Abstract
East Coast fever, caused by the tick-borne intracellular apicomplexan parasite Theileria parva, is a highly fatal lymphoproliferative disease of cattle. The pathogenic schizont-induced lymphocyte transformation is a unique cancer-like condition that is reversible with parasite removal. Schizont-infected cell-directed CD8+ cytotoxic T lymphocytes (CTL) constitute the dominant protective bovine immune response after a single exposure to infection. However, the schizont antigens targeted by T. parva-specific CTL are undefined. Here we show the identification of five candidate vaccine antigens that are the targets of MHC class I-restricted CD8+ CTL from immune cattle. CD8+ T cell responses to these antigens were boosted in T. parva-immune cattle resolving a challenge infection and, when used to immunize naïve cattle, induced CTL responses that significantly correlated with survival from a lethal parasite challenge. These data provide a basis for developing a CTL-targeted anti-East Coast fever subunit vaccine. In addition, orthologs of these antigens may be vaccine targets for other apicomplexan parasites.
Keywords: cattle, East Coast fever, immunoscreening, protozoan parasite, vaccination
A single inoculation with a potentially lethal dose of Theileria parva sporozoites and simultaneous treatment with a long-acting oxytetracycline induces solid immunity to homologous and, in certain instances, heterologous parasite challenge (1, 2). This methodology has been adopted as a live vaccine for the control of East Coast fever (ECF) (3). The long-lasting immunity to ECF contrasts with the partial immunity to malaria that develops after only several years of exposure to T. parva-related Plasmodium spp. (4). Manufacture and delivery of the live ECF vaccine is difficult to sustain, but it has enabled elucidation of the dominant protective immune response against the disease. Kinetic and adoptive cell transfer studies (5, 6) have demonstrated that protection of cattle is mediated by MHC class I-restricted CD8+ cytotoxic T lymphocytes (CTL) that destroy schizont-infected lymphocytes, the pathogenic life-cycle stage of T. parva. In addition, there is a strong correlation between the specificity of the CTL response and cross-immunity profiles of distinct parasite strains (2). The identification of schizont antigens targeted by CTL from T. parva-immune cattle has been elusive but should pave the way for the development of a subunit vaccine against ECF and provide a long-term solution to a socioeconomically important constraint to livestock agriculture in Africa (7).
We adopted two approaches to antigen identification, both dependent on screening of transiently transfected antigen-presenting cells with fully characterized CTL (8, 9) from live vaccine-immunized cattle of diverse bovine leukocyte antigen (BoLA) MHC class I genotypes. First, in a targeted gene approach, we immunoscreened genes that were predicted by using preliminary sequence data from one of the four T. parva chromosomes (10) to contain a secretion signal. The approach was based on the observation that the schizont lies free in the host cell cytoplasm (11) whereby signal peptide-containing parasite proteins would directly access the host cell MHC class I antigen processing and presentation pathway. Second, we used an approach involving a random immunoscreen of schizont cDNA clones because secretion of proteins that do not contain signal sequences has been reported (12). We describe here the antigens identified from these immunoscreens and the immunological characterization conducted to validate these molecules as CTL targets. We also report on the initial evaluation of these antigens as vaccine candidates and show successful induction of CTL responses that correlated with reduced disease severity after challenge infection.
Results and Discussion
Identification of CTL Target Antigens by Targeted and Random Immunoscreens.
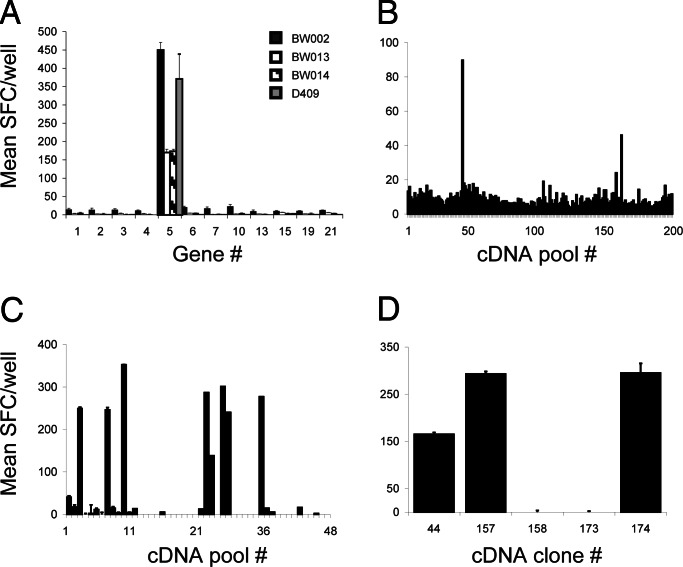
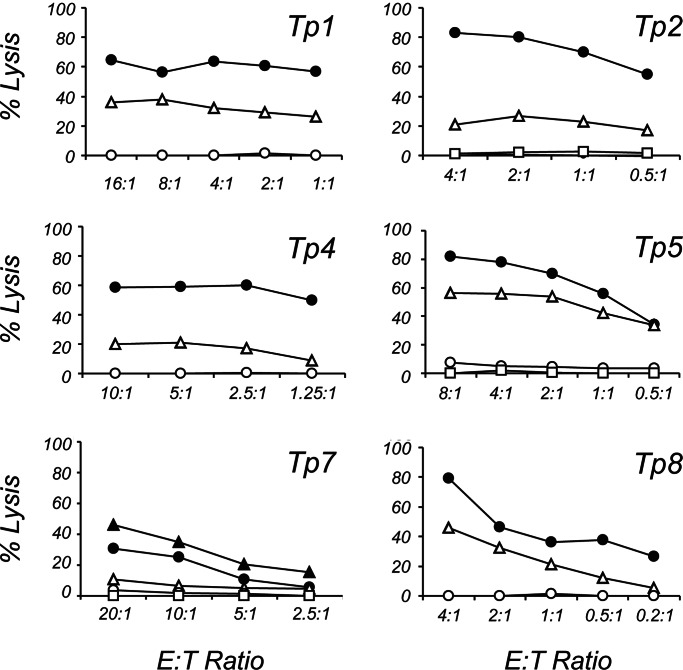
In the targeted approach, CTL from four immunized cattle showed IFN-γ ELISpot responses to immortalized skin fibroblasts (iSF) transfected with gene no. 5 (hereafter referred to as Tp2) (Fig. 1A), whereas, in the random approach, CTL from these and an additional six animals recognized targets in several cDNA pools. Sibling analysis was used to identify positive clones (Fig. 1 B–D) whose sequencing and database searches revealed Tp2 and five additional antigens (Tp1, Tp4, Tp5, Tp7, and Tp8; Table 1). Thus, Tp2 was identified as an antigen by means of two independent approaches. The CTL lines exhibited specific lytic activity against transfected autologous iSF and COS-7 cells (Fig. 2), albeit lower than that against autologous schizont-infected cells because of low transfection efficiencies and polyclonality of CTL. Antigen-presenting cells transfected with an irrelevant T. parva gene encoding a schizont surface protein called PIM (13) or allogeneic iSF transfected with the Tp genes were not lysed. The low levels of lysis against Tp7-transfected iSF were significantly enhanced when iSF were pulsed with the minimal-length antigenic peptide 206EFISFPISL214 (Fig. 2), which was mapped by using Tp7 synthetic peptides (data not shown). These data demonstrate that the six antigens are targets of BoLA MHC class I-restricted, IFN-γ-secreting, and lytic CD8+ T cells and indicate that the live vaccine primes a CTL response against them.
Fig. 1.
Targeted and random approaches to T. parva antigen identification. (A) Screening of targeted genes with T. parva-specific CTL from four cattle showed specific reaction to iSF transfected with targeted gene 5 (Tp2) as assessed by IFN-γ ELISpot. (B) Representative results of screening pools of random schizont cDNA with T. parva-specific CTL. By using the IFN-γ ELISpot assay, CTL from animal BV115 were used to screen COS-7 cells cotransfected with BoLA-N*01301 cDNA and pools of parasite cDNA to reveal positive pools. (C) Screening of resolved cDNA pools allowed the identification of five cDNA clones. (D) Confirmatory assays revealed specific reaction against three identical cDNA clones encoding Tp1. Responses are presented as mean numbers of SFC per well.
Table 1.
Summary of antigens identified through targeted and random screening with CTL derived from immune cattle
| Antigen | Animal nos. | Antigen-presenting cell | GenBank accession no. (chromosome location) | No. of amino acids | Secretion signal | Gene annotation |
|---|---|---|---|---|---|---|
| Tp1 | BV115 | iSF and Cos/BoLA-N*01301 | XP_762973 (3) | 543 | SP | Hypothetical |
| Tp2 | BW002 | iSF | XP_765583 (1) | 174 | SP | Hypothetical |
| BW013 | ||||||
| BW014 | ||||||
| D409 | ||||||
| Tp4 | BX063 | iSF and Cos/BoLA-N*00101 | XP_763228 (3) | 579 | None | ε-TCP1 |
| BX064 | ||||||
| BX065 | ||||||
| Tp5 | BV050 | iSF | XP_765334 (2) | 155 | None | eIF-1A |
| Tp7 | BW012 | iSF | XP_764810 (2) | 721 | None | Hsp90 |
| Tp8 | BX063 | iSF | XP_764709 (2) | 440 | SA | Cysteine proteinase |
| BX064 |
Shown are the full-length size of protein predicted by each gene sequence and whether the sequence contained a signal peptide (SP) or signal anchor (SA) sequence at the N terminus. ε-TCP1, ε-subunit of T complex protein 1; eIF-1A, translation elongation initiation factor 1A; hsp90, heat-shock protein 90.
Fig. 2.
CTL activity against transfected targets. Lysis was determined against autologous iSF transfected with Tp1, Tp2, Tp4, Tp5, and Tp7 or COS-7 cotransfected with BoLA-N*00101 and Tp8. Filled circles, autologous schizont-infected cells; open triangles, Tp gene-transfected autologous iSF or COS-7; open circles, irrelevant PIM gene-transfected autologous iSF; open squares, Tp gene-transfected allogeneic iSF; filled triangles, Tp7 peptide-pulsed autologous iSF.
Comparison of the identified cDNA sequence data with predicted T. parva gene models (10) revealed that the clones encoded full-length Tp1, Tp4, and Tp5 proteins and all but the first 22 amino acid residues of Tp8, whereas Tp7 cDNA encoded only 1–291 of 721 residues. Tp1 and Tp2 were annotated as hypothetical proteins (10), whereas Tp4, Tp5, Tp7, and Tp8 were predicted to encode the ε-subunit of T complex protein 1, elongation translation initiation factor 1A, heat-shock protein 90, and cysteine proteinase, respectively. More significantly, Tp4, Tp5, and Tp7 do not contain a predicted secretion signal and would not have been identified by the targeted approach. Potential orthologs of the antigens we have identified exist in Theileria annulata, Cryptosporidium parvum, and Plasmodium falciparum (Table 4, which is published as supporting information on the PNAS web site).
CTL Target Antigens Recognized by T Cells from Immune Cattle Resolving a Challenge Infection.
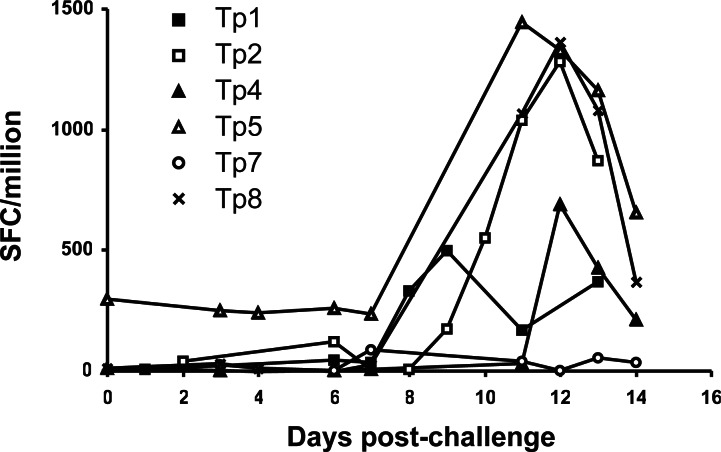
We assessed whether the protective CD8+ T cells elicited in live vaccine-immunized cattle after challenge with a lethal dose of sporozoites included T cells specific to the six antigens (Fig. 3). Responses to Tp1, Tp2, Tp5, and Tp8 increased from prechallenge levels from day 8 and peaked between days 9 and 13 after challenge with ≈500–2,000 spot-forming cells (SFC) per million CD8+ T cells. The kinetics of these responses correlated well with those previously described for protective schizont-specific CTL in efferent lymph and peripheral blood after challenge of immune animals (5, 6), indicating that these antigens are recognized by CD8+ T cells during the resolution of the infection. By contrast, the response to Tp4 was delayed and observed only from day 12 after challenge, which may reflect delayed expression of Tp4 during the parasite's development in the bovine host. The lack of boosting of Tp7-specific CD8+ T cells may relate to a low frequency of responder cells.
Fig. 3.
Antigen specificity of CD8+ T cell responses after challenge of T. parva-immune cattle. IFN-γ responses of ex vivo CD8+ T cells were measured in the presence of pools of overlapping peptides. Results are presented as medium corrected mean numbers of SFC per 1 × 106 CD8+ T cells. Some time points were omitted because of a background >100 SFC per 1 × 106 CD8+ T cells.
Immunogenicity of CTL Target Antigens After Prime/Boost Immunization and Parasite Challenge.
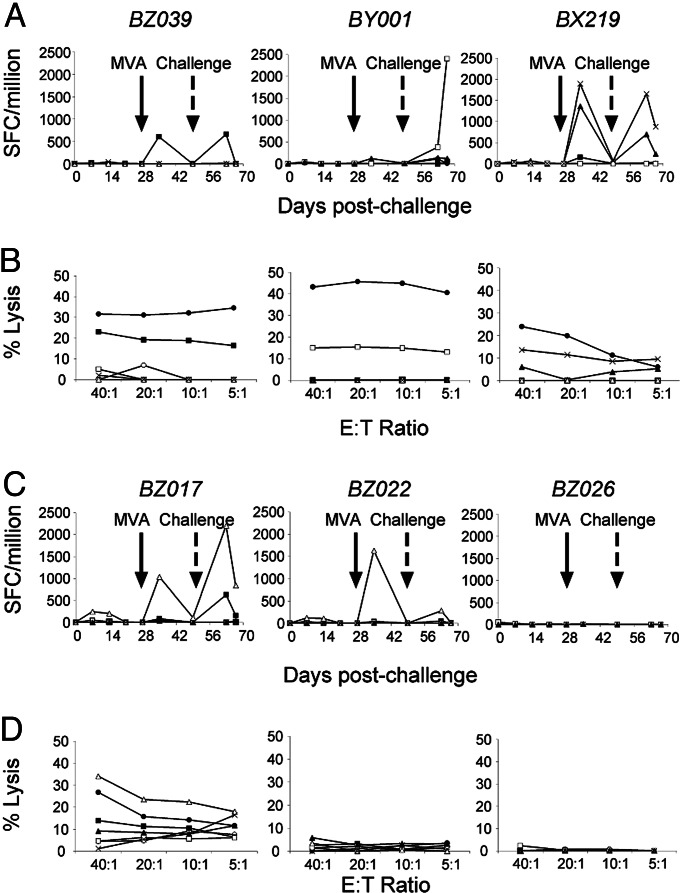
Based on these ex vivo data and our knowledge of the protective bovine immune response, we sought to determine whether vaccination with Tp1, Tp2, Tp4, Tp5, and Tp8 induced CTL that would contribute to immunity against ECF. Plasmid DNA/modified vaccinia virus Ankara strain (MVA) or canarypox virus (CP)/MVA prime/boost vaccination regimes were used to test the antigens in cattle. These regimes are efficient and potent inducers of CD8+ T cell-mediated IFN-γ and lytic responses in rodents (14) and, to a lesser extent, in humans (15). In cattle, such prime/boost regimes have induced T cell responses (16, 17), although an antigen-delivery strategy to elicit protective antigen-specific CD8+ CTL is lacking. Two groups of 12 cattle, of appropriate BoLA types, were subjected to DNA/MVA or CP/MVA immunization with all five antigens (Table 2). Three weeks after the booster, vaccinates and a group of four naïve cattle were inoculated with a lethal dose of sporozoites. Antigen-specific CD4+ and CD8+ T cell-mediated IFN-γ responses and CD8+ CTL activity against infected cells and peptide-pulsed lymphoblasts were measured over the course of the experiment. Results are discussed as a single group because there were no significant differences in any parameter between the two vaccination regimes (Table 2). CTL activity was detected in 29% of the vaccinates; four cattle before challenge (data not shown) and 2 weeks after challenge, the same four cattle and an additional three were positive in the assays (Fig. 4 and Table 2). By contrast, 19 (79%) of the vaccinated cattle exhibited antigen-specific CD8+ T cell IFN-γ responses (Fig. 4 and Table 2), and responses were boosted in all but four animals after sporozoite challenge. These responses were quantitatively similar to those reported for humans after prime/boost vaccination with P. falciparum antigens (15). The antigen specificity of the lytic response in the animals paralleled the specificity of their IFN-γ responses. Surprisingly, marginal CD4+ T cell responses were detected in nine (38%) of the vaccinated cattle, and these did not correlate with CD8+ T cell reactivity (data not shown). The general failure to induce adequate helper CD4+ T cell responses may be a consequence of some of the CTL target antigens lacking CD4+ T cell epitopes. It is possible that the IFN-γ-releasing and lytic CD8+ T cells detected in the assays may represent different subpopulations, as reported in studies on human T cell subsets (18), with unique helper CD4+ T cell requirements.
Table 2.
Summary of details of cattle and their responses to heterologous prime/boost immunization with CTL target antigens and challenge with T. parva
| Group | Animal no. (breed†) | Age, months | BoLA allele | CD8+ T cell IFN-γ | CTL | ECF index | Killing, days after challenge |
|---|---|---|---|---|---|---|---|
| CP/MVA | BX215 (B) | 22.33 | N*00101 | + | − | 4.63 | |
| BZ027 (HF) | 8.26 | N*01301 | + | + | 6.31 | ||
| BZ017 (HF) | 6.36 | T5 | + | + | 7.26 | ||
| BY142 (C) | 8.69 | N*01201 | − | − | 7.42 | ||
| BY206 (C) | 13.51 | N*01201 | + | + | 7.45 | ||
| BZ039 (HF) | 15.67 | N*01301 | + | + | 7.85 | ||
| BX223 (B) | 24.89 | N*00101 | + | − | 7.97 | 20 | |
| BX216 (B) | 21.93 | N*00101 | + | − | 8.45 | 17 | |
| BZ026 (HF) | 9.74 | N*01201 | − | − | 8.64 | 17 | |
| BZ016 (HF) | 6.36 | T5 | + | − | 8.70 | 16 | |
| BZ018 (HF) | 4.33 | N*01301 | + | − | 8.93 | 15 | |
| BZ022 (HF) | 10.03 | T5 | + | − | 8.93 | 16 | |
| DNA/MVA | BY001 (C) | 15.93 | N*01201 | + | + | 3.39 | |
| BX219 (B) | 24.69 | N*00101 | + | + | 6.28 | ||
| BZ023 (HF) | 7.02 | N*01301 | + | + | 6.96 | ||
| BY138 (HF) | 20.45 | N*01201 | − | − | 7.09 | 20 | |
| BZ013 (HF) | 18.39 | T5 | + | − | 7.28 | ||
| BX225 (B) | 21.87 | N*00101 | + | − | 7.61 | 16 | |
| BX220 (B) | 22.23 | N*00101 | − | − | 8.44 | 20 | |
| BZ019 (HF) | 4.20 | T5 | + | − | 8.59 | 16 | |
| BZ020 (HF) | 12.07 | N*01201 | − | − | 8.60 | 17 | |
| BY100 (C) | 10.92 | N*01301 | + | − | 8.78 | 20 | |
| BZ025 (HF) | 9.64 | T5 | + | − | 8.83 | 15 | |
| BZ040 (HF) | 7.34 | N*01301 | + | − | 8.91 | 15 | |
| Control | BY029 (C) | 13.05 | N*01201 | − | − | 7.88 | |
| BY071 (HF) | 26.82 | T5 | − | − | 8.50 | 16 | |
| BZ038 (HF) | 16.69 | N*01301 | − | − | 8.76 | 17 | |
| BX218 (B) | 24.82 | N*00101 | − | − | 8.90 | 20 |
Ages of cattle at the time of priming immunization are presented. Calves were selected on the basis of four BoLA class I alleles defined to restrict CTL responses to the five antigens: BoLA-N*01301, Tp1; BoLA-N*01201, Tp2; BoLA-T5, Tp5; BoLA-N*00101, Tp4 and Tp8. Cattle were defined as CD8+ T cell IFN-γ and CTL responders (+) or nonresponders (−) by criteria defined in Materials and Methods.
†B, Boran; HF, Holstein–Friesian; C, crossbreed.
Fig. 4.
Antigen specificity of CD8+ T cells in vaccinated cattle. (A and C) Representative IFN-γ responses of five responder cattle illustrate reactivity to each of the antigens and are compared with a nonresponder calf. Responses presented are the medium corrected number of SFC per 1 × 106 CD8+ T cells. (B and D) Four of the five IFN-γ responder cattle exhibited lytic responses and are compared with two nonresponder cattle. Filled squares, Tp1; open squares, Tp2; filled triangles, Tp4; open triangles, Tp5; crosses, Tp8; filled circles, schizont-infected cells; open circles, unpulsed T cell blasts.
Association Between CTL Responses and Outcome to Challenge.
The significance of the lytic CD8+ T cell responses became apparent when they were correlated with the outcome to challenge (Tables 2 and 3). All of the challenge controls developed severe ECF, with three animals killed. Of the 19 cattle that made CD8+ T cell IFN-γ responses, nine survived, whereas all seven cattle that made lytic CD8+ T cell responses survived. There was a highly significant association between the ability to mount a lytic response and survival (P < 0.001). Two of these cattle suffered mild-to-moderate ECF, and five suffered moderate-to-severe ECF as judged by their ECF reaction index, a statistical measurement of disease severity (19). Their mean ECF reaction index was significantly lower than that of nonresponder and control cattle (P < 0.01) (Table 3). The high number of cytokine responders that succumbed to the challenge infection indicates that ex vivo IFN-γ responses do not serve as a predictive marker of protection against severe disease or fatality. This observation is notable in the context of previous studies, which demonstrated an inhibitory effect of IFN-γ on the development of the early schizont stage in vitro (20). Our data suggest that vaccine-induced CD8+ lytic responses may contribute to survival from a lethal challenge infection and highlight possible distinct roles of IFN-γ-secreting and lytic CD8+ T cells in immunity against ECF. The data validate these antigens for an in-depth study on improving the current vaccination regimes to induce protective lytic CD8+ T cell responses.
Table 3.
Association between CTL responses and outcome to challenge
| Outcome to challenge | Vaccinated cattle |
Control cattle |
|
|---|---|---|---|
| CTL responder* (n = 7) | CTL nonresponder (n = 17) | CTL nonresponder† (n = 4) | |
| Survived | 7‡ | 3 | 1 |
| Killed | 0 | 14 | 3 |
| ECF index | 6.04‡ | 8.04 | 8.49 |
*CTL responders were defined by criteria described in Materials and Methods.
†CTL responses were not detected in the challenge control group.
‡Survival (P < 0.001) and ECF index (P < 0.01) of CTL responder cattle were significantly different from those of CTL nonresponders and control cattle.
The application of a rational strategy to identify schizont antigens and the induction of antigen-specific lytic CD8+ T cell responses that correlate with immunity represent key milestones in subunit vaccine development for ECF. Availability of homologs of the T. parva antigens and correlates of immunity may be of value in initiatives to develop vaccines against tropical theileriosis, heartwater, coccidiosis, malaria, and tuberculosis caused by intracellular pathogens against which CTL contribute to immunity.
Materials and Methods
Cloning of Targeted Genes.
Analysis of proteins encoded by the predicted T. parva genes (10) on chromosome 1 for the presence of signal peptides (signalp-2.0; www.cbs.dtu.dk/services/SignalP-2.0) and transmembrane domains (tmhmm, www.cbs.dtu.dk/services/TMHMM) generated a set of 55 candidate antigen genes that were selected for cloning and screening (Table 5, which is published as supporting information on the PNAS web site). ORFs were amplified by using a OneStep RT-PCR kit (Qiagen, Crawley, U.K.) with RNA purified from T. parva (Muguga) schizont-infected cells. Amplified genes were cloned into eukaryotic expression vector pTargeT (Promega).
Construction of Purified T. parva Muguga Schizont cDNA Library.
A unidirectional cDNA library was constructed in the eukaryotic expression plasmid vector pcDNA3 (Invitrogen) essentially as described in ref. 21. Briefly, double-stranded cDNA was generated from 2 μg of total poly(A)+-RNA (FastTrack 2.0 kit, Invitrogen) isolated from purified schizonts (22) by using the SuperScript Choice System (Invitrogen). Plasmid DNA minipreps were prepared from 1,000 pools of bacterial cultures each containing 50 colonies and another 1,000 pools each containing 10 colonies for high-throughput immunoscreening. To resolve a positive pool of 50 cDNAs, 256 colonies were pooled into 48 clusters of 16 colonies by using a three-way matrix and screened to identify the individual clone in three unique clusters. For the resolution of a positive pool of 10 cDNA, plasmid DNA was prepared from 48 colonies and tested to identify the individual clone. Positive cDNA clones were end-sequenced, and sequence information was used to identify the complete ORFs from the T. parva genome sequence database (www.ncbi.nlm.nih.gov/sutils/blast_table.cgi?taxid=Protozoa).
Infection and Treatment Immunization of Cattle and Generation of CTL.
All animal experimentation was reviewed and approved by the International Livestock Research Institute (ILRI) Institutional Animal Care and Use Committee. Four Boran (Bos indicus), one Jersey (Bos taurus), one Holstein–Friesian (B. taurus), and four crossbred animals, selected on the basis of BoLA type, were immunized against T. parva Muguga stock and then challenged after 3 months (2). Schizont-specific polyclonal CTL lines and clones were established and exhibited 30–80% lytic activity against autologous schizont-infected targets at an effector:target ratio <10:1, in line with published data (2, 8, 9).
Transfection of iSF and COS-7 Cells.
iSF were immortalized by using standard procedures (23) with modifications (unpublished observations). iSF were transfected in 96-well plates with clones or pools of schizont cDNA (100 ng per well) by using FuGENE 6 (Roche Diagnostics, Mannheim, Germany) and cultured for 24 h. COS-7 cells were cotransfected with 100 ng of each clone per well or pooled together with 50 ng of pcDNA3 BoLA-N*00101 (24) or BoLA-N*01301 cDNA per well.
IFN-γ ELISpot and 51Chromium Release Assays.
Transfectants were cocultured with schizont-specific CTL, and recognition was assessed by using an IFN-γ ELISpot assay (16). Transfectants and schizont-infected cells were labeled with 51Chromium (Amersham Pharmacia Biosciences), and lysis by schizont-specific CTL lines was assessed (9).
Ex Vivo Detection of Antigen-Specific T Cell Responses.
Immune cattle donors for schizont-specific CTL used to identify Tp1, Tp2, Tp4, Tp5, Tp7, and Tp8 were challenged with a lethal dose of T. parva (Muguga) sporozoites and bled on selected days after challenge. CD8+ T cells and monocytes were purified from peripheral blood mononuclear cells by magnetic cell sorting according to the manufacturer's instructions (Miltenyi Biotec). CD8+ T cells were stained with anti-CD8 mAb (IL-A105) (ILRI) followed by incubation with anti-mouse IgG microbeads (Miltenyi Biotec). Monocytes were stained directly with CD14 microbeads (Miltenyi Biotec). Magnetically sorted CD8+ T cells (2.5 × 105 per well) and CD14+ monocytes (2.5 × 104 per well) were added to IFN-γ ELISpot plates containing pools of overlapping synthetic peptides (16 mers offset by 4 residues) spanning the lengths of Tp1, Tp2, Tp4, Tp5, Tp7, and Tp8 (1 μg/ml each peptide, Pepscan Systems, Lelystad, The Netherlands) and were incubated and developed essentially as described in ref. 16.
Preparation of Recombinant Plasmid DNA and Viruses.
Tp1, Tp2, Tp4, Tp5, and Tp8 ORFs, excluding the stop codons, were amplified by OneStep RT-PCR from schizont RNA. The coding sequence of a mouse B cell epitope of SV5 P/V protein, Pk-Tag, was ligated at the 3′ end of each gene. These antigen gene Tag fragments were cloned into pSG2 DNA vaccine plasmids (25), and endotoxin-free DNA was prepared (PlasmidFactory, Bielefeld, Germany). MVA and CP were generated by homologous recombination of the wild-type viruses MVA and ALVAC, respectively, with the corresponding shuttle vectors MVA-GFP and pC5H6p, incorporating antigen gene Tag inserts, in accordance with published protocols (26). Expression was confirmed by immunostaining with anti-Pk-Tag mAb (MCA1360P, Serotec) and CTL recognition of virus-infected iSF. All vaccine preparations were screened for the presence of bovine viral diarrhea virus (27) and mycoplasma (Mycoplasma Detection kit version 2.0; American Type Culture Collection).
Heterologous Prime/Boost Immunization and Parasite Challenge of Cattle.
The experiment was designed in a double-blind fashion in which animals were retagged between immunization and challenge, and the code was managed by the biometrics staff. Twenty-eight Boran (B. indicus), Holstein–Friesian (B. taurus), and crossbred calves, 4–24 months old and expressing one of four MHC class I alleles, were randomized into three groups (Table 2). Recombinant DNA, CP, and MVA constructs expressing single antigens (Tp1, Tp2, Tp4, Tp5, and Tp8) were prepared separately and inoculated at different sites. Cattle were primed with 0.5 mg of each of the five recombinant DNA plasmids by intradermal injection or 1 × 108 plaque-forming units of each of the five recombinant CP viruses by s.c. injection. After 4 weeks, cattle were inoculated s.c. with 5 × 108 plaque-forming units of each of the five MVA recombinants. Control cattle received s.c. inoculations with the vaccine diluent alone. Three weeks after boost, cattle were challenged with a lethal dose of T. parva (1:20 dilution of T. parva Muguga stock sporozoite stabilate 4133; ILRI). Reactions to challenge were calculated as an ECF reaction index by combination of 13 parameters, including temperature, hematological measurements, and parasitological measurements, by using first-principle-component analysis as described in ref. 19. Cattle were assessed daily by two independent veterinarians, and animals displaying clinical signs of severe ECF were killed by captive bolt. The experiment was terminated 21 days after challenge. Antigen-specific CD4+ and CD8+ T cell IFN-γ responses were measured against synthetic peptides by ELISpot assay every 2 weeks over the course of immunization and challenge. CD4+ T cells were sorted by magnetic cell sorting indirectly by using a mAb specific for bovine CD4 (IL-A12; ILRI), CD8+ T cells and CD14+ monocytes were sorted, and ELISpot assays were conducted as described above. Antigen-specific CTL responses were recalled by subjecting peripheral blood mononuclear cells to three rounds of restimulation with T. parva-infected cells, and cytotoxicity against infected cells and uninfected cells pulsed with pools of synthetic peptides were assayed as described above. Cattle were classified as IFN-γ responders when the number of SFC in response to antigen was three times greater than to medium alone and as CTL responders when lysis of T. parva-infected lymphoblasts and/or antigen peptide-pulsed uninfected lymphoblasts was three times greater and at least 15% higher than that of unpulsed uninfected lymphoblasts. For all responder cattle, responses were >2 standard deviations (95% confidence interval) above the mean SFC of unstimulated cells or mean lysis against unpulsed uninfected lymphoblasts.
Statistical Analysis.
Analysis of variance was used for the analysis of fixed effects on different traits by using sas release 8.2 (SAS Institute, Cary, NC). A Kruskal–Wallis test was carried out to compare the ECF reaction index for CTL responders and nonresponders. A χ2 test was used to compare the differences in survival for CTL responders and nonresponders.
Supplementary Material
Acknowledgments
We thank the staff of the ILRI large animal and tick units for excellent animal care and provision of parasites; the ILRI Biometrics Unit for advice on experimental design and statistical analysis; Shirley Ellis (Institute for Animal Health, Compton, U.K.) for providing the BoLA-N*01301 cDNA; and The Institute for Genomic Research Board of Trustees, J. Craig Venter, and Hank Fitzhugh for support of the T. parva genome project. Support by Thierry Boon (Ludwig Institute for Cancer Research) for the screening of cDNA libraries is greatly appreciated. This work was funded in part by the Animal Health Program of the U.K. Department for International Development for the benefit of developing countries (Project No. R8042). Y.H. was supported in part by the Ministry of Foreign Affairs of Japan. This work is ILRI publication no. 200502.
Abbreviations
- CTL
cytotoxic T lymphocyte(s)
- BoLA
bovine leukocyte antigen
- iSF
immortalized skin fibroblast
- MVA
modified vaccinia virus Ankara strain
- CP
canarypox virus
- ILRI
International Livestock Research Institute
- ECF
East Coast fever
- SFC
spot-forming cell.
Footnotes
References
- 1.Radley D. E., Brown C. G. D., Cunningham M. P., Kimber C. D., Musisi F. L., Payne R. C., Purnell R. E., Stagg S. M., Young A. S. Vet. Parasitol. 1975;1:35–41. [Google Scholar]
- 2.Taracha E. L. N., Goddeeris B. M., Morzaria S. P., Morrison W. I. Infect. Immun. 1995;63:1258–1262. doi: 10.1128/iai.63.4.1258-1262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uilenberg G. Trop. Med. Int. Health. 1999;4:A12–A20. doi: 10.1046/j.1365-3156.1999.00446.x. [DOI] [PubMed] [Google Scholar]
- 4.Good M. F. Trends Parasitol. 2005;21:29–34. doi: 10.1016/j.pt.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Morrison W. I., Goddeeris B. M., Teale A. J., Groocock C. M., Kemp S. J., Stagg D. A. Parasite Immunol. 1987;9:563–578. doi: 10.1111/j.1365-3024.1987.tb00530.x. [DOI] [PubMed] [Google Scholar]
- 6.McKeever D. J., Taracha E. L., Innes E. L., MacHugh N. D., Awino E., Goddeeris B. M., Morrison W. I. Proc. Natl. Acad. Sci. USA. 1994;91:1959–1963. doi: 10.1073/pnas.91.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norval R. A. I., Perry B. D., Young A. S. The Epidemiology of Theileria in Africa. London: Academic; 1992. [Google Scholar]
- 8.Taracha E. L. N., Goddeeris B. M., Teale A. J., Kemp S. J., Morrison W. I. J. Immunol. 1995;155:4854–4860. [PubMed] [Google Scholar]
- 9.Goddeeris B. M., Morrison W. I. J. Tissue Cult. Methods. 1988;11:101–110. [Google Scholar]
- 10.Gardner M. J., Bishop R., Shah T., de Villiers E. P., Carlton J. M., Hall N., Ren Q., Paulsen I. T., Pain A., Berriman M., et al. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- 11.Shaw M. K. Trends Parasitol. 2003;19:2–6. doi: 10.1016/s1471-4922(02)00015-6. [DOI] [PubMed] [Google Scholar]
- 12.Nacer A., Berry L., Slomianny C., Mattei D. Int. J. Parasitol. 2001;31:1371–1379. doi: 10.1016/s0020-7519(01)00253-3. [DOI] [PubMed] [Google Scholar]
- 13.Toye P. G., Goddeeris B. M., Iams K., Musoke A. J., Morrison W. I. Parasite Immunol. 1991;13:49–62. doi: 10.1111/j.1365-3024.1991.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 14.Schneider J., Gilbert S. C., Hannan C. M., Degano P., Prieur E., Sheu E. G., Plebanski M., Hill A. V. Immunol. Rev. 1999;170:29–38. doi: 10.1111/j.1600-065x.1999.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 15.McConkey S. J., Reece W. H., Moorthy V. S., Webster D., Dunachie S., Butcher G., Vuola J. M., Blanchard T. J., Gothard P., Watkins K., et al. Nat. Med. 2003;9:729–735. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 16.Taracha E. L. N., Bishop R., Musoke A. J., Hill A. V. S., Gilbert S. C. Infect. Immun. 2003;71:6906–6914. doi: 10.1128/IAI.71.12.6906-6914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vordermeier H. M., Rhodes S. G., Dean G., Goonetilleke N., Huygen K., Hill A. V., Hewinson R. G., Gilbert S. C. Immunology. 2004;112:461–470. doi: 10.1111/j.1365-2567.2004.01903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aandahl E. M., Sandberg J. K., Beckerman K. P., Tasken K., Moretto W. J., Nixon D. F. J. Immunol. 2003;170:2349–2355. doi: 10.4049/jimmunol.170.5.2349. [DOI] [PubMed] [Google Scholar]
- 19.Rowlands G. J., Musoke A. J., Morzaria S. P., Nagda S. M., Ballingall K. T., McKeever D. J. Parasitology. 2000;120:371–381. doi: 10.1017/s0031182099005600. [DOI] [PubMed] [Google Scholar]
- 20.Preston P. M., Brown C. G. D., Richardson W. Parasite Immunol. 1992;14:125–141. doi: 10.1111/j.1365-3024.1992.tb00456.x. [DOI] [PubMed] [Google Scholar]
- 21.De Plaen E., Lurquin C., Lethe B., van der Bruggen P., Brichard V., Renauld J. C., Coulie P., Van Pel A., Boon T. Methods. 1997;12:125–142. doi: 10.1006/meth.1997.0462. [DOI] [PubMed] [Google Scholar]
- 22.Baumgartner M., Tardieux I., Ohayon H., Gounon P., Langsley G. Microbes Infect. 1999;1:1181–1188. doi: 10.1016/s1286-4579(99)00244-0. [DOI] [PubMed] [Google Scholar]
- 23.Jha K. K., Banga S., Palejwala V., Ozer H. L. Exp. Cell Res. 1998;245:1–7. doi: 10.1006/excr.1998.4272. [DOI] [PubMed] [Google Scholar]
- 24.Bensaid A., Kaushal A., Baldwin C. L., Clevers H., Young J. R., Kemp S. J., MacHugh N. D., Toye P. G., Teale A. J. Immunogenetics. 1991;33:247–254. doi: 10.1007/BF00230502. [DOI] [PubMed] [Google Scholar]
- 25.McShane H., Brooks R., Gilbert S. C., Hill A. V. Infect. Immun. 2001;69:681–686. doi: 10.1128/IAI.69.2.681-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccini A., Perkus M. E., Paoletti E. Methods Enzymol. 1987;153:545–563. doi: 10.1016/0076-6879(87)53077-4. [DOI] [PubMed] [Google Scholar]
- 27.Weinstock D., Bhudevi B., Castro A. E. J. Clin. Microbiol. 2001;39:343–346. doi: 10.1128/JCM.39.1.343-346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.