Abstract
Rapid quantitative methods for characterizing small molecules, peptides, proteins, or RNAs in a broad array of cellular assays would allow one to discover new biological activities associated with these molecules and also provide a more comprehensive profile of drug candidates early in the drug development process. Here we describe a robotic system, termed the automated compound profiler, capable of both propagating a large number of cell lines in parallel and assaying large collections of molecules simultaneously against a matrix of cellular assays in a highly reproducible manner. To illustrate its utility, we have characterized a set of 1,400 kinase inhibitors in a panel of 35 activated tyrosine-kinase-dependent cellular assays in dose–response format in a single experiment. Analysis of the resulting multidimensional dataset revealed subclusters of both inhibitors and kinases with closely correlated activities. The approach also identified activities for the p38 inhibitor BIRB796 and the dual src/abl inhibitor BMS-354825 and exposed the expected side activities for Glivec/STI571, including cellular inhibition of c-kit and platelet-derived growth factor receptor. This methodology provides a powerful tool for unraveling the cellular biology and molecular pharmacology of both naturally occurring and synthetic chemical diversity.
Keywords: drug discovery, high-throughput screening, tyrosine kinase
The ability to simultaneously interrogate the activities of a library of molecules against a large panel of cellular assays would provide a rapid efficient means to begin to characterize and correlate the biological properties of both natural and synthetic chemical diversity. For example, libraries of noncoding RNAs, mutant growth factors, small molecule kinase inhibitors, or even existing drugs could be assayed for their potency and selectivity in pathway-based or receptor screens or toxicity and metabolic stability in diverse cell types to discover a new biological activity or optimize the pharmacological properties of a molecule (1–3). Although whole-cell systems represent an attractive milieu to characterize gene and small-molecule function, no robust and systematic method exists to correlate chemical structure and biological activity across a large number of molecules and cellular assays. To address this problem, we have developed an approach that affords rapid cost-effective broad-based cellular profiling in parallel against molecular libraries. An industrial-scale automated compound profiling (ACP) system has been designed, which consists of an automated tissue culture system for propagating cell lines, integrated with a system for automatically performing miniaturized cell-based assays in 384- or 1,536-well microplates. The ACP can rapidly test thousands of arrayed molecules, in replicates, in dose–response format against hundreds of unique cellular assays in a single experiment.
To demonstrate this capability, we focused on the problem of identifying selective small-molecule inhibitors of protein tyrosine kinases. Tyrosine kinases play a key role in many cellular processes, including development, differentiation, and proliferation; misregulation of tyrosine kinase expression and activity leads to a number of disorders, most notably cancer (4, 5). For example, spontaneous translocations in which tyrosine kinase domains become fused to other genes (such as Bcr, Tel, and NPM) have been identified as the etiological basis for multiple B cell lymphomas, including the Philadelphia chromosome Bcr-Abl and chronic myelogenous leukemia (6, 7). It remains a significant challenge to develop selective inhibitors for tyrosine kinases given their homology and potential structural plasticity. To this end, we have established a panel of tyrosine-kinase-dependent cellular assays by creating stably expressed Tel-tyrosine kinase fusions representing each branch of the tyrosine kinase phylogeny. A library of >1,000 kinase-directed heterocycles was then profiled against this panel of cellular assays. The resulting dataset was examined for global structure–activity relationships (SARs), potency and selectivity correlations, and opportunistic side activities.
Results and Discussion
Construction of an ACP System.
A robotic system was designed that combines cell culture processes with compound screening capabilities to create a massively parallel compound profiling system. This system mechanically integrates all steps involved in cell line maintenance, including propagation (passaging, splitting, cell density, and viability determination) with the steps involved in performing compound screens (cell dispensing into microtiter plates, compound addition, incubation, detection, reagent addition, and plate reading/imaging), as described in Materials and Methods and as depicted in Fig. 1. The design specifications include the ability to rapidly profile thousands of compounds in dose–response format in miniaturized 1,536-well plate format against hundreds of robotically maintained cell-based assays in a highly reproducible way. The focus on cell maintenance required strict environmental control of humidity, temperature, sterility, and cell line cross-contamination, completely distinct from those confronted using biochemical/protein assays. The combination of automated tissue culture and robotic assay technologies enables small-molecule screens to be performed on an unprecedented scale in cell-based formats. This system can also be adapted to screen other molecular libraries, including secreted peptides and proteins, antibodies, cDNAs, and siRNAs against collections of cellular assays targeting either specific gene families (kinases, G-protein-coupled receptors, proteases, nuclear hormone receptors, etc.), signaling pathways (using reporter gene, phenotypic, and image-based readouts), or pharmacological properties (metabolic stability, cellular toxicity, or transport).
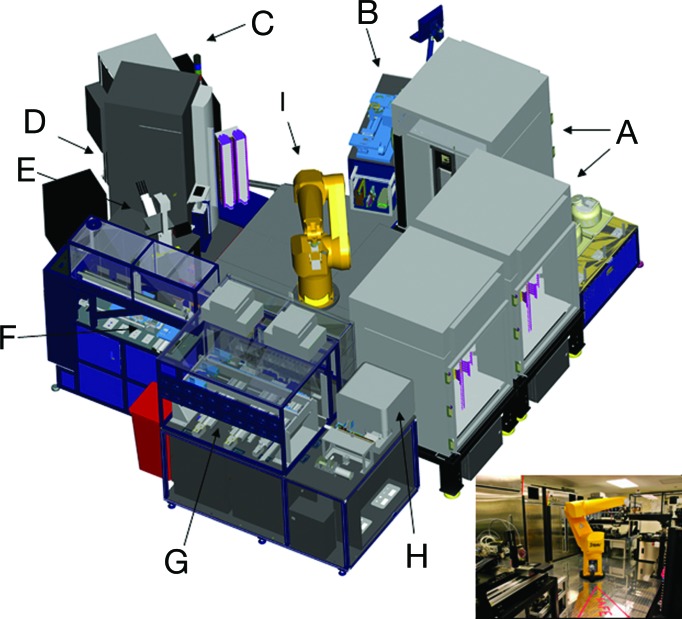
Fig. 1.
The components of the ACP system. (A) Three custom environmentally controlled (temperature, CO2, and humidity) 486-plate or flask capacity incubators. (B) Combination flask to 1,536-well plate cell and reagent dispenser. (C) Control station consisting of a computer running custom scheduling software. (D) Perkin–Elmer ViewLux plate reader. (E) Olympus (Melville, NY) inverted microscope with an automated stage and Molecular Devices metamorph software. (F) Compound transfer station (typically 5- to 50-nl transfer volumes). (G) Proprietary tissue culture (TC) station. (H) BD Biosciences FACSArray Bioanalyzer with custom sampling tray. (I) Staubli RX130 anthropomorphic robot with a custom end effector (gripper).
Generation and Validation of a Tel-Fused Tyrosine Kinase Panel in Ba/F3 Cells.
To illustrate the utility of ACP, we chose to profile a panel of kinase-dependent cellular assays against a collection of kinase-directed heterocycles. Although parallel interrogations of the kinome against chemical space have been performed by using in vitro biochemical and phage-display assays, the cell-based format described here assays the physiological conformation of the kinase in the presence of other cellular components, cell permeability, and nonselective cellular toxicity (6, 7). To profile tyrosine kinases in a cell-based format, we made use of the well established fact that kinases can be constitutively activated by genomic rearrangements leading to the juxtaposition of a fusion partner to a kinase (8). Several chimeric tyrosine kinases, including Bcr-Abl, NPM-Alk, and ETV6-NTRK3, have been described and shown to be causative to human cancer and hematopoietic malignancies. Frequently found fusion partners include structural proteins as well as transcription factors or genes of unknown functions. The most frequent fusion partner for tyrosine kinases is ETV6/Tel, a gene that has been found in chimeras with both cytosolic and receptor tyrosine kinases (Abl, NTRK3, PDGFR, and Jak2) (9–12), suggesting that fusion to ETV6/Tel might be a generally applicable strategy to activate tyrosine kinases.
To generate a cDNA library of kinases fused to ETV6/Tel, a retroviral plasmid based on a pMSCV backbone was constructed that allows rapid in-frame cloning of a kinase domain upstream of a Tel cassette and downstream of a Myc tag using a Gateway cassette system (Fig. 2a). A total of 81 Tel fusion vectors representing individual kinases, as well as the control non-Tel fusion Bcr-Abl, were established and used to transform Ba/F3 cells as described in Material and Methods. Ba/F3 is a hematopoietic cell line that depends on IL-3 for proliferation and survival but is rendered IL-3-independent by transformation with known tyrosine kinase oncogenes, e.g., Bcr-Abl, Flt3, and NPM-Alk (13–15). This cell line model has frequently been used to demonstrate the differential cytotoxicity of kinase inhibitors against transformed IL-3-independent cells versus parental Ba/F3 cells grown in the presence of IL-3 (16–18).
Fig. 2.
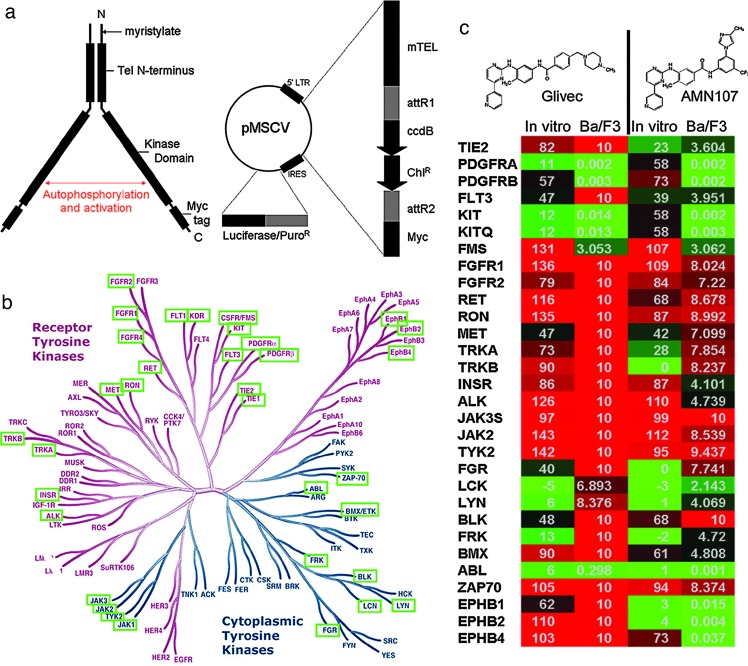
Ba/F3 construction. (a) Retroviral construct for Ba/F3-TK library. pTK is based on the pMSCV (Clontech) backbone. The first 1 kb of ETV6, with an artificial myristoylation sequence (mTel), is located upstream of the Gateway reading frame B cassette (Invitrogen), which is followed by the Myc tag sequence EQKISEEDL and a stop codon. This is linked by the HCV IRES to puromycin in which the stop codon has been replaced by the luciferase coding sequence from pLucN2 (Clontech). The expressed protein, a functional dimer, is detailed to the left. Dimerization of the mTel domains bring the two fused kinase domains into proximity, leading to autophosphorylation and activation. (b) The tyrosine kinome. The tyrosine kinase-dependent Ba/F3 lines used in this screen are highlighted in green boxes against a phylogeny of all 90 tyrosine kinases reproduced courtesy of Cell Signaling Technology (Danvers, MA). (c) Cellular and biochemical assays. The Bcr-Abl-targeted kinase inhibitors Glivec and AMN107 were tested against the kinase cell line panel (Ba/F3) and the purified enzyme (in vitro). Data for Ba/F3 are listed as the IC50 in μM, whereas the in vitro data are listed as the percent enzymatic activity remaining at 10 μM compound. The boxes are colored in green (potent inhibition), black (mild inhibition), or red (little to no inhibition).
After selection for puromycin and IL-3 withdrawal, 35 of the 81 fusion kinase constructs gave rise to unique cell populations exhibiting growth factor independence. Fig. 2b summarizes the identity of the kinases found to induce growth factor independence in Ba/F3 cells. All cells were further validated as described (Materials and Methods) by Western blot and RT-PCR/sequencing of the respective fusion and by using sets of reference compounds for each individual kinase. In the latter case, two independently generated cultures of IL-3-independent cells were required to show similar responses to reference compounds. To determine whether kinase inhibitor activity was accurately reflected by the Tel-kinase-transformed Ba/F3 cells, a collection of ≈80 tyrosine kinase inhibitors with known activities and selectivities was tested in dose–response format against the cell panel. For example, the Sugen c-Met inhibitor PHA-665752 inhibited the growth of Ba/F3/Tel-Met with an IC50 of 80 nM and showed no toxicity toward the rest of the panel, in agreement with published results (19). Likewise, the kinase-insert domain receptor (KDR) inhibitor AAL993 (20) shows selective nanomolar potency against Ba/F3/Tel-KDR and the homologous VEGF receptor family member FLT4. Known BCR-ABL inhibitors, STI571 and AMN107, were also tested against the Ba/F3 Tel-TK panel and a panel of recombinant enzymes. As shown in Fig. 2c, both STI571 and AMN107 inhibited Bcr-Abl, c-Kit, and PDGF-RA/B with IC50s between 1 and 100 nM as expected [data are shown as percent activity of the enzyme versus control at a single concentration of 10 μM and was compared to the inhibitory concentrations (IC50) generated by using Ba/F3 cell proliferation assays]. In addition, AMN107 also inhibited the proliferation of Ba/F3 cells expressing Tel fusions of the receptor kinases EphB1, B2, and B4, suggesting that the increased potency of AMN107 compared with STI571 results in somewhat decreased selectivity. Interestingly, several discrepancies between the cellular and enzymatic assays were found. For example, although very potent on both PDGF-RA/B in cellular assays, both STI571 and AMN107 only partially inhibited PDGF-RA/B in the enzymatic assays. A similar observation was made for EphB4 and AMN107. This is best explained by the fact that both inhibitors were shown to bind the inactive conformations of kinases, which might constitute only a fraction of the enzyme preparations used. Finally, some of the activities seen in the enzymatic assays could not be reproduced using cellular assays. IC50 measurements for these enzymes would be required to draw more detailed conclusions.
Kinase Profiling Experiment.
A chemical library totaling 1,400 unique small molecules targeting tyrosine kinases was selected that includes purines, pyrimidines, benzoimidazoles, and quinazolines (21). The library was supplemented with an additional 10 known tyrosine kinase inhibitors as controls and plated into 1,536-well microtiter plates prediluted into dose–response format ranging from 10 μM down to 3 nM. Each individual population of Ba/F3-TK cells was seeded on the automated tissue culture station, propagated for two to three passages; checked for cell viability, growth rates, and cell density; and then plated into 1,536-well assay plates on the robotic platform. The preplated compounds were transferred to assay plates in register (1:1) by using a 1,536-pin transfer device, incubated overnight, then assayed for cell viability as described in Materials and Methods. The experiment was repeated in triplicate resulting in ≈1.5 million data points. To compare the data quality generated on the robotic system with data generated manually with workstations, a subset (936) of the compounds was tested against 20 of the Ba/F3-TK cells in 384-well assay plate format in duplicate. Analysis of the resulting datasets showed that for both the ACP and the manual method, >95% of the data had coefficients of variation of <10%. By defining outliers as those IC50s with >3-fold or less than one-third of the average IC50, only two outliers of 936 data points were observed in the ACP dataset relative to the manual dataset.
Global Profile Data Analysis.
An examination of the 1,400 compounds tested demonstrated that only 30 (2.1%) were toxic to wild-type IL-3-dependent Ba/F3 cells <1 μM; ≈400 compounds showed slight of toxicity at micromolar concentrations. Interestingly, 282 (20.1%) compounds did not affect the activity of any TK-dependent cell line <5 μM; however, every kinase on the panel was inhibited by at least one compound. Finally, 83 (5.9%) compounds selectively inhibited a single kinase in the panel.
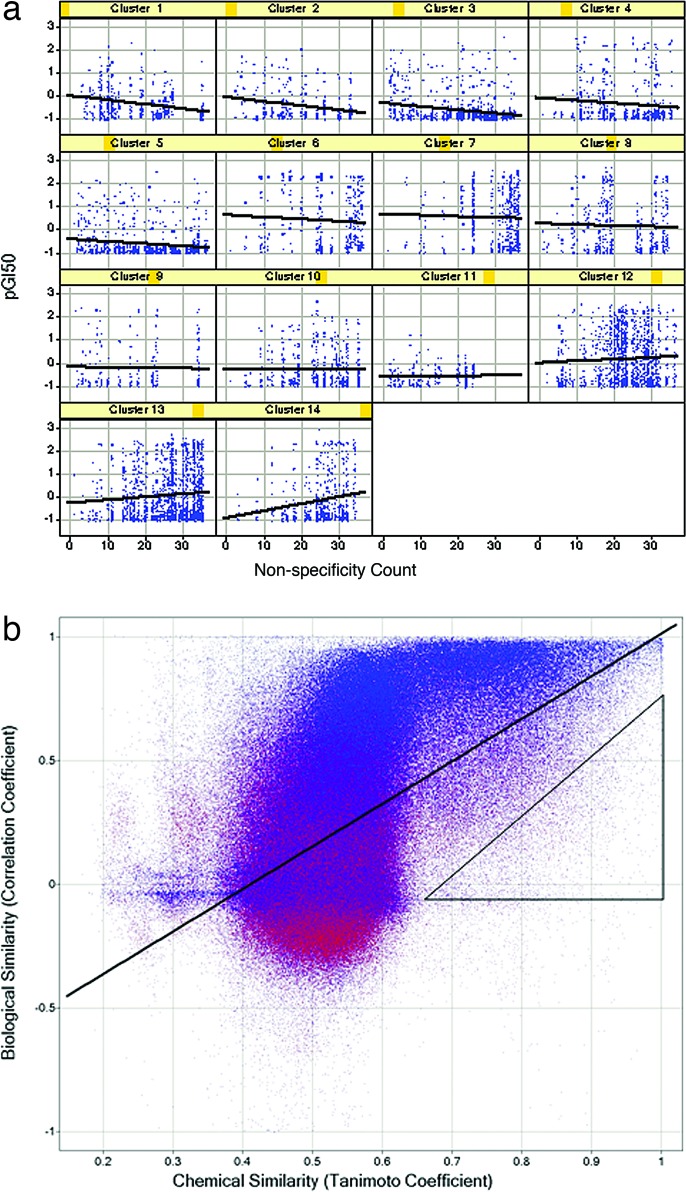
Typically, as the potency of a compound increases, parallel gains in selectivity occur (22, 23). Regression analysis was used to determine whether the profiling data are consistent with this premise. Compounds were classified according to specificity by counting the number of the 36 assays in which each compound displayed a 50% growth inhibition (GI50) < 10 μM, giving a “non-specificity count.” Every test point for which the GI50 was <10 μM was plotted with the negative log of the GI50 (pGI50) on the ordinate and the non-specificity count of that compound on the abscissa. Although the global dataset of 935 nontoxic compounds was uninformative, inspection of clusters of structurally related compounds revealed 9 of 14 classes that showed a modest correlation between increases in potency and selectivity (Fig. 3a).
Fig. 3.
Global data analysis. (a) Selectivity/potency correlation. The compounds were clustered based on chemical structure (Pipeline Pilot, Scitegic, San Diego) to yield 94 clusters. The test points for 14 clusters (those with 20 or more compounds) were plotted broken out by cluster to assess the specificity–potency relationships for focused structural classes. The clusters are numbered from 1 to 14, where blue dots represent compounds, and are distributed by specificity (abscissa) and potency (GI50; ordinate). (b) Relationship of structural similarity and biological similarity. Each marker represents a pair of compounds, with the pairwise chemical distance (Tanimoto coefficient) on the abscissa and the pairwise biological profile distance (Pearson correlation coefficient) on the ordinate. The line shown is the ordinary least-squares fit. The triangle encloses the outliers, which are discussed in the text. Markers are colored according to an alternative measure of biological profile similarity, the cosine (or “uncentered”) correlation coefficient.
Next, we asked whether chemical similarity was a predictor of biological activity within this dataset. For each pair of compounds, the chemical similarity was computed by the Tanimoto similarity of 512-bit Daylight fingerprints. The similarity in biological response between two compounds was calculated as the ordinary Pearson correlation of the vectors composed of the pGI50 values across the 36 assays. The plot of these two metrics displays a very strong relationship between the similarity in chemical structure and similarity in biological activity (Fig. 3b). Although the relationship between chemical and biological similarity is strong, it is clearly nonlinear and noisy. One source of this nonlinearity is the folded nature of Daylight fingerprint bitmaps, which causes the similarity for unrelated compounds to cluster around a Tanimoto coefficient of 0.5. The most interesting outliers are those that have a high chemical but a low biological similarity (enclosed by the triangle in Fig. 3b). An inspection of the biological profiles of these outliers reveals three general classifications. There are outliers in which the biological profile vector has low variance for one or both compounds in the pair, usually because the compound has little or no activity in all of the kinase assays. Such low variance causes correlation-based distance measures to be brittle, responding dramatically to slight changes in the measured GI50 for a single assay. Another group of outliers are compound pairs in which a small structural change leads to a slight general cytotoxicity. Because this cytotoxicity is reflected in the GI50 for all 36 kinase assays, the cumulative effect is to produce large differences in biological profile. Finally, there are a smaller number of outliers that appear to be genuine exceptions to the SAR hypothesis, in which small changes of chemical structure lead to large changes in biological profile. These three categories are comingled in Fig. 3b, and inspection of the individual profiles (not shown, but analogous to those in Figs. 2c and 4b) is necessary to distinguish them.
Fig. 4.
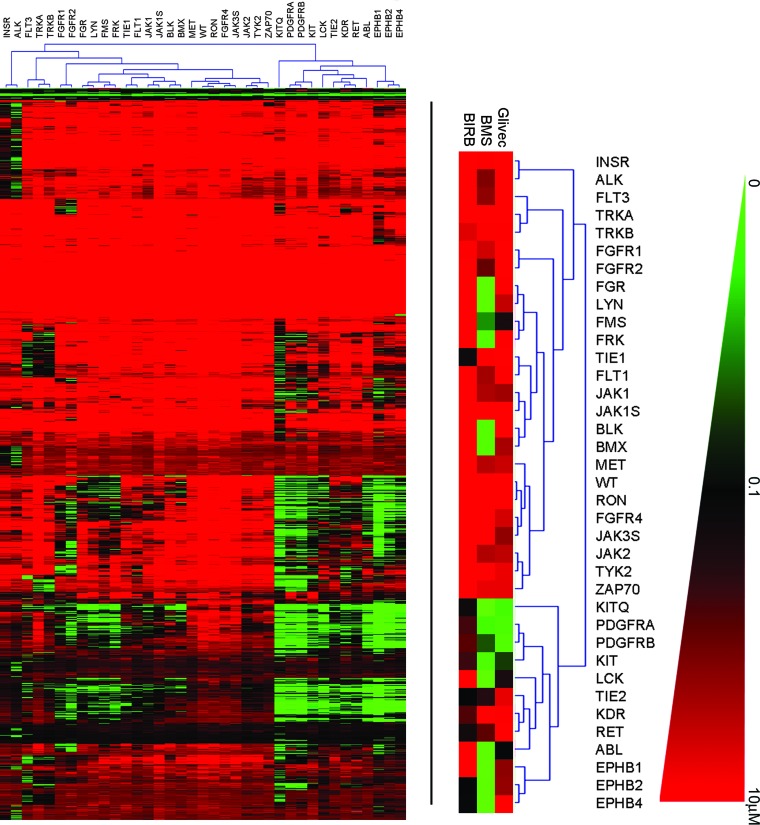
New targets. (a) Heat map of ACP data. IC50 values from the triplicate screens were averaged and clustered by using The Institute of Genome Research Multiple Array Viewer according to the average linking method of the Euclidian distance hierarchical clustering metric to generate the heat map shown in the lower left. The scale ranges from 0 (green) to 10 μM (red), as shown in the right-hand color triangle. The dendrogram associated with the heat map, showing kinase linkage, is enlarged in b, and selected inhibitors referred to in the text are expanded underneath. (b) Activities. Selected compounds from the ACP profiling experiment suggest alternate targets for several compounds. Here, the pan-p38 kinase inhibitor BIRB796 also inhibits Tie1 and Tie2. The dual src-abl inhibitor BMS-354825 also inhibits the Ephrins. Glivec (STI571) inhibits BCR-ABL, c-kit, and PDGF-RA/B.
An SAR dendrogram was created to relate kinase similarity as a function of compound activity. Distances between kinase pairs in “profile space” were calculated as the Euclidean distance between the vectors composed of the pGI50 values of the 935 nontoxic compounds, with inactive compounds being assigned a surrogate GI50 of 10 μM. This distance matrix was subjected to agglomerative clustering by using the complete linkage method. The data are represented as a dendrogram (Fig. 5, which is published as supporting information on the PNAS web site). As expected, highly homologous kinases are most often inhibited comparably by small molecules. The most notable exception is the close proximity of Flt3 to the Trk kinases in the SAR dendrogram versus the sequence-based tree. This is likely because these kinases share a critical “gatekeeper” phenylalanine side chain in the ATP-binding center (see Fig. 2b). In general, the SAR dendrograms can be used as tools to guide knowledge-based target selection and analyze multiparameter datasets obtained from compound profiling. Targets in close proximity on the dendrogram are more likely to exhibit comparable inhibition by a particular group of inhibitors than targets that are more distant. The dendrogram can also be used to select and pursue targets that are inhibited by similar chemotypes.
Previously Uncharacterized Activities.
The data were next scanned to identify whether any known drugs and well characterized compounds from the chemical collection displayed any previously unknown activities. The data were configured into a heat map by hierarchical clustering for ease of analysis (Fig. 4a). The data were examined to find targets for existing molecules and to understand the activity of molecules on related targets. For Glivec, activity against Bcr-Abl, platelet-derived growth factor, and c-kit were all manifest (see Fig. 4b). Interesting cross-activities for other less well characterized kinase inhibitors currently under clinical investigation were also discovered. For example, BIRB796, a kinase inhibitor for all p38 kinase isoforms (24, 25), was also shown to inhibit Tie2 and to a lesser extent Tie1 (Fig. 4b). These activities were confirmed to be target-related and not dependent on inhibition of p38 by Western blot of both Tel-Tie1 and Tel-Tie2 immunoprecipitated from Ba/F3 cells and probed with antiphosphotyrosine antibodies (data not shown). Tie2 has recently been implicated in angiogenesis as well as stem cell quiescence and mobilization during chemotherapy (24–27). Therefore, expansion to Tie2 as a target could potentially increase the utility of this compound.
Although BIRB796 displayed high selectivity versus other tyrosine kinases in our experiment, BMS-354825, a dual inhibitor of Src and Abl (28), was shown to inhibit multiple other kinases, including several Ephrin receptors (B1, B2, and B4). Ephrin receptors have been implicated in both tumor angiogenesis and growth and survival of tumor cells (29–31). BMS-354825 is currently being tested in humans for its potential to overcome Glivec-resistant Bcr-Abl-positive chronic mylogenous and acute lymphoblastic leukemia (32). Our results indicate the potential for expansion into other tumor types. In summary, these data demonstrate the power of the aforementioned ACP in identifying novel indications for known drugs and drug candidates, a fact that will become increasingly important given that market sizes for molecularly targeted therapies based on genetic lesions might generally be rather small.
Conclusion
Herein, we have profiled a collection of 1,400 small-molecule kinase inhibitors in a dose–response format against an array of 35 cell-based tyrosine kinase assays in a single experiment using a highly efficient profiling technology. The ACP provides a mechanism for systematic 2D combinatorial screening of chemical space against biological space. Automated kinase profiling expands SAR from one target against a set of compounds to many targets against many compounds, thus providing a more comprehensive dataset. In the case of the kinome, this type of information facilitates the process of “kinase hopping,” to determine which scaffolds are most likely to have activity on new targets of interest.
Opportunistic cellular profiling is the preclinical corollary to serendipitous clinical profiling, which led to discoveries such as Viagra’s use in erectile dysfunction (despite its originally intended use in heart disease) or the demonstration that cholesterol synthesis inhibitors, statins, reduce CD69 T cell antigen levels in T cells, which may extend the statin’s benefits to immune regulation (33–35). As demonstrated here, the ACP experiment identified PDGFR and c-kit as side activities for Glivec, which are under investigation for alternative treatments of asthma and gastrointestinal stromal disorder (GIST). Similarly, the activities identified for the p38 kinase inhibitor BIRB796 and dual src/abl inhibitor BMS-354825 may prove useful as tools to validate Tie2 and the Ephrins as drug targets in angiogenesis.
ACP profiling of molecular libraries against diverse cellular assays can be applied to many other problems as well. For example, it may be possible to identify novel ligands for whole panels of orphan G-protein-coupled receptors by profiling collections of diverse lipid, metabolite, and neuropeptide hormone libraries. It may also be possible to identify combinations of drugs that act synergistically against panels of patient-derived tumor cell lines. For pharmacogenomics, disease-associated SNPs identified by haplotype mapping can be engineered into SNP-dependent cellular assays and profiled against panels of preclinical drug candidates to prospectively match patient variants with treatment. The configuration of the ACP also allows screens for ligands with enhanced potency, selectivity, stability, or expression levels from evolved protein libraries.
Materials and Methods
Automated Cellular Assays.
The assay portion of the ACP consists of the same control software as the automated tissue culture station, a noncontact 1,536-well reagent dispenser (dispense range of 250 nl to 5 μl at 5% coefficient of variation (CV), dispense time of <1 min/plate at 5 μl), a 1,536-pin transfer device (validated at 9 nl and >10% CVs), room-temperature hotels, the same incubators as described above, and a Perkin–Elmer ViewLux plate reader. To request cells for an assay, the operator inputs the desired final number of assay plates, cell density, and well volume. The automated tissue culture station calculates the correct number of daughter flasks for pooling (if the number of cells required is unavailable, the cell propagation protocol is enacted). When daughter flasks have incubated for a defined amount of time, they are pooled in a matrix fashion into several empty recipient flasks. An equal volume from each daughter cell flask will be deposited into each recipient flask. A sample from one pooled flask will be counted (only one sample is necessary, because all of the flasks have been adjusted to the same cell density by the pooling process). All of the flasks will have their volume adjusted to the desired cell density for plating. This pooling function, although complex, obviates the requirement for a common pooling container, thus decreasing the possibility of contamination. The flasks are then moved to the cell dispenser, where cells are pulled from flasks and dispensed into 1,536-well assay plates, typically 5 μl/well. The plates of cells are placed into an incubator and the empty flasks discarded. For the assay described in this paper, compounds are introduced to the system in 1,536-well plates through the room-temperature hotels. Cell plates are requested at 105 cells/ml, 5 μl/well, in 1,536-well plates, for each of the 35 Ba/F3 cell lines. The cell requests are processed as described above. After cell plating, preplated compounds in single-dose or dose–response format are transferred from the compound plates to the assay plates by using the 1,536 pintool. The assay plates are incubated in the environmentally controlled (37°C, 95% humidity, 5% CO2) incubators for 48 h (a custom lid design minimizes edge effects from evaporation and enables 5-μl cell-based assays to be incubated for up to 5 days). After incubation, 5 μl/well Cell Titer Glo (Promega) is added with the reagent dispenser, incubated for 10 min at room temperature, and luminescence quantitated with the Perkin–Elmer ViewLux. A luminescence value less than the control value indicates a decrease in either the number of cells or cell viability.
Supporting Text.
Protocols detailing the automatic propagation of cell lines using the ACP, as well as the construction of the Ba/F3/Tel-kinase library, are presented in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Abbreviations
- ACP
automated compound profiling
- SAR
structure–activity relationship
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Plavec I., Sirenko O., Privat S., Wang Y., Dajee M., Melrose J., Nakao B., Hytopoulos E., Berg E. L., Butcher E. C. Proc. Natl. Acad. Sci. USA. 2004;101:1223–1228. doi: 10.1073/pnas.0308221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabow A. A., Shoemaker R. H., Sausville E. A., Covell D. G. J. Med. Chem. 2002;45:818–840. doi: 10.1021/jm010385b. [DOI] [PubMed] [Google Scholar]
- 3.Butcher E. C., Berg E. L., Kunkel E. J. Nat. Biotechnol. 2004;22:1253–1259. doi: 10.1038/nbt1017. [DOI] [PubMed] [Google Scholar]
- 4.Blume-Jensen P., Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Hunter T. Harvey Lect. 1998;94:81–119. [PubMed] [Google Scholar]
- 6.Seidler J., McGovern S. L., Doman T. N., Shoichet B. K. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 7.Knight Z. A., Shokat K. M. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Krause D. S., Van Etten R. A. N. Engl. J. Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 9.Golub T. R., Barker G. F., Lovett M., Gilliland D. G. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos P., Ridge S. A., Boucher C. A., Stocking C., Wiedemann L. M. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 11.Lacronique V., Boureux A., Valle V. D., Poirel H., Quang C. T., Mauchauffe M., Berthou C., Lessard M., Berger R., Ghysdael J., et al. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 12.Eguchi M., Eguchi-Ishimae M., Tojo A., Morishita K., Suzuki K., Sato Y., Kudoh S., Tanaka K., Setoyama M., Nagamura F., et al. Blood. 1999;93:1355–1363. [PubMed] [Google Scholar]
- 13.Daley G. Q., Baltimore D. Proc. Natl. Acad. Sci. USA. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai R. Y., Dieter P., Peschel C., Morris S. W., Duyster J. Mol. Cell. Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa F., Towatari M., Kiyoi H., Tanimoto M., Kitamura T., Saito H., Naoe T. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 16.Weisberg E., Boulton C., Kelly L. M., Manley P., Fabbro D., Meyer T., Gilliland D. G., Griffin J. D. Cancer Cell. 2002;1:433–443. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 17.Shah N. P., Tran C., Lee F. Y., Chen P., Norris D., Sawyers C. L. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 18.Weisberg E., Manley P. W., Breitenstein W., Bruggen J., Cowan-Jacob S. W., Ray A., Huntly B., Fabbro D., Fendrich G., Hall-Meyers E., et al. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Christensen J. G., Schreck R., Burrows J., Kuruganti P., Chan E., Le P., Chen J., Wang X., Ruslim L., Blake R., et al. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 20.Manley P. W., Bold G., Bruggen J., Fendrich G., Furet P., Mestan J., Schnell C., Stolz B., Meyer T., Meyhack B., et al. Biochim. Biophys. Acta. 2004;1697:17–27. doi: 10.1016/j.bbapap.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Ding S., Gray N. S., Ding Q., Schultz P. G. J. Org. Chem. 2001;66:8273–8276. doi: 10.1021/jo016010f. [DOI] [PubMed] [Google Scholar]
- 22.Gallion S. L., Qian D. Curr. Opin. Drug Discov. Dev. 2005;8:638–645. [PubMed] [Google Scholar]
- 23.Gill A. L., Frederickson M., Cleasby A., Woodhead S. J., Carr M. G., Woodhead A. J., Walker M. T., Congreve M. S., Devine L. A., Tisi D., et al. J. Med. Chem. 2005;48:414–426. doi: 10.1021/jm049575n. [DOI] [PubMed] [Google Scholar]
- 24.Regan J., Breitfelder S., Cirillo P., Gilmore T., Graham A. G., Hickey E., Klaus B., Madwed J., Moriak M., Moss N., et al. J. Med. Chem. 2002;45:2994–3008. doi: 10.1021/jm020057r. [DOI] [PubMed] [Google Scholar]
- 25.Pargellis C., Tong L., Churchill L., Cirillo P. F., Gilmore T., Graham A. G., Grob P. M., Hickey E. R., Moss N., Pav S., Regan J. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 26.Oliner J., Min H., Leal J., Yu D., Rao S., You E., Tang X., Kim H., Meyer S., Han S. J., et al. Cancer Cell. 2004;6:507–516. doi: 10.1016/j.ccr.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi H., Lin P. C. Front. Biosci. 2005;10:666–674. doi: 10.2741/1561. [DOI] [PubMed] [Google Scholar]
- 28.Lombardo L. J., Lee F. Y., Chen P., Norris D., Barrish J. C., Behnia K., Castaneda S., Cornelius L. A., Das J., Doweyko A. M., et al. J. Med. Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 29.Kullander K., Klein R. Nat. Rev. Mol. Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 30.Brantley-Sieders D. M., Chen J. Angiogenesis. 2004;7:17–28. doi: 10.1023/B:AGEN.0000037340.33788.87. [DOI] [PubMed] [Google Scholar]
- 31.Brantley-Sieders D., Parker M., Chen J. Curr. Pharm. Des. 2004;10:3431–3442. doi: 10.2174/1381612043383160. [DOI] [PubMed] [Google Scholar]
- 32.Shah N. P. Hematology. 2005;1:183–187. doi: 10.1182/asheducation-2005.1.183. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel E. J., Plavec I., Nguyen D., Melrose J., Rosler E. S., Kao L. T., Wang Y., Hytopoulos E., Bishop A. C., Bateman R., et al. Assay Drug Dev. Technol. 2004;2:431–441. doi: 10.1089/adt.2004.2.431. [DOI] [PubMed] [Google Scholar]
- 34.Boolell M., Allen M. J., Ballard S. A., Gepi-Attee S., Muirhead G. J., Naylor A. M., Osterloh I. H., Gingell C. Int. J. Impot. Res. 1996;8:47–52. [PubMed] [Google Scholar]
- 35.Padma-Nathan H. Am J. Cardiol. 1999;84:18N–23N. doi: 10.1016/s0002-9149(99)00115-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.