. Britton G., Liaaen-Jensen S., Pfander H., editors. Basel: Birkhäuser; 1995. pp. 295–316. [Google Scholar]
- 3.Johnson E. A., Schroeder W. A. Adv. Biochem. Eng. Biotechnol. 1996;53:119–178. doi: 10.1007/BFb0102327. [DOI] [PubMed] [Google Scholar]
- 4.Liaaen-Jensen S. In: Carotenoids: Biosynthesis and Metabolism. Britton G., Liaaen-Jensen S., Pfander H., editors. Vol. 3. Basel: Birkhäuser; 1998. pp. 217–247. [Google Scholar]
- 5.Yoshii Y., Hanyuda T., Wakana I., Miyaji K., Arai S., Ueda K., Inouye I. J. Phycol. 2004;40:1170–1177. [Google Scholar]
- 6.Adams W. W., III, Demmig-Adams B., Lange O. L. Oecologia. 1993;94:576–584. doi: 10.1007/BF00566975. [DOI] [PubMed] [Google Scholar]
- 7.Schagerl M., Pichler C. Aquat. Bot. 2000;67:117–129. [Google Scholar]
- 8.Marquardt J., Hanelt D. Eu. J. Phycol. 2004;39:285–292. [Google Scholar]
- 9.Partensky F., Hoepffner N., Li W., Ulloa O., Vaulot D. Plant Physiol. 1993;101:285–296. doi: 10.1104/pp.101.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stickforth P., Steiger S., Hess W. R., Sandmann G. Arch. Microbiol. 2003;179:409–415. doi: 10.1007/s00203-003-0545-4. [DOI] [PubMed] [Google Scholar]
- 11.Tian L., DellaPenna D. Arch. Biochem. Biophys. 2004;430:22–29. doi: 10.1016/j.abb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Tian L., DellaPenna D. Plant Mol. Biol. 2001;47:379–388. doi: 10.1023/a:1011623907959. [DOI] [PubMed] [Google Scholar]
- 13.Sun Z., Gantt E., Cunningham F. X., Jr J. Biol. Chem. 1996;271:24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]
- 14.Tian L., Magallanes-Lundback M., Musetti V., DellaPenna D. Plant Cell. 2003;15:1320–1332. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D. Proc. Natl. Acad. Sci. USA. 2004;101:402–407. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasco F., Kauffmann I., Schmid R. D. Appl. Microbiol. Biotechnol. 2004;64:671–674. doi: 10.1007/s00253-003-1529-7. [DOI] [PubMed] [Google Scholar]
- 17.Emanuelsson O., Nielsen H., von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogson B., McDonald K. A., Truong M., Britton G., DellaPenna D. Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A. I., Thornber J. P. Plant Physiol. 1995;107:565–574. doi: 10.1104/pp.107.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lokstein H., Tian L., Polle J. E., DellaPenna D. Biochim. Biophys. Acta. 2002;1553:309–319. doi: 10.1016/s0005-2728(02)00184-6. [DOI] [PubMed] [Google Scholar]
- 21.Klimmek F., Ganeteg U., Ihalainen J. A., van Roon H., Jensen P. E., Scheller H. V., Dekker J. P., Jansson S. Biochemistry. 2005;44:3065–3073. doi: 10.1021/bi047873g. [DOI] [PubMed] [Google Scholar]
- 22.Alfonso M., Montoya G., Cases R., Rodriguez R., Picorel R. Biochemistry. 1994;33:10494–10500. doi: 10.1021/bi00200a034. [DOI] [PubMed] [Google Scholar]
- 23.Croce R., Morosinotto T., Castelletti S., Breton J., Bassi R. Biochim. Biophys. Acta. 2002;1556:29–40. doi: 10.1016/s0005-2728(02)00304-3. [DOI] [PubMed] [Google Scholar]
- 24.Cunningham F. X., Jr., Pogson B., Sun Z., McDonald K. A., DellaPenna D., Gantt E. Plant Cell. 1996;8:1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogson B. J., Rissler H. M. Philos. Trans. R. Soc. London B. Biol. Sci. 2000;355:1395–1403. doi: 10.1098/rstb.2000.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoch G. A., Yano J. K., Wester M. R., Griffin K. J., Stout C. D., Johnson E. F. J. Biol. Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 27.Scott E. E., He Y. A., Wester M. R., White M. A., Chin C. C., Halpert J. R., Johnson E. F., Stout C. D. Proc. Natl. Acad. Sci. USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaminsky L. S., Guengerich F. P. Eur. J. Biochem. 1985;149:479–489. doi: 10.1111/j.1432-1033.1985.tb08950.x. [DOI] [PubMed] [Google Scholar]
- 29.Kelley R. W., Reed J. R., Backes W. L. Biochemistry. 2005;44:2632–2641. doi: 10.1021/bi0477900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutton D. R., McMillen S. K., Sonderfan A. J., Thomas P. E., Parkinson A. Arch. Biochem. Biophys. 1987;255:316–328. doi: 10.1016/0003-9861(87)90399-7. [DOI] [PubMed] [Google Scholar]
- 31.Burbulis I. E., Winkel-Shirley B. Proc. Natl. Acad. Sci. USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 33.Wentworth M., Ruban A. V., Horton P. Biochemistry. 2004;43:501–509. doi: 10.1021/bi034975i. [DOI] [PubMed] [Google Scholar]
- 34.Thayer S., Bjorkman O. Photosynth. Res. 1990;23:331–343. doi: 10.1007/BF00034864. [DOI] [PubMed] [Google Scholar]
- 35.Demming-Adams B., Adams W., III Plant Cell Environ. 1992;15:411–419. [Google Scholar]
- 36.Koniger M., Harris G., Virgo A., Winter K. Oecologia. 1995;104:280–290. doi: 10.1007/BF00328362. [DOI] [PubMed] [Google Scholar]
- 37.Barth C., Krause G. H., Winter K. Plant Cell Environ. 2001;24:163–176. [Google Scholar]
- 38.Krause G. H., Koroleva O. Y., Dalling J. W., Winter K. Plant Cell Environ. 2001;24:1345–1352. [Google Scholar]
- 39.Krause G. H., Grube E., Koroleva O. Y., Barth C., Winter K. Funct. Plant Biol. 2004;31:743–756. doi: 10.1071/FP03239. [DOI] [PubMed] [Google Scholar]
- 40.Till B. J., Reynolds S. H., Greene E. A., Codomo C. A., Enns L. C., Johnson J. E., Burtner C., Odden A. R., Young K., Taylor N. E., et al. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak K. J. User Bulletin No. 2: ABI Prism 7700 Sequence Detection System. Foster City, CA: Applied Biosystems; 1997. pp. 11–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.