Abstract
Given that retroposed copies of genes are presumed to lack the regulatory elements required for their expression, retroposition has long been considered a mechanism without functional relevance. However, through an in silico assay for transcriptional activity, we identify here >1,000 transcribed retrocopies in the human genome, of which at least ≈120 have evolved into bona fide genes. Among these, ≈50 retrogenes have evolved functions in testes, more than half of which were recruited as functional autosomal counterparts of X-linked genes during spermatogenesis. Generally, retrogenes emerge “out of the testis,” because they are often initially transcribed in testis and later evolve stronger and sometimes more diverse spatial expression patterns. We find a significant excess of transcribed retrocopies close to other genes or within introns, suggesting that retrocopies can exploit the regulatory elements and/or open chromatin of neighboring genes to become transcribed. In direct support of this hypothesis, we identify 36 retrocopy–host gene fusions, including primate-specific chimeric genes. Strikingly, 27 intergenic retrogenes have acquired untranslated exons de novo during evolution to achieve high expression levels. Notably, our screen for highly transcribed retrocopies also uncovered a retrogene linked to a human recessive disorder, gelatinous drop-like corneal dystrophy, a form of blindness. These functional implications for retroposition notwithstanding, we find that the insertion of retrocopies into genes is generally deleterious, because it may interfere with the transcription of host genes. Our results demonstrate that natural selection has been fundamental in shaping the retrocopy repertoire of the human genome.
Keywords: origin of new genes, retroposition, transcription
The emergence of new genes is fundamental to the evolution of lineage- or species-specific traits (1). The major mechanism providing raw material for the origin of new genes is gene duplication (2). Gene duplication initially generates gene copies with the same sequences and often similar expression patterns that may diversify during evolution (3). Duplication of chromosomal segments (segmental duplications containing genes) represents one mechanism of gene duplication (4). Because this mechanism is DNA-based, it tends to produce daughter copies that inherit the genetic features (exon/intron organization and core promoters) of their parental genes. Thus, these gene copies usually not only show the same protein functions but also exhibit very similar expression patterns in their early evolution.
This mechanism contrasts with an alternative mode of gene duplication mediated by L1 retrotransposons: L1-derived enzymes can reverse-transcribe mRNAs from “parental” genes and subsequently insert the resulting cDNA into the genome, thus creating intronless gene copies (retrocopies; see refs. 1 and 5). This retroduplication mechanism is commonly thought to generate nonfunctional gene copies (retropseudogenes), because the inserted cDNA is generally expected to lack regulatory elements that could promote retrocopy transcription (6). However, in recent genome-wide studies and individual characterizations of retrocopies, a significant number of transcribed and functional retrocopies (retrogenes) were uncovered in primate and rodent genomes (7–12). In addition, three recent studies using EST data (13, 14) and tiling-microarray data from chromosome 22 (15) indicated that retrocopy transcription may be widespread, although these surveys were limited, and potential functional implications were not addressed.
To further explore the functional significance of retroposition in the human genome, we systematically screened for signatures of selection related to retrocopy transcription. Our results suggest that retrocopy transcription is not rare and that the pattern of transcription of human retrocopies has been profoundly shaped by natural selection, acting both for and against transcription.
Results
Transcribed Retrocopies.
We identified 3,590 retrocopies in the human genome using a refinement of our previous procedure (see Materials and Methods and ref. 12). To analyze transcription of these retrocopies, we used ESTs and full-length cDNAs (both termed ESTs here for convenience), because they enable better discrimination between close paralogs than data from hybridization-based technologies or short-tag expression sequences (16, 17). To map ESTs to retrocopies, we used a rigorous procedure that excludes erroneous assignment to parental genes or other paralogs (see Materials and Methods). This analysis revealed that approximately one-third of retrocopies (1,080 of 3,590 or 30.1%) matched to at least one EST (Data Set 1, which is published as supporting information on the PNAS web site), which suggests that a large proportion of retrocopies are transcribed and processed to mature mRNAs (“transcription” refers to EST evidence for mature mRNAs throughout the text).
Retrocopy Transcription and Signatures of Selection.
To test whether the transcription of these retrocopies has functional implications or, alternatively, reflects spurious transcriptional activity in the human genome, we first compared the evidence for transcription of the 575 (≈16%) potentially functional retrocopies with an intact ORF to that of the 3,015 retropseudogene copies (with reading-frame disruptions, such as frameshifts and stop codons, that may preclude gene function).
We find that the proportion of intact retrocopies with at least one EST (271 of 575 or ≈47.1%) is >1.7 times larger than that of retropseudogenes (809 of 3,015 or ≈26.8%), a highly significant difference (P < 10−20, Fisher’s exact test). On the basis of this 20.3% excess of transcription for intact retrocopies, we estimate that the human genome carries ≈117 (20.3% of 575 intact retrocopies) bona fide retrogenes.
However, this estimate represents a lower bound for several reasons. First, it is likely that some lowly transcribed retrogenes have not been detected in EST libraries. Second, unambiguous EST assignment may not be possible for young retrocopies with very high similarity to their parental gene. Related to this issue, we note that because intact retrocopies tend to be younger than disabled ones (P < 10−4, Mann–Whitney U test), EST assignment for intact copies is more limited, leading to an underestimation of transcription for such copies. Finally, retrocopies with disruptions in their ORFs are treated as pseudogenes in this analysis, although new retrogenes (that function as protein coding or RNA genes) (18, 19) may emerge from truncated coding regions (1, 20).
An overview of the most highly transcribed retrocopies (as measured by the number of matching ESTs) also provides compelling evidence for the correlation of transcription and functionality (Table 1). Among the 50 most highly transcribed copies (the ≈5% of transcribed retrocopies with the largest number of ESTs), the vast majority (42 of 50 or 84.0%) are intact, whereas only a minority (533 of 3,540 or 15.1%) of the remaining retrocopies are intact (P < 10−25, Fisher’s exact test). A similar result is obtained in an extended analysis with the top 100 transcribed retrocopies (70 of 100 or 70.0% vs. 505 of 3,490 or 14.5%; P < 10−33, Fisher’s exact test). Consistently, a number of retrocopies with many ESTs have been previously described as functional genes (e.g., HNRPF) (see Data Set 1 and ref. 21).
Table 1.
Top 50 transcribed retrocopies
| ID | Retrocopy | Parent | Origin | KA/KS | KS | ESTs | Type |
|---|---|---|---|---|---|---|---|
| 1 | HNRPF | HNRPH1 | 5 | 0.132 | 1.028 | 810 | E |
| 2 | PCBP1 | PCBP2 | 12 | 0.098 | 0.778 | 802 | |
| 3 | HSPA1A | HSPA8 | 11 | 0.021 | 4.511 | 790 | |
| 4 | RHOB | RHOA | 3 | 0.034 | 3.539 | 710 | |
| 5 | RRAGA | RRAGB | X | 0.003 | 3.626 | 425 | |
| 6 | RPL36AL | RPL36A | X | 0.009 | 0.519 | 417 | E |
| 7 | HSPA2 | HSPA8 | 11 | 0.018 | 4.520 | 336 | E |
| 8 | SFN | YWHAZ | 8 | 0.053 | 3.778 | 324 | |
| 9 | TAF9 | TAF9L | X | 0.108 | 0.857 | 319 | I, F |
| 10 | HNRPH2 | HNRPH1 | 5 | 0.029 | 0.727 | 299 | E |
| 11 | NUP62 | — | X | 0.113 | 3.713 | 288 | I, F |
| 12 | TAF7 | TAF7L | X | 0.312 | 0.847 | 282 | |
| 13 | SLC35A4 | SLC35A3 | 1 | 0.235 | 3.995 | 251 | E |
| 14 | HSPA1B | HSPA8 | 11 | 0.021 | 4.518 | 248 | |
| 15 | RHOG | RAC1 | 7 | 0.066 | 3.705 | 219 | E |
| 16 | HMGN4 | HMGN2 | 1 | 0.206 | 0.389 | 175 | E |
| 17 | TACSTD2 | TACSTD1 | 2 | 0.109 | 3.977 | 161 | |
| 18 | ARF6 | ARF3 | 12 | 0.073 | 3.439 | 160 | E |
| 19 | SOD3 | SOD1 | 21 | 0.170 | 3.454 | 152 | E |
| 20 | NXT1 | NXT2 | X | 0.079 | 1.961 | 133 | E |
| 21 | ALDH1B1 | ALDH2 | 12 | 0.062 | 2.927 | 121 | E |
| 22 | MIP-2A | TRAPPC2 | X | 0.166 | 0.064 | 104 | I, F |
| 23 | HSPA6 | HSPA8 | 11 | 0.032 | 4.572 | 89 | |
| 24 | CNO | BCAS4 | 20 | 0.137 | 3.309 | 88 | |
| 25 | — | RCN1 | 11 | 0.479 | 0.016 | 80 | |
| 26 | — | FGL1 | 8 | 0.145 | 3.674 | 79 | F |
| 27 | GSPT2 | GSPT1 | 16 | 0.151 | 0.406 | 75 | |
| 28 | RHOH | RAC1 | 7 | 0.143 | 3.631 | 72 | E |
| 29 | FAM50B | FAM50A | X | 0.037 | 3.521 | 70 | E |
| 30 | — | VAPA | 18 | 0.531 | 0.068 | 67 | |
| 31 | KLHL9 | KLHL13 | X | 0.049 | 0.710 | 65 | |
| 32 | — | TOMM20 | 1 | 1.038 | 0.026 | 64 | |
| 33 | — | C20orf81 | 20 | 0.228 | 1.456 | 62 | E |
| 34 | — | RNF4 | 4 | 0.256 | 1.381 | 60 | |
| 35 | SPIN2 | SPIN | 9 | 0.212 | 0.562 | 59 | E |
| 36 | COX7B2 | COX7B | X | 0.241 | 0.732 | 54 | E |
| 37 | FAM11B | FAM11A | X | 0.081 | 0.820 | 53 | |
| 38 | RANBP6 | RANBP5 | 13 | 0.159 | 0.745 | 49 | |
| 39 | UTP14C | UTP14A | X | 0.685 | 0.069 | 46 | I, F |
| 40 | — | RPL32 | 3 | 0.608 | 0.194 | 44 | F |
| 41 | TKTL2 | TKT | 3 | 0.059 | 3.886 | 41 | |
| 42 | DNAJB7 | DNAJB6 | 7 | 0.357 | 0.806 | 40 | I |
| 43 | PPP3R2 | PPP3R1 | 2 | 0.044 | 2.279 | 39 | I |
| 44 | WDR5B | WDR5 | 9 | 0.051 | 1.031 | 38 | |
| 45 | CALML3 | CALM3 | 19 | 0.029 | 3.214 | 35 | |
| 46 | EID3 | C10orf86 | 10 | 0.438 | 0.915 | 34 | I |
| 47 | — | RPL23A | 17 | 0.597 | 0.140 | 33 | E |
| 48 | HSPA1L | HSPA8 | 11 | 0.024 | 4.899 | 33 | |
| 49 | PGK2 | PGK1 | X | 0.108 | 0.587 | 32 | |
| 50 | — | RPL10 | X | 0.714 | 0.080 | 31 |
Retrocopy IDs correspond to IDs given in Data Set 1. IDs of retrocopies with intact ORF are in bold. Retrocopy and parental gene names are from the Swiss-Prot database (CNO and EID3), literature (MIP2A, see ref. 24), and Hugo database (remaining genes). Dashes indicate sequences without established gene names. The origin of a retrocopy is given as the chromosome where the parental gene is located. ESTs indicates the number of ESTs mapped to a retrocopy. Type indicates the retrocopy types: I, intronic; E, exon-acquisition; and F, fusion.
As another indicator of functionality, we used the ratio of nonsynonymous substitutions over synonymous substitutions per site (KA/KS) for retrocopy/parental pairs. For a given pair, KA/KS < 0.5 is indicative of purifying selection (KA/KS < 1) on the retrocopy and parental gene (11). The top 50 transcribed retrocopies show significantly lower KA/KS values (median, 0.111) than the remaining copies (median, 0.570), suggesting that they were predominantly shaped by purifying selection throughout their evolution (P < 10−5, Mann–Whitney U test). Similarly, the top 100 transcribed retrocopies show KA/KS values significantly lower (median, 0.165) than the remaining retrocopies (median, 0.571; P < 10−5, Mann–Whitney U test), which confirms that the majority of retrocopies with numerous ESTs represent functional genes evolving under selective constraints.
Promoter Acquisition.
To unravel the source of the high transcriptional activity that allowed a substantial number of functional retrogenes to emerge, we systematically screened the genomic environment of retrocopies for potential donors of promoters and other regulatory elements, because we hypothesized that retrogenes (in addition to other possible mechanisms; see Discussion) might use the regulatory elements of other genes.
To test this hypothesis, we first compared the extent of transcription of retrocopies located inside and outside other genes. Among the 1,099 retrocopies that inserted into introns of “host” genes, 463 (42.1%) show evidence for transcription (i.e., at least one EST), which is higher than the proportion of transcribed intergenic retrocopies (617 of 2,491 or 24.8%). This difference is highly significant (P < 10−24, Fisher’s exact test) and suggests that transcribed retrocopies often rely on transcriptional mechanisms of host genes that surround their insertion site.
To test whether transcription of intergenic retrocopies may also rely on nearby genes, we compared the distance of transcribed and untranscribed intergenic retrocopies to the nearest gene. The distance of transcribed intergenic retrocopies to their closest neighbor (median, 22.9 kb) is significantly smaller (P < 10−4, Mann–Whitney U test) than that of transcriptionally silent retrocopies (median, 38.7 kb).
Finally, we find that regions surrounding transcribed retrocopies are transcriptionally more active (median number of ESTs, 44) than regions surrounding silent retrocopies (median number of ESTs, 10; P < 10−4, Mann–Whitney U test).
Taken together, these observations suggest that retrocopy transcription is often driven by nearby genes. We propose two major mechanisms by which retrocopy transcription may profit from close-by genes. First, retrocopies may directly “hitchhike” on regulatory elements of genes in their vicinity, for example, by gene fusion (see below). Second, retrocopies may insert into chromosomal domains that favor transcription. Such domains may facilitate retrocopy transcription because of widely open, actively transcribed chromatin (22). In a second type of domain (“regulatory landscapes”) (23), transcription of newly inserted retrocopies may be facilitated by long-range regulatory sequences.
Chimeric Genes.
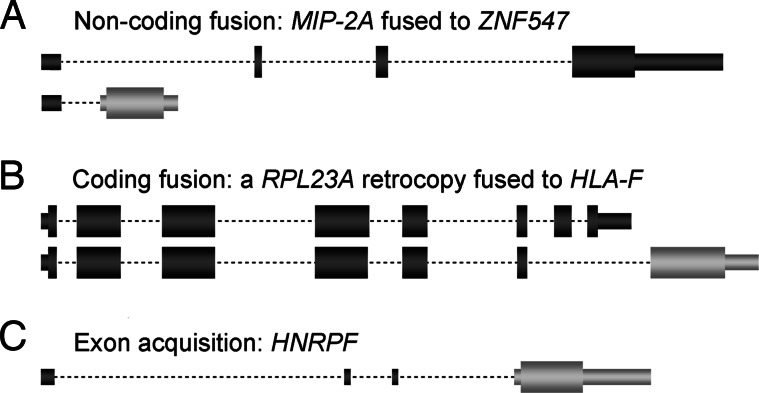
To delve further into the relationship between retrocopy transcription and their genomic environment, we screened our data for fusion transcripts of retrocopies and host genes, which would provide direct evidence for a hitchhiking scenario. Indeed, we find 36 retrocopies that are fused to neighboring host exons. Among these cases, 19 represent retrocopy fusions with the UTR of the host gene (“noncoding fusion,” where the coding sequence stems from the retrocopy) (Fig. 1A), whereas 17 involve coding exon fusions (“coding fusion,” where the coding sequence stems both from the retrocopy and the host gene) (Fig. 1B). Generally, gene fusions have significantly more ESTs (mean, 33.7) than transcribed single-exon retrocopies (mean, 7.8; P < 10−5, Mann–Whitney U test), which illustrates that this mechanism facilitates high transcription but also functionality of retrocopies (see below).
Fig. 1.
Examples of retrocopy types. (A) Noncoding fusion. (B) Coding fusion. (C) Retrocopy with newly acquired exons/introns. Retrocopy-derived sequences are light gray. Large boxes represent coding sequences. Small boxes indicate UTR sequences. Dotted lines represent introns. Transcript orientation is from left (5′) to right (3′). Transcripts are not drawn to scale.
Among the top 50 expressed retrocopies, there are six (17 in the top 100) retrocopy–host gene fusions (Table 1 and Data Set 1), including chimeric genes that emerged in primates (Data Set 1 and Table 1, IDs 22 and 79). An interesting example is a retrogene [MIP-2A (24) formerly called SEDLP1 (25)] (Table 1 and Data Set 1, ID 22) that is located in the first intron of a zinc finger gene (ZNF547) and captured the first exon of this host gene, using it as a UTR (Fig. 1A). The MIP-2A-encoded protein was shown to be involved in cell-growth regulation through an interaction with the c-myc promoter-binding protein 1 (24). Interestingly, MIP-2A shows a low divergence at synonymous sites (KS ≈ 0.06) from its parent (TRAPPC2) on chromosome X and has no ortholog in the mouse genome, which suggests that it has an origin in primates. Indeed, experimental dating reveals that it originated on the primate lineage leading to humans (A. C. Marques and H.K., unpublished data). This example illustrates that promoter hitchhiking by capturing exons from the host provides a means for retrocopies to become highly transcribed and functional within a short evolutionary time span.
Interestingly, we also identified coding fusions that emerged on the primate lineage leading to humans. For example, a retrocopy that derived from the ribosomal protein gene RPL23A (Data Set 1, ID 57) inserted close to the 3′ end of HLA-F (a class I MHC gene) and fused to an alternative transcript of this host gene (Fig. 1B).
Acquisition of New Exons.
In addition to host gene fusions, we identified 27 other retrocopies transcribed together with additional exons. Our analyses show that these copies did not fuse to other resident genes but acquired new exons/introns de novo. A possible scenario for how this might have occurred is that, during evolution, new introns were created in the original transcripts that included the retrocopy sequence (26, 27). We find a striking overrepresentation of such cases among highly expressed copies. For example, we identified 17 exon acquisition cases among the 50 most highly transcribed retrocopies (21 among the top 100), a >4,200% excess relative to the 0.4 cases expected based on the overall data (P < 10−15, χ2 test). Several additional lines of evidence revealed that most of these retrocopies are not only highly expressed but represent bona fide genes.
First, several of the most highly transcribed retrocopies of this type have been functionally characterized in other studies [e.g., HMGN4 (28) and SOD3 (29)]. Notably, these genes were often not recognized as retrogenes because of their multiexonic gene structures. For example HNRPF (Table 1, ID 1), which is supported by >800 ESTs, is known to be involved in RNA processing and transport (20) but was not recognized as a (intronless) retrogene, probably because it recruited three 5′-UTR exons (Fig. 1C). Second, most of these cases have an ortholog in the mouse genome with intact ORFs (Data Set 1). Third, pairwise KA/KS analyses of parents/retrogenes show that most retrocopies with new exons/introns have been subject to strong purifying selection (Table 1 and Data Set 1). The fact that such retrocopies are usually functional but old suggests that they probably emerged as single-exon genes and evolved more complex gene structures with time, which probably facilitated a more efficient and potentially also more sophisticated expression.
Out of the Testes.
To explore spatial transcription patterns of retrocopies, we analyzed the tissue distribution of their ESTs using the evoc ontology (30). Based on several individual cases, we previously obtained suggestive evidence that retrogenes are often transcribed and functional in testes (12). To test this hypothesis on a more global scale, we first compared the amount of testis expression between retrocopies and annotated multiexon genes in the human genome. The proportion of testis ESTs that mapped to retrocopies (713 of 10,536 or 6.8%) is over twice that of multiexon genes (105,548 of 3,592,280 or 2.9%).
To assess the functional relevance of this observation, we compared the proportion of retrocopies and retropseudogenes transcribed in testis. This analysis revealed that more than twice as many intact retrocopies are expressed in testis relative to retropseudogenes (89 of 227 or 39.2% vs. 107 of 656 or 16.3%; P < 10−11, Fisher’s exact test). This 22.9% excess of intact copies with testis ESTs corresponds to ≈52 (22.9% of 227) retrogenes that evolved a function in testis. Thus, the general testis bias among new retrocopies does not merely reflect transcriptional noise due to “hypertranscription” in this tissue (31, 32) but has profound functional implications, generalizing our previous notion (12).
We also observe that young transcribed retrocopies that are absent in the mouse tend to be transcribed at a low level (median number of ESTs, 2), with a large proportion of their ESTs found in testis (10.7%). In contrast, older transcribed retrocopies that have an ortholog in the mouse have significantly more ESTs (median, 21; P < 10−5, Mann–Whitney U test), with a smaller bias toward testis transcription (5.4%). These observations suggest a scenario where the majority of retrocopies are initially transcribed in testis because of the facilitated transcription in this tissue. This effect enabled many retrocopies (≈52) to obtain a functional role in this tissue, which is known to evolve rapidly (12, 33). Other retrogenes evolved broader expression patterns and functions by evolving stronger and more diverse regulatory elements. Thus, it seems that, generally, functional retrogenes emerge out of the testis.
Out of the X.
In human, mouse, and fly, genes located on the X chromosome have generated an excess of autosomal retrogene counterparts (10, 11). These counterparts probably serve as substitutes for their X-linked parents that are silenced during male meiosis. Strikingly, we find 14 X chromosome-derived retrogenes among the top 50 transcribed copies, although only 1.9 are expected based on the proportion (3.84%) of potential parental genes on the X chromosome. This excess is highly significant as assessed by a resampling test (P < 10−6) (see Materials and Methods). The excess among the 100 most highly transcribed retrocopies is still >600% (23 observed vs. 3.8 expected; P < 10−6) (see Materials and Methods). If we consider all retrocopies and compare the proportions of X chromosome-derived transcribed (79 of 1,080 or ≈7.3%) and transcriptionally silent (121 of 2,510 or ≈4.8%) retrocopies, we find that transcribed retrocopies show an excess of X chromosome-derived retrocopies that corresponds to ≈27 (2.5% of 1,080) retrogenes that emerged from the X chromosome. This number is ≈2.5 times higher than the 11 cases estimated in our earlier study of annotated retrogenes (11). However, this estimate probably still represents a lower bound, because the out-of-X “movement” is an ongoing process (12) and because unambiguous EST mapping to very young X chromosome-derived copies is not always possible.
The X-chromosome-replacement hypothesis posits that autosomal substitutes sustain essential functions during male X chromosome inactivation (34). Indeed, several X chromosome-derived genes that can be considered essential have been previously described (11). RPL36AL, which derives from a parental gene encoding a ribosomal protein (RPL36A) is an interesting example in our data (Table 1 and Data Set 1, ID 6). RPL36AL is a retrogene (with previously uncharacterized UTR exons) supported by >400 ESTs and shows a strong signature of purifying selection in a pairwise comparison with its parent (KA/KS ≈ 0.01). An extended analysis shows that this retrogene is not only present and conserved in the mouse but also in the rat and dog genomes (Data Set 1). RPL10L, another highly transcribed ribosomal retrogene that stems from the X chromosome (Data Set 1, ID 56) is similarly conserved in mouse, rat, and dog. Thus, these genes are clearly functionally preserved and provide evidence against the common belief that ribosomal gene duplicates are not viable because of gene dosage constraints during ribosome assembly (35–37).
Retrogenes and Disease.
The out-of-X mouse retrogene UTP14B was recently demonstrated to play an essential role in spermatogenesis and reported as the first mammalian retrogene causing a disease phenotype when disabled (38). An independent retroposition event from the same parent gave rise to a similar retrocopy in humans (UTP14C), which was suggested to be functionally equivalent to the mouse retrogene (38). Our data shows that the human UTP14C retrocopy is among the 50 most highly expressed retrocopies (Table 1, ID 39). As with UTP14B, it has evolved a multiexonic gene structure by fusing to its host gene, thus supporting its functionality and potential implication in human disease phenotypes.
To identify human disease-associated retrogenes, we searched the transcribed retrocopies against the available literature and uncovered a retrogene responsible for a human recessive disorder: Disabling mutations in the TACSTD2 gene on chromosome 1 cause gelatinous drop-like corneal dystrophy, an autosomal recessive disorder characterized by severe corneal amyloidosis leading to blindness (39). Our data reveals that TACSTD2 is an intronless retrogene derived from the eight-intron-containing parent, TACSTD1, on chromosome 2 (Table 1, ID 17).
Selection Against Retrocopies.
We have shown that retrocopies are frequently transcribed and often profit from their genomic environment for transcription and functionality. However, we hypothesized that retrocopy insertion may generally be detrimental to host genes because of interference with host gene expression. First, highly transcribed retrocopies (e.g., carrying their own promoters) may interfere with the transcriptional machinery of the host gene. Second, the sequence of a retrocopy itself may directly interfere with proper splicing and/or polyadenylation of the host gene by providing an aberrant exon or a premature polyadenylation site, akin to observations from retrotransposable elements (40). Therefore, natural selection may often act against retrocopy insertion, retrocopy transcription, or both.
To test these predictions, we first compared the proportion of intronic retrocopies among highly transcribed retrocopies and those transcribed to a lesser extent. This analysis shows that intronic retrocopies are significantly underrepresented among the 100 most expressed copies (19 of 100 or 19.0%) compared with the remaining transcribed copies (1,080 of 3,490 or 30.9%; P < 0.02, Fisher’s exact test). Therefore, although intronic retrocopies can hitchhike on host gene promoters, highly transcribed retrocopies tend to be deleterious to their hosts.
To test whether natural selection acts against retrocopy insertion into other genes per se, we compared the frequency of retrocopies on the sense and antisense strand relative to the direction of host gene transcription. We predicted that the processing signals of retrocopies inserted on the sense strand are more likely to interfere with proper RNA maturation of the host gene. Indeed, we identified fewer copies on the sense (462) than antisense (661) strand, a highly significant difference (P < 10−8, χ2 test). This finding suggests that the retrocopy sequence itself may interfere with polyadenylation and/or proper splicing of the host gene. The proportion of sense retrocopies with ESTs (47.2%) is significantly higher than that of antisense retrocopies (38.9%; P < 10−2, Fisher’s exact test), which supports the idea that sense retrocopies are indeed more likely to be spliced as exons and thus interfere with proper mRNA formation of the host gene.
Based on the proportions of transcribed retrocopies among intact (91 of 170 or 53.5%) relative to disabled (372 of 929 or 40.0%) intragenic retrocopies, we estimate that, nevertheless, 23 (13.5% of 170) intragenic retrogenes resisted evolutionary pressures of host gene maintenance. However, the same analysis for intergenic copies (intact, 180 of 405 or 44.4%, vs. disabled, 437 of 2,086 or 20.9%) yields 95 (23.5% of 405) retrogenes, a significantly higher number (P < 10−2, Fisher’s exact test). This result further illustrates that an overlap of retrogenes and host genes is usually disfavored by natural selection.
Discussion
Here we have shown that transcription (and splicing) of retrocopies is not rare by using a targeted approach based on EST mapping. The finding of extensive retrocopy transcription is in line with recent genome-wide transcription studies that revealed that an unexpectedly large proportion of the genome is transcribed (19, 41–44). We unraveled some sources for the transcriptional activity of retrocopies. Many retrocopies seem to rely on regulatory elements from other genes, for example, by directly fusing to host genes or by otherwise hitchhiking on the regulatory elements (e.g., enhancers) of nearby genes.
However, we envision at least two additional mechanisms pertaining to the origin of core promoters of retrocopies. First, retrocopies may directly inherit promoters from their parental transcripts if these transcripts carry alternative promoters of the parental gene, as has been observed for individual cases (45). This direct inheritance mechanism of promoters might be common for retrocopies, because a recent global study of promoters in the human genome revealed that many genes carry multiple promoters (46). Second, retrotransposons, such as Alu elements and LINEs (long interspersed nuclear element), which are abundant in the genome and were shown to affect the transcription of genes (40, 47), may provide retrocopies with promoter elements. Assessing the contribution of these mechanisms to retrocopy transcription warrants further work. It would also be interesting to analyze when retrocopies acquire regulatory elements, because they may initially be transcriptionally silent and obtain promoters and, hence, functionality long after the retroposition event (48, 49).
In this study, we have obtained compelling evidence that much of the extensive transcriptional activity of retrocopies does not represent transcriptional “noise” but has been profoundly shaped by natural selection. Although selection generally acted against the insertion and high transcription of retrocopies located inside other genes, it favored the emergence of a substantial number of new genes with diverse gene structures and functions during the evolution of the human genome.
Materials and Methods
Retrocopy Screen.
To identify retrocopies for this study, we modified our previous procedure (12). We required that a minimum of two introns from the parental gene be absent in alignable regions with the retrocopy. Retrocopies with a KS of >2 were required to lack at least three parental introns. These criteria were used to ensure that the old identified copies truly emerged by retroposition (and are not the result of intron gain events in other types of duplicate genes). However, other intronless genes (e.g., olfactory receptor genes) or genes that feature an unusually large (internal) exon may also have emerged by retroposition (50).
Mapping to the Ensembl Annotation.
We mapped all retrocopies to Ensembl (version 29) transcripts. For each retrocopy that mapped to a transcript, we analyzed whether the retrocopy overlapped with introns and/or exons of the transcript. Because the retrocopy coordinates identified in our procedure are limited to regions with homology to coding sequences of the parental gene and do not include UTRs, we also used blast (51) hits (E < 0.001) of parental transcripts around the genomic site of the retrocopy (retrocopy and 10-kb flanking sequences). When a retrocopy and/or a blast hit from its parental transcript overlapped with an Ensembl exon, we considered this exon to be retrocopy-derived. Using this information, we identified retrocopies that where fused to exogenous exon(s) in a chimeric transcript. Among these, we further distinguished between de novo exon/intron acquisition and host gene fusion cases. For this procedure, we assumed that chimeric transcripts from genes that have non-retrocopy-derived coding exons represent retrocopy–host gene fusions. Other chimeric transcripts were regarded as retrocopies that acquired new exons.
Transcription Analysis.
Coordinates of all ESTs that mapped to the human genome were downloaded from the University of California, Santa Cruz, database (assembly May 2004). For each EST, this mapping contains the best Blat hit in the genome as well as other hits that have a nucleotide identity value that falls within 0.5% of the best hit and have at least 96% nucleotide identity with the genomic sequence. To properly discriminate between parental genes and retrocopy transcription, we proceeded as follows. (i) We only considered ESTs that mapped to a unique location according to the University of California, Santa Cruz, database criteria; that produced an alignment with the genomic sequence of >100 bp; and that showed a nucleotide identity of >97%. (ii) Among these ESTs, we identified all ESTs that align with a genomic sequence overlapping a retrocopy. (iii) To ensure that ESTs were not erroneously mapped to retrocopies because of their lack of introns (which may produce better Blat scores), we built a database containing all Ensembl transcripts and genomic sequences of retrocopies that include 4 kb of flanking sequences. All ESTs that mapped to retrocopies in step ii were then blasted against this database and retained if their best hit was the genomic sequence of a retrocopy or an Ensembl transcript containing a retrocopy. In addition, we used ESTs to support multiexonic transcripts (retrocopies with new exons and gene fusions). We considered an EST as supportive evidence if it aligned both with a retrocopy-derived and a non-retrocopy-derived exon.
Level of Transcriptional Activity Surrounding Retrocopies.
Excluding ESTs that align with retrocopies or their 2-kb flanking sequences, we counted the number of EST(s) that mapped within 40 kb of the retrocopy boundaries. We assumed that the counts obtained were indicative of the level of transcriptional activity in the genomic region surrounding retrocopies.
Distance to Closest Gene (Intergenic Retrocopies).
We used retrocopy and Ensembl transcript coordinates (start and end) and computed the minimal distance between a retrocopy and its neighboring gene. We did not consider transcript orientations for this analysis (transcripts that were on the opposite strand of a retrocopy and transcripts on the same strand where treated equally). Ensembl transcripts that corresponded to annotated retrocopies where disregarded in this analysis.
Presence/Absence of Retrocopies in Mouse.
We used human/mouse chained alignments available at the University of California, Santa Cruz (hg17 vs. Mm6), to identify the presence/absence of orthologs of human retrocopies in the mouse genome. We first extracted the best alignments that overlap with the genomic location of retrocopies and that are >15 kb (this length ensures that the alignment also covers surrounding, non-retrocopy-derived sequences in the two species). If no such alignments could be identified, presence/absence in mouse was not determined. We then scanned the alignments for an aligned block that overlapped with the retrocopy. If such a block was found, the retrocopy was considered to have emerged before the human/mouse split. When no such block was found, the retrocopy was assumed to have emerged after human/mouse split.
Spatial Transcription Patterns.
The transcription patterns of retrocopies were established by linking mapped ESTs to the anatomical system ontology of evoc (30). This procedure enabled us to establish the sample source (testis or other tissues) of the ESTs mapping to retrocopies.
Statistical Tests.
For most statistical analysis, we used standard Fisher’s exact, χ2, and Mann–Whitney U tests. In addition, we used a resampling test to assess the statistical significance of the excess of X-chromosome-derived retrocopies among the top 50 (100) transcribed retrocopies. We built a set of 3,590 retrocopies with 138 X-chromosome-derived retrocopies according to the proportion (≈3.84%) of potential X-chromosome-linked parental genes. Random samples (106) containing 50 retrocopies from this set all showed a lower number of X-chromosome-derived retrocopies than the 14 (23 in the top 100 retrocopies) observed.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their valuable comments on the manuscript; Max Ingman, Ana Claudia Marques, Lukasz Potrzebowski, and Lia Rosso for constructive discussions; J. Chamary for comments on the manuscript; and the Vital-IT team at the University of Lausanne for computational support. This research was supported by funds available to H.K. from the Center for Integrative Genomics (University of Lausanne), European Union Grant PKB140404, the EMBO Young Investigator Program, and Swiss National Science Foundation Grant 3100A0-104181.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Long M., Betran E., Thornton K., Wang W. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 3.Li W. H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 4.Samonte R. V., Eichler E. E. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 6.Mighell A. J., Smith N. R., Robinson P. A., Markham A. F. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 7.Betran E., Long M. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betran E., Wang W., Jin L., Long M. Mol. Biol. Evol. 2002;19:654–663. doi: 10.1093/oxfordjournals.molbev.a004124. [DOI] [PubMed] [Google Scholar]
- 9.Burki F., Kaessmann H. Nat. Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- 10.Betran E., Thornton K., Long M. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson J. J., Kaessmann H., Betran E., Long M. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 12.Marques A., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison P. M., Zheng D., Zhang Z., Carriero N., Gerstein M. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano Y., Saito R., Yoshida N., Yoshiki A., Wynshaw-Boris A., Tomita M., Hirotsune S. J. Mol. Med. 2004;82:414–422. doi: 10.1007/s00109-004-0550-3. [DOI] [PubMed] [Google Scholar]
- 15.Zheng D., Zhang Z., Harrison P. M., Karro J., Carriero N., Gerstein M. J. Mol. Biol. 2005;349:27–45. doi: 10.1016/j.jmb.2005.02.072. [DOI] [PubMed] [Google Scholar]
- 16.Harbers M., Carninci P. Nat. Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 17.Reinartz J., Bruyns E., Lin J. Z., Burcham T., Brenner S., Bowen B., Kramer M., Woychik R. Brief Funct. Genomic Proteomic. 2002;1:95–104. doi: 10.1093/bfgp/1.1.95. [DOI] [PubMed] [Google Scholar]
- 18.Hirotsune S., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- 19.Brosius J. Trends Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Wang P. J., Page D. C. Hum. Mol. Genet. 2002;11:2341–2346. doi: 10.1093/hmg/11.19.2341. [DOI] [PubMed] [Google Scholar]
- 21.Honore B., Rasmussen H. H., Vorum H., Dejgaard K., Liu X., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J. E. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N. P., Bickmore W. A. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Spitz F., Herkenne C., Morris M. A., Duboule D. Nat. Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A. K., Majumder M., Steele R., White R. A., Ray R. B. Mol. Cell. Biol. 2001;21:655–662. doi: 10.1128/MCB.21.2.655-662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gecz J., Hillman M. A., Gedeon A. K., Cox T. C., Baker E., Mulley J. C. Genomics. 2000;69:242–251. doi: 10.1006/geno.2000.6326. [DOI] [PubMed] [Google Scholar]
- 26.Brosius J. Trends Genet. 1999;15:304–305. doi: 10.1016/s0168-9525(99)01783-7. [DOI] [PubMed] [Google Scholar]
- 27.Brosius J., Gould S. J. Proc. Natl. Acad. Sci. USA. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birger Y., Ito Y., West K. L., Landsman D., Bustin M. DNA Cell Biol. 2001;20:257–264. doi: 10.1089/104454901750232454. [DOI] [PubMed] [Google Scholar]
- 29.Zelko I. N., Mariani T. J., Folz R. J. Free Radical Biol. Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 30.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien S., Smedley D., Otgaar D., Greyling G., Jongeneel C. V., McCarthy M. I., et al. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt E. E. Curr. Biol. 1996;6:768–769. doi: 10.1016/s0960-9822(02)00589-4. [DOI] [PubMed] [Google Scholar]
- 32.Kleene K. C., Mulligan E., Steiger D., Donohue K., Mastrangelo M. A. J. Mol. Evol. 1998;47:275–281. doi: 10.1007/pl00006385. [DOI] [PubMed] [Google Scholar]
- 33.Swanson W. J., Vacquier V. D. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 34.McCarrey J. R., Thomas K. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- 35.Yoshihama M., Uechi T., Asakawa S., Kawasaki K., Kato S., Higa S., Maeda N., Minoshima S., Tanaka T., Shimizu N., et al. Genome Res. 2002;12:379–390. doi: 10.1101/gr.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uechi T., Tanaka T., Kenmochi N. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z., Harrison P., Gerstein M. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradley J., Baltus A., Skaletsky H., Royce-Tolland M., Dewar K., Page D. C. Nat. Genet. 2004;36:872–876. doi: 10.1038/ng1390. [DOI] [PubMed] [Google Scholar]
- 39.Tsujikawa M., Kurahashi H., Tanaka T., Nishida K., Shimomura Y., Tano Y., Nakamura Y. Nat. Genet. 1999;21:420–423. doi: 10.1038/7759. [DOI] [PubMed] [Google Scholar]
- 40.Medstrand P., van de Lagemaat L. N., Mager D. L. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T. R. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 43.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 44.Semon M., Duret L. Trends Genet. 2004;20:229–232. doi: 10.1016/j.tig.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Feral C., Guellaen G., Pawlak A. Nucleic Acids Res. 2001;29:1872–1883. doi: 10.1093/nar/29.9.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Lagemaat L. N., Landry J. R., Mager D. L., Medstrand P. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Krull M., Brosius J., Schmitz J. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 49.Singer S. S., Mannel D. N., Hehlgans T., Brosius J., Schmitz J. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 50.Brosius J. Cytogenet Genome Res. 2005;110:8–24. doi: 10.1159/000084934. [DOI] [PubMed] [Google Scholar]
- 51.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.