Abstract
The stem–loop-binding protein (SLBP) is involved in multiple aspects of histone mRNA metabolism. To characterize the modification status and sites of SLBP, we combined mass spectrometric bottom-up (analysis of peptides) and top-down (analysis of intact proteins) proteomic approaches. Drosophilia SLBP is heavily phosphorylated, containing up to seven phosphoryl groups. Accurate Mr determination by Fourier transform ion cyclotron resonance (FTICR)-MS and FTICR-MS top-down experiments using a variety of dissociation techniques show there is removal of the initiator methionine and acetylation of the N terminus in the baculovirus-expressed protein, and that T230 is stoichiometrically phosphorylated. T230 is highly conserved; we have determined that this site is also completely phosphorylated in baculovirus-expressed mammalian SLBP and extensively phosphorylated in both Drosophila and mammalian cultured cells. Removal of the phosphoryl group from T230 by either dephosphorylation or mutation results in a 7-fold reduction in the affinity of SLBP for the stem–loop RNA.
Keywords: Fourier transform ion cyclotron resonance mass spectrometry, histone mRNA
The metazoan replication-dependent histone mRNAs are the only eukaryotic mRNAs that are not polyadenylated. Instead, they end in a 26-nt sequence, which includes a stem–loop consisting of a six-base stem and four-base loop (1). The 3′ end of histone mRNA is formed by an endonucleolytic cleavage. The 3′ end of histone mRNA is required for translation of the mRNA (2) and participates in the regulation of histone mRNA half life (3). The major protein that interacts with the conserved 26-nt sequence is the stem–loop-binding protein (SLBP), a 31-kDa protein that is involved in multiple steps of histone mRNA metabolism (4). SLBP contains a novel RNA-binding domain (RBD), present only in SLBPs, and RBD is the only region of SLBP conserved among diverse metazoans (Caenorhabditis elegans, Drosophila, and vertebrates). SLBP binds to the nascent pre-mRNA transcript and helps recruit the U7 snRNP, resulting in cleavage of the pre-mRNA (5). SLBP then accompanies the mRNA to the cytoplasm (6, 7), where it is required for efficient histone mRNA translation as well as protecting the mRNA from degradation. In mammalian cells, SLBP is regulated during the cell cycle (8) and is responsible for the posttranscriptional component of the cell cycle regulation of histone mRNA (9).
SLBP is a phosphoprotein, and phosphorylation is required for its degradation at the end of S-phase in mammalian cells (9). In Drosophila SLBP (dSLBP), stoichiometric phosphorylation of four serines at the C terminus (10) is necessary for efficient histone pre-mRNA processing (5) and plays a role in stabilizing the structure of the dSLBP (11). SLBP is also extensively phosphorylated during vertebrate oocyte maturation (12, 13) and when protein synthesis is inhibited (6). The function of these phosphorylations is not known.
MS is useful for characterization of posttranslational modifications (PTM) in proteins because of its combination of sensitivity and specificity and its ability to obtain structural and sequence information, which allows the detection and identification of PTM sites in proteins at physiological levels (14). Currently, bottom-up approaches, in which peptides are obtained by proteolytic digestion of proteins, are used for identification of PTMs (15). The MS analysis of peptides has several advantages over the analysis of proteins. First, MS analyses of peptides are, in general, more sensitive than analyses of proteins. Second, because MS has only limited tolerance for salts and detergents, these contaminants need to be removed (or replaced for solubility reasons) before the MS analysis, especially if electrospray ionization MS is performed, which is itself more challenging and demanding for proteins than for peptides.
Comprehensive analyses of PTMs, however, also include quantitative information about the modification status of the protein and the stoichiometry of each modification site. This can be done by bottom-up approaches, often by using isotopically labeled peptides as internal standards (16). With recent improvements in Fourier transform ion cyclotron resonance (FTICR) MS technology and the development of new MS dissociation techniques, top-down proteomics offers a practical alternative (17–22). In top-down experiments, sequence information is obtained from analysis of the intact protein, and quantitative information about the stoichiometry and proportion of modifications can be obtained. The ultrahigh resolution and mass accuracy of FTICR-MS (23) allow interpretation of the complex mixture of nonenzymatic peptide fragments in the mass spectrum and unambiguous identification of the high molecular-weight peptide fragments obtained by MS sequencing of proteins (24, 25). As in the example shown in this paper, a large multiply phosphorylated peptide or protein may be detectable where a smaller multiply phosphorylated peptide may not be able to be ionized. This is because the additional nonphosphorylated residues mitigate the electronegativity of the phosphoryl group(s). Similarly, differences in proton affinities between the phosphorylated and unphosphorylated forms are also minimized by looking at an intact protein instead of a peptide, so semiquantitative information can be obtained by comparison of the peak heights or peak areas of the unmodified and variously phosphorylated forms of a large protein. FTICR-MS is further capable of performing multiple MS staging (MSn) by conducting several fragmentation techniques in series, which permits determination of the exact location of PTMs. Thus “top-down” sequencing is especially useful for large peptides or multiply phosphorylated peptides that do not contain convenient enzymatic cleavage sites and do not give good tandem MS (MS/MS) data with conventional ionization techniques.
In this study, we used top-down and bottom-up proteomic approaches to analyze the PTMs of SLBP, combining the advantages of both techniques. Using top-down proteomics, we determined the modification status and modification stoichiometry of dSLBP and human SLBP (hSLBP) expressed in baculovirus. These results directed the analysis of both SLBPs isolated from cultured cells, by bottom-up approaches, making use of the higher sensitivity and reduced requirement for protein purification of the bottom-up approach. The combined technologies indicated stoichiometric phosphorylation of T230 (numbering of dSLBP) in the highly conserved TPNK motif in the RBD. This modification contributes to high-affinity RNA binding.
Results
Identification of the Stoichiometrically Phosphorylated Threonine in the Conserved TPNK Motif.
We previously showed that dSLBP is stoichiometrically phosphorylated on multiple sites, both in embryos and cultured Drosophila cells, based on the change in mobility on gel electrophoresis after phosphatase treatment (26). Moreover, from previous Mr determinations of intact and truncated dSLBP proteins expressed in baculovirus, it was clear that there are multiple phosphorylation sites at the C terminus (5). We identified four serines in the C terminus that were phosphorylated (10). However, the Mr determined by quadrupole TOF-MS of these dSLBP proteins (including full-length dSLBP) was ≈9 Da lower than the calculated Mr (5). To explain this discrepancy and to determine all of the phosphorylation sites on each phosphorylated isoform, we performed top-down proteomics by using an FTICR mass spectrometer with ultra-high resolution and mass accuracy. In top-down experiments, intact proteins are intensively analyzed in the mass spectrometer; this means that no prior enzymatic or chemical protein digestions are required (27).
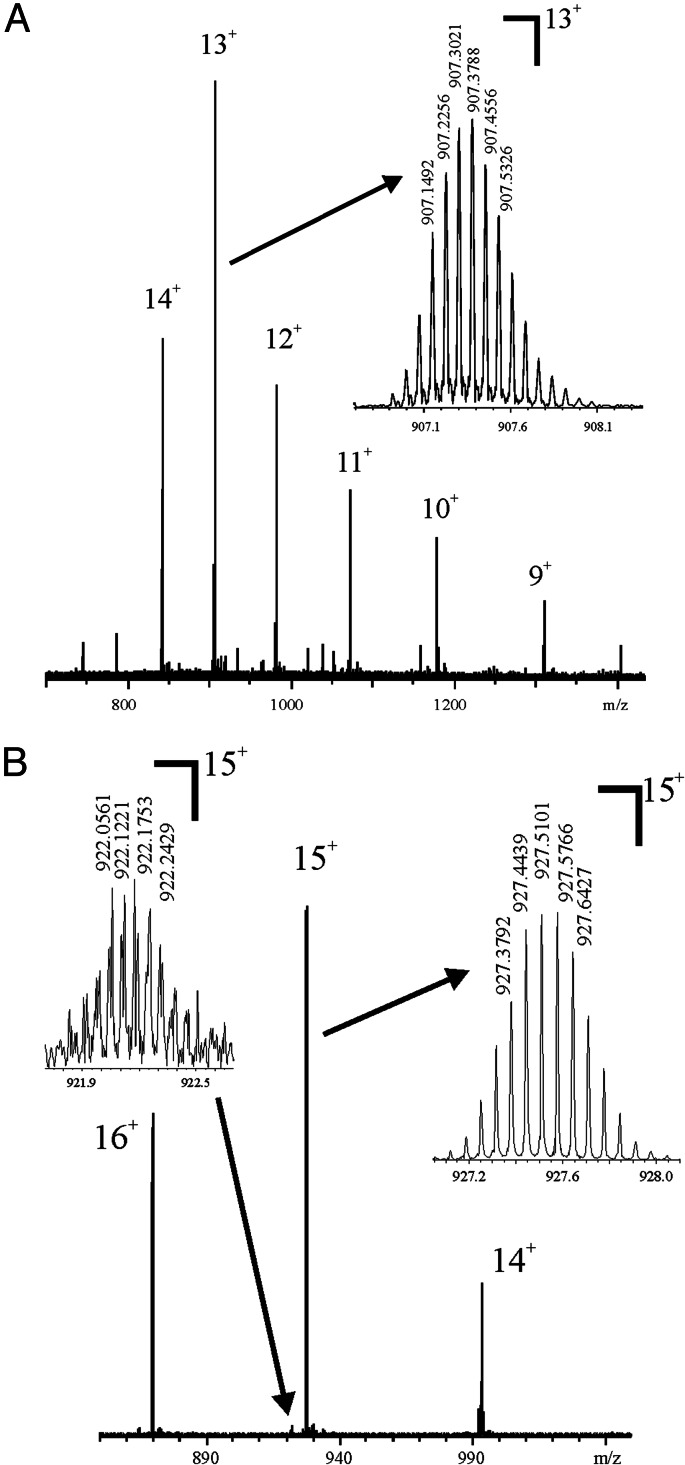
To determine the phosphorylation sites at the C terminus of dSLBP and to explain the mass discrepancy of dSLBP, baculovirus proteins containing the RBD (dSLBP-RBD, 98 amino acids, including the his-tag added by the vector), and proteins possessing the RNA-binding site plus the last 16 C-terminal amino acid residues [dSLBP-RNA-processing domain (RPD), 114 amino acids] were analyzed. The accurate Mr determination by FTICR-MS of dSLBP-RBD gave a Mr 8.997 Da lower than that predicted from the amino acid sequence (Fig. 1, Table 1). The Mr analysis of dSLBP-RPD showed two ion series corresponding to a Mr 239.94 (minor component) and 319.96 Da (major component) higher than the predicted Mr, corresponding to the incorporation of three and four phosphoryl groups, with the same discrepancy of 8.99 Da, thus suggesting there are three or four phosphoryl groups present in the last 16 C-terminal amino acid residues (5, 10).
Fig. 1.
dSLBP-RPD and -RBD are each a single major molecular species. The Mr determination of truncated his-tagged dSLBP proteins expressed in insect cells by FTICR-MS of (A) dSLBP-RBD and (B) dSLBP-RPD. The minor peak at m/z 922.2 has one fewer phosphoryl group than the major peak at m/z 927.5.
Table 1.
Accurate Mr determination of truncated dSLBP proteins by FTICR-MS
| Mrcal | Mrdet | Δm1 | Δm2 | Δm2/MrHPO3 | |
|---|---|---|---|---|---|
| dSLBP RBD | 11,791.82 | 11,782.83 | −8.99 | 0 | |
| dSLBP-RPD | 13,586.56 | 13,817.51 | 230.95 | 239.94 | 3.00 |
| dSLBP-RPD | 13,586.56 | 13,897.53 | 310.97 | 319.96 | 4.00 |
Δm1 = Mrdet − Mrcal. Δm2 = Mrdet − Mrcal − Δm1dSLBP RBD.
The hybrid portion of the (Qq) of the Qq-FTICR mass spectrometer allows selective fragmentation of the different phosphorylated isoforms. To determine the location of the phosphoryl sites and to explain the 9-Da discrepancy, ion signals corresponding to the putatively four times-phosphorylated dSLBP-RPD were selected for top-down experiments by collision-induced fragmentation in the quadrupole of the Qq-FTICR mass spectrometer followed by FTICR Mr determination of the fragment ions. Inspection of the lower Mr fragment ions revealed y19, y19-H3PO4, y19-2(H3PO4), confirming the presence of four phosphoryl groups at the C terminus. In addition, sequence tags corresponding to the N terminus of the truncated protein were detected that did not correspond to the b ions predicted from the amino acid sequence (Fig. 2A, Table 2). These fragment ions were further fragmented in triple mass spectrometry experiments by using electron capture dissociation (ECD) (data not shown) or skimmer-induced fragmentation followed by collision-induced dissociation (CID), and FTICR-MS in tandem (Fig. 2B). Analysis of these spectra by de novo sequencing revealed that the N-terminal methionine had been removed, and that the new N-terminal serine was acetylated. In addition, both C- and N-terminal ECD-generated fragments containing the TPNK region had higher than expected molecular weights, revealing an additional phosphorylation site identified as T230 of dSLBP from the z ion z47 (Fig. 2C).
Fig. 2.
Top-down analysis of dSLBP-RPD by FTICR-MS. (A) De novo sequencing of the 15+ charge state (m/z 927.5) of phosphorylated dSLBP-RPD, by CID outside the FTICR cell. The fragment ions did not match the predicted sequence, but de novo sequencing showed that they corresponded to a modified N terminus. (B) FTICR-MS3 analysis of dSLBP-RPD by skimmer-induced fragmentation and CID. The first MS stage was performed by skimmer-induced fragmentation, and ions with the mass corresponding to the doubly charged b13 ion (see Table 2) were selected for CID. biotools software assignments show that the N-terminal methionine has been removed (MSYYHHHHHHDYDI…), and the new N terminus is acetylated (Ac-SYYHHHHHHDYDI…). (C) ECD analysis of dSLBP-RPD. The phosphorylation site T230 was directly confirmed by the observation of the z ion z47. N-terminal (b ions by CID, c ions by ECD), C-terminal (y ions by CID, z ions by ECD), and abundant internal fragment ions are assigned.
Table 2.
Fragment ions from the top-down experiment of dSLBP RPD (Mrdet: 13,817.51 Da) localizing and identifying posttranslational modifications
| Fragment ions | Sequence | Δm1, ppm |
|---|---|---|
| y19 | PPTQARDTAKDSNSDSDSD* | 1.5 |
| y19-H3PO4 | [PPTQARDTAKDSNSDSDSD*] | 1.7 |
| y19-2x H3PO4 | [PPTQARDTAKDSNSDSDSD*] | 0.42 |
| b10 | Ac-SYYHHHHHHD | −0.48 |
| b12 | Ac-SYYHHHHHHDYD | −0.30 |
| b13 | Ac-SYYHHHHHHDYDI | −0.96 |
| b15 | Ac-SYYHHHHHHDYDIPT | −0.05 |
| b17 | Ac-SYYHHHHHHDYDIPTTE | −0.09 |
| b19 | Ac-SYYHHHHHHDYDIPTTENL | −0.60 |
| b20 | Ac-SYYHHHHHHDYDIPTTENLY | −0.60 |
| b21 | Ac-SYYHHHHHHDYDIPTTENLYF | −1.1 |
| b22 | Ac-SYYHHHHHHDYDIPTTENLYFQ | −0.63 |
*Four times phosphorylated.
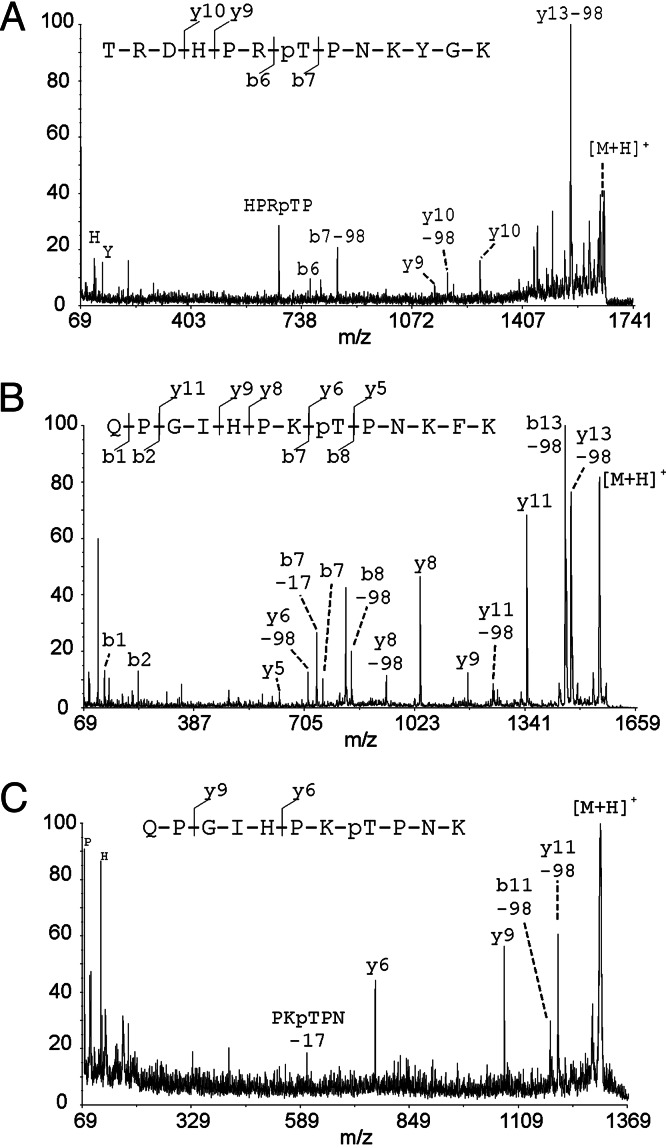
The observed discrepancy of 9 Da corresponds to the sum of the loss of the N-terminal methionine (−131) and the acetylation of the new N-terminal amino acid, serine (+42 Da), plus an additional phosphorylation (+80 Da). This phosphorylation site (T230) in dSLBP had been previously identified by bottom-up proteomics, but the techniques used in those experiments did not allow us to evaluate the extent of phosphorylation at this site (28). We confirmed that the additional phosphorylation was on T230 by analysis of tryptic digests of dSLBP. Based on the results from the FTICR intact Mr determination and top-down proteomics, we conclude that T230 is stoichiometrically phosphorylated (Fig. 1A) in baculovirus-expressed dSLBP, because no other ion series were detected. To determine whether T230 was phosphorylated in vivo, we digested the dSLBP-RPD purified from baculovirus and also full-length dSLBP purified from Drosophila cultured cells with trypsin. In both cases, a peptide was detected corresponding to phosphorylation of T230 (Fig. 3A shows the peptide from protein purified from cultured cells). The sequence was confirmed by sequencing with MALDI-MS/MS, whereas the corresponding unphosphorylated peptide was not detected (data not shown) in the digest of dSLBP purified from cultured cells or baculovirus, consistent with the dSLBP being heavily phosphorylated in T230 in cultured Drosophila cells.
Fig. 3.
Bottom-up analysis by MALDI-MS/MS of Drosophila and human SLBP reveals conserved phosphorylation at threonine in the TPNK motif. (A) MALDI-MS/MS spectrum of m/z 1649.8 (amino acids 224–236) from a tryptic digest of dSLBP isolated from Drosophila cultured cells. (B) MALDI-MS/MS spectrum of m/z 1571.8 (amino acids 164–176) from an Arg-C digest of hSLBP expressed in baculovirus (this peptide displayed anomalous cleavage by Arg-C after K176). (C) MALDI-MS/MS spectrum of m/z 1296.6 (amino acids 164–174) from an in-gel tryptic digest of hSLBP purified from HeLa cells.
Because T230 is present in a highly conserved TPNK sequence present in all SLBPs, we analyzed hSLBP to determine the phosphorylation status of the corresponding T171 in the TPNK region. The Mr determination of the hSLBP-RPD expressed in baculovirus by quadrupole TOF-MS also showed the 9-Da mass discrepancy (data not shown), suggesting that the N-terminal is acetylated, and that there is a stoichiometric phosphorylation. Because the N-terminal His tag is identical to His-tagged dSLBP, it is not surprising that it has the same N-terminal modification. Using bottom-up approaches by MS analysis of the Arg-C proteolytic digest of baculovirus-expressed hSLBP, we detected ion signals at m/z 1,571.8, corresponding to the mass of the phosphorylated hSLBP peptide 164QPGIHPKTPNKFK176 (resulting from anomalous cleavage at K176). This assignment was confirmed by sequencing analysis by using MALDI-MS/MS (Fig. 3B). The sequence-specific y- and b-ion fragment ions shown in the spectrum unambiguously localized the phosphorylation to T171. We did not observe the unphosphorylated form of this peptide in the digests of baculovirus-expressed hSLBP, consistent with only a single isoform of the protein being present in the baculovirus-expressed hSLBP.
We also determined the phosphorylation status of T171 in hSLBP purified from a HeLa cell lysate. We identified the phosphopeptide 164QPGIHPKTPNK174 in a tryptic digest using MALDI-MS/MS. The resulting spectrum displayed a similar fragmentation pattern to the MS/MS spectrum from the corresponding peptide from the baculovirus-expressed protein (Fig. 3C). Consistent with our results from baculovirus, we were unable to detect the unphosphorylated form of this peptide, suggesting that T171 is extensively phosphorylated in human as well as insect cells.
Effect of Phosphorylation at T230 on RNA Binding.
We have shown above that T230 in dSLBP (T171 in hSLBP) is phosphorylated in Drosophila and mammalian cultured cells, as well as in these proteins expressed in baculovirus. To determine whether this conserved modification plays a role in RNA binding, we expressed the full-length hSLBP, the 93-aa hSLBP-RPD, as well as full-length dSLBP, the T230A mutant dSLBP, and the dSLBP-RPD in baculovirus. The hSLBP-RPD contains a single phosphoryl group on T171, whereas the full-length human SLBP contains multiple phosphoryl groups (data not shown). To determine the effect of phosphorylation on RNA binding, we treated the hSLBPs with calf intestinal phosphatase. To assess the effect of the T230 phosphorylation in dSLBP, we used the T230A mutant, which lacks only the T230 phosphate, because the dSLBP also contains phosphoryl groups on the C-terminal portion of the RPD (5).
The hSLBP-RPD and dSLBP proteins bound the histone mRNA stem–loop from the mouse histone H4–12 gene (29) in a gel mobility-shift assay (Fig. 4B). Dephosphorylation of the SLBP proteins had no discernible effect on the extent of RNA complex formation, although there was a change of mobility in the human RPD complex as a result of dephosphorylation (Fig. 4B, lanes 2 and 3), which allowed us to conclude that dephosphorylation was complete. We measured the affinity of the SLBP proteins for the stem–loop RNA by using a filter-binding assay. Our affinity measurements are in good agreement with values previously reported by Battle and Doudna for xSLBP1 determined under similar conditions (17). The Kd of baculovirus expressed phosphorylated hSLBP for stem–loop RNA is 1.5 ± 0.2 nM, and that of dSLBP is 1.6 ± 0.3 nM. In addition, all determinants for RNA binding reside within the RPD for both hSLBP and dSLBP, because both RPDs bind with similar affinity to the stem–loop as the full-length protein (Table 3). Baculovirus-expressed dephosphorylated forms of hSLBP had a 7-fold lower affinity for the stem–loop than the proteins with T230A phosphorylated (Fig. 4C, Table 3). The Kd values for dephosphorylated hSLBP proteins were also similar to the Kd value determined for the dSLBP T230A–RNA interaction (Table 3), indicating that the T230 phosphate contributes 1- to 1.2-kcal·mol−1 to the overall affinity of the complex.
Fig. 4.
Effect of phosphorylation on the affinity of SLBP for stem–loop RNA. (A) Schematic of proteins used for RNA-binding measurements. (B) An electrophoretic mobility-shift assay of hSLBP and dSLBP proteins with a 26-nt hairpin from mouse histone H4–12 mRNA. Lane 1 is free probe. Lanes 2–5 are reactions with baculovirus-expressed hSLBP-RPD, dephosphorylated hSLBP-RPD, dSLBP, and T230A dSLBP, respectively. (C) Representative plots of fraction of protein bound to RNA as a function of increasing SLBP concentration are shown. The data were fit to a standard two-state binding model, as described in Materials and Methods.
Table 3.
Dissociation constants and binding free energies of human and Drosophila SLBP proteins for the stem–loop RNA
| Protein | KD, nM | KDmut/KDwt* | ΔΔGbind* (kcal·mol−1) |
|---|---|---|---|
| Human SLBP | |||
| Phosphorylated full-length wild type | 1.52 ± 0.16 | — | — |
| Dephosphorylated full- length wild type | 10.53 ± 0.18 | 6.93 | 1.15 |
| Phosphorylated RPD | 1.25 ± 0.29 | 0.82 | 0.18 |
| Dephosphorylated RPD | 11.36 ± 0.33 | 7.47 | 1.19 |
| Drosophila SLBP | |||
| Phosphorylated full-length wild type | 1.55 ± 0.32 | — | — |
| T230A full-length | 8.99 ± 0.42 | 5.8 | 1.04 |
| RPD | 2.00 ± 0.19 | 1.29 | 0.15 |
*The KDmut/KDwt and the ΔΔG values reported are calculated for mutant proteins relative to the corresponding wild-type isoform at 25°C.
†RPD refers to the RNA-binding and processing domain extending from residues V124 to A218 of hSLBP and residues H174 to D276 of dSLBP. Both proteins have a C-terminal extension that is required for efficient RNA processing in vitro. The dSLBP RBD refers to the minimal RNA-binding domain extending from residues E189 to P259 that lacks the phosphorylated C terminus.
Thus, both hSLBP and dSLBP are extensively phosphorylated at the conserved TPNK site in the RBD. This phosphorylation affects both the RNA-binding properties of hSLBP and dSLBP as well as the structural stability of dSLBP (11).
Discussion
Biological Implications of T230 Phosphorylation.
Phosphorylation on T230 is also essential for the function of dSLBP in vivo, because a null mutant of dSLBP, which is not viable, is not rescued by T230A dSLBP (30). The dephosphorylated form of hSLBP binds the 3′ end of histone mRNA with ≈10-fold lower affinity than the phosphorylated form. This lower affinity likely results in a reduced efficiency of histone premRNA processing in vivo. Mutations in the stem–loop of a mammalian histone gene that reduce the affinity of the mRNA for SLBP by 10-fold or more (17) result in a much lower expression of histone mRNA from that gene (31). These same mutations in the stem–loop also decreased processing of histone pre-mRNA in an in vitro system from mammalian cells (32). This suggests that phosphorylation of the threonine in the RBD is required for efficient histone pre-mRNA processing both in vivo and in vitro.
Our studies of sea urchin SLBP also emphasize the role of the amino acids around the TPNK in RNA binding. The sea urchin SLBP, which has a different sequence around the TPNK than Drosophila or mammalian SLBP, does not bind the stem–loop RNA with high affinity when it is expressed in baculovirus or in reticulocyte lysates (33). Possibly the sea urchin SLBP is not phosphorylated by the kinase, as yet unknown, present in insect cells that modifies the mammalian and Drosophila TPNK.
Mechanism of SLBP–RNA Binding.
In our previous studies of the SLBP–RNA interaction, we reported that the minimal dSLBP-RPD that is necessary and sufficient for high-affinity interaction with the histone mRNA hairpin is a molten globule that is stabilized via interactions with histone stem–loop RNA (11). In addition, thermal denaturation studies using circular dichroism together with chemical-shift perturbations in the 31P NMR spectrum of the phosphorylated dSLBP–RNA complex suggested that one or more of the dSLBP phosphoryl groups contributes favorably to the overall stability of the SLBP–RNA complex. However, because the precise sites of phosphorylation had not yet been characterized, we were not able to ascertain the energetic contributions of the individual phosphoryl groups to RNA complex formation. We have now demonstrated there are five phosphoryl groups on the dSLBP-RPD: the four serines in the C-terminal 17 amino acids as well as T230 in the RBD (10). The exact role of threonine phosphorylation on the structure of the SLBP–RNA complex remains to be elucidated.
In this study, we have combined top-down and bottom-up proteomics and have used the advantages of both approaches to obtain information on modification sites. Using both approaches, we have identified a stoichiometric phosphorylation, which is necessary for high-affinity binding to RNA, in a conserved motif of SLBP.
Materials and Methods
Protein Isolation, Expression, and Purification.
Full-length human and Drosophila SLBP wild-type full-length T230A mutant dSLBP, the dSLBP RNA-binding domain, and the hSLBP and dSLBP-RPD were expressed in Sf9 cells by using a baculovirus pFastBac HTa (GIBCO/BRL) expression system, as described (5). All proteins have an N-terminal His tag and were purified by using standard Ni2+-affinity chromatography (IMAC) on Ni-NTA agarose (Qiagen, Germantown, MD). Before MS analysis, proteins were purified on microcolumns packed with POROS R1 (C4) resin (Applied Biosystems) (34). Endogenous hSLBP and dSLBP were purified from HeLa cell extracts or S2 cell extracts by affinity purification by using a biotinylated stem–loop, as described (35).
MS Analysis.
The Mr of intact proteins was determined on an apex-Qe 94 Qq-FTICR (Bruker Daltonics, Billerica, MA) equipped with a 9.4-T actively shielded magnet and an Apollo (Bruker Daltonics) electrospray ionization source. ECD experiments were performed on a similarly configured apex-Qe at a 12-T magnetic field strength. Proteins were eluted from the POROS R1 column with 50:50 methanol:2% acetic acid (vol:vol) and diluted 1:10 with water, resulting in a protein concentration of ≈0.1 mg/ml. FTICR analyses were carried out by direct infusion by using a syringe pump with a flow rate of ≈1 μl/min. A total of 50 scans at a scan rate of 3 s per scan were collected.
For top-down analysis by Qq-FTICR, multiply charged ions of the intact protein were selected (by the mass-selective quadrupole, Q) as precursor ions for CID, which was carried out in the collision cell “q” by using argon as the collision gas at a pressure of 1 × 10−3 mbar. Mass analysis of the fragment ions was carried out in the FTICR region of the mass spectrometer by using 3-s acquisitions. MS3 top-down experiments by Qq-FTICR were performed on fragment ions obtained by skimmer-induced fragmentation of the protein followed by CID and FTICR analysis, as described above. For ECD of the intact dSLBP mutants, a hollow dispenser cathode was operated at 1.7 A, and the bias was gated from +5 V (quiescent) to −1.35 V during the electron irradiation event, which lasted 500 ms. To maximize the ion population before the irradiation event, gated trapping was used, and the cell was filled with 12 iterations of ion accumulation from the external collision cell of the isolated precursor. This experimental design minimized the storage time at high pressure, which could have led to charge reduction. MS spectra were analyzed by using the biotools data software package from Bruker Daltonics.
hSLBP-RPD expressed in baculovirus (≈3.5 μg) was analyzed by bottom-up approaches after purification of the protein by C4 Zip-tip (Millipore) and digestion with endoprotease Arg-C (Sigma–Aldrich) in 50 mM ammonium bicarbonate at an enzyme/substrate ratio of 1:10 at 36°C for 16 h. The resulting peptides were desalted with a C18 Zip-tip, eluted with 50% MeCN/0.1% trifluoroacetic acid, and ≈3% of the eluate (≈100 ng) was analyzed by MALDI-MS and MS/MS by using an ABI Voyager (Applied Biosystems) 4700 MALDI-TOF/TOF mass spectrometer, as described (36). Purified endogenous hSLBP was separated by SDS/PAGE and stained with Coomassie, and the protein (<1 μg) was in-gel-digested with trypsin (36). The extracted phosphopeptides were analyzed by MALDI-MS and -MS/MS, as described (37).
Nitrocellulose Filter-Binding Assays for Measurement of Equilibrium Dissociation Constants.
Equilibrium-binding reactions were performed by incubating serial dilutions of the protein with labeled RNA (17). The reactions were incubated overnight at 4°C to equilibrate. The protein–RNA reaction mixtures were passed through a 0.45-mm nitrocellulose filter to capture the SLBP–RNA complex followed by a Hybond-n + positively charged filter to capture the free RNA. The filters were washed twice, allowed to air dry, and exposed to a Molecular Dynamics PhosphorImager. The spots were quantified by using the program imagequant, normalized to background, and analyzed by using Microsoft excel and kaleidagraph. Kd values were obtained by least squares fitting the data to the equation
 |
where f is the fraction of RNA bound, fmax is the filter-binding capacity, P is the concentration of active protein, and c is the x-intercept. The fit assumes a 1:1 stoichiometry. The amount of active protein was determined by using an SLBP activity assay, as described (17). All Kd values reported are an average of three independent experiments with different protein samples plus or minus the standard deviation.
Acknowledgments
This study was supported by a gift from an anonymous donor; a Ford Foundation Diversity Fellowship (to M.P.T.); and National Institutes of Health Grants GM29832 and GM58921 (to W.F.M.) and 2P30ES010126, 5P30CA016086, and 1S10RR19889 (to C.H.B.).
Abbreviations
- CID
collision-induced dissociation
- ECD
electron capture dissociation
- FTICR
Fourier transform ion cyclotron resonance
- MS/MS
tandem MS
- PTM
posttranslational modification
- SLBP
stem–loop-binding protein
- dSLBP
Drosophila SLBP
- hSLBP
human SLBP
- RBD
RNA-binding domain
- RPD
RNA processing domain.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Dominski Z., Marzluff W. F. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez R., Marzluff W. F. Mol. Cell. Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey N. B., Marzluff W. F. Mol. Cell. Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzluff W. F., Duronio R. J. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 5.Dominski Z., Yang X.-C., Raska C. S., Santiago C., Borchers C. H., Duronio R. J., Marzluff W. F. Mol. Cell. Biol. 2002;22:6648–6660. doi: 10.1128/MCB.22.18.6648-6660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitfield M. L., Kaygun H., Erkmann J. A., Townley-Tilson W. H. D., Dominski Z., Marzluff W. F. Nucleic Acids Res. 2004;32:4833–4842. doi: 10.1093/nar/gkh798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erkmann J. A., Wagner E. J., Dong J., Zhang Y., Kutay U., Marzluff W. F. Mol. Biol. Cell. 2005;16:2960–2971. doi: 10.1091/mbc.E04-11-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield M. L., Zheng L. X., Baldwin A., Ohta T., Hurt M. M., Marzluff W. F. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng L., Dominski Z., Yang X. C., Elms P., Raska C. S., Borchers C. H., Marzluff W. F. Mol. Cell. Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres M. P., Marzluff W. F., Borchers C. H. J. Proteome Res. 2005;4:1628–1635. doi: 10.1021/pr050129d. [DOI] [PubMed] [Google Scholar]
- 11.Thapar R., Marzluff W. F., Redinbo M. R. Biochemistry. 2004;43:9401–9412. doi: 10.1021/bi036315j. [DOI] [PubMed] [Google Scholar]
- 12.Allard P., Champigny M. J., Skoggard S., Erkmann J. A., Whitfield M. L., Marzluff W. F., Clarke H. J. J. Cell Sci. 2002;115:4577–4586. doi: 10.1242/jcs.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller B., Link J., Smythe C. J. Biol. Chem. 2000;275:24284–24293. doi: 10.1074/jbc.M003253200. [DOI] [PubMed] [Google Scholar]
- 14.Kuster B., Mann M. Curr. Opin. Struct. Biol. 1998;8:393–400. doi: 10.1016/s0959-440x(98)80075-4. [DOI] [PubMed] [Google Scholar]
- 15.Nemeth-Cawley J. F., Tangarone B. S., Rouse J. C. J. Proteome Res. 2003;2:495–505. doi: 10.1021/pr034008u. [DOI] [PubMed] [Google Scholar]
- 16.Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle D. J., Doudna J. A. RNA. 2001;7:642–643. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogorzalek-Loo R. R., Hayes R., Yang Y., Hung F., Ramachandran P., Kim N., Gunsalus R., Loo J. A. Int. J. Mass Spectrom. 2005;240:317–325. [Google Scholar]
- 19.Strader M. B., VerBerkmoes N. C., Tabb D. L., Connelly H. M., Barton J. W., Bruce B. D., Pelletier D. A., Davison B. H., Hettich R. L., Larimer F. W., et al. J. Proteome Res. 2004;3:965–978. doi: 10.1021/pr049940z. [DOI] [PubMed] [Google Scholar]
- 20.Bergquist J. Curr. Opin. Mol. Ther. 2003;5:310–314. [PubMed] [Google Scholar]
- 21.VerBerkmoes N. C., Bundy J. L., Hauser L., Asano K. G., Razumovskaya J., Larimer F., Hettich R. L., Stephenson J. L., Jr J. Proteome Res. 2002;1:239–252. doi: 10.1021/pr025508a. [DOI] [PubMed] [Google Scholar]
- 22.Bogdanov B., Smith R. D. Mass Spectrom. Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 23.Borchers C. H., Marquez V. E., Schroeder G. K., Short S. A., Snider M. J., Speir J. P., Wolfenden R. Proc. Natl. Acad. Sci. USA. 2004;101:15341–15345. doi: 10.1073/pnas.0406781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith R. D. Comp. Funct. Genom. 2002;3:143–150. doi: 10.1002/cfg.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Masselon C. D., Anderson G. A., Pasa-Tolic L., Lee S.-W., Shen Y., Zhao R., Lipton M. S., Conrads T. P., Tolic N., et al. Anal. Chem. 2001;73:3312–3322. doi: 10.1021/ac010192w. [DOI] [PubMed] [Google Scholar]
- 26.Lanzotti D. J., Kaygun H., Yang X., Duronio R. J., Marzluff W. F. Mol. Cell. Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelleher N. L., Lin H. Y., Valaskovic G. A., Aaserud D. J., Fridriksson E. K., McLafferty F. W. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 28.Raska C. S., Parker C. E., Dominski Z., Marzluff W. F., Glish G. L., Pope R. M., Borchers C. H. Anal. Chem. 2002;74:3429–3433. doi: 10.1021/ac0111199. [DOI] [PubMed] [Google Scholar]
- 29.Meier V. S., Boehni R., Schuemperli D. Nucleic Acids Res. 1989;17:795. doi: 10.1093/nar/17.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanzotti D. J., Kupsco J. M., Yang X.-C., Dominski Z., Marzluff W. F., Duronio R. J. Mol. Biol. Cell. 2004;15:1112–1123. doi: 10.1091/mbc.E03-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey N. B., Williams A. S., Sun J. H., Brown V. D., Bond U., Marzluff W. F. Mol. Cell. Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominski Z., Zheng L. X., Sanchez R., Marzluff W. F. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson A. J., Howard J. T., Dominski Z., Schnackenberg B. J., Sumerel J. L., McCarthy J. J., Coffman J. A., Marzluff W. F. Nucleic Acids Res. 2004;32:811–818. doi: 10.1093/nar/gkh193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kast J., Parker C. E., van der Drift K., Dial J. M., Milgram S. L., Wilm M., Howell M., Borchers C. H. Rapid Commun. Mass Spectrom. 2003;17:1825–1834. doi: 10.1002/rcm.1114. [DOI] [PubMed] [Google Scholar]
- 35.Dominski Z., Yang X.-C., Kaygun H., Dadlez M., Marzluff W. F. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 36.Parker C. E., Warren M. R., Loiselle D. R., Dicheva N. N., Scarlett C. O., Borchers C. H. In: Methods in Molecular Biology. Patterson W. C., Cyr D. M., editors. Vol. 301. Patterson, NJ: Humana; 2005. pp. 117–151. [DOI] [PubMed] [Google Scholar]
- 37.Hall M. C., Warren E. N., Borchers C. H. Cell Cycle. 2004;3:1278–1284. doi: 10.4161/cc.3.10.1153. [DOI] [PubMed] [Google Scholar]