Abstract
The ATP-dependent integral membrane protease FtsH is universally conserved in bacteria. Orthologs exist in chloroplasts and mitochondria, where in humans the loss of a close FtsH-homolog causes a form of spastic paraplegia. FtsH plays a crucial role in quality control by degrading unneeded or damaged membrane proteins, but it also targets soluble signaling factors like σ32 and λ-CII. We report here the crystal structure of a soluble FtsH construct that is functional in caseinolytic and ATPase assays. The molecular architecture of this hexameric molecule consists of two rings where the protease domains possess an all-helical fold and form a flat hexagon that is covered by a toroid built by the AAA domains. The active site of the protease classifies FtsH as an Asp-zincin, contrary to a previous report. The different symmetries of protease and AAA rings suggest a possible translocation mechanism of the target polypeptide chain into the interior of the molecule where the proteolytic sites are located.
Keywords: AAA, protease, protein degradation, x-ray
ATP-dependent proteases play crucial roles in protein quality control and regulation (for reviews, see refs. 1 and 2). One of these is FtsH, initially described as a temperature-sensitive and cell-division-defective mutant, which is also called HflB, named after a high-frequency of lysogenization locus of bacteriophage λ. FtsH is an integral membrane protease found in bacteria, chloroplasts, and mitochondria (reviewed in ref. 3) (Fig. 6, which is published as supporting information on the PNAS web site). In bacteria, FtsH malfunction causes severe phenotypes like cell division defects and growth arrest (4, 5) (reviewed in ref. 3). Deletion of the human mitochondrial homolog paraplegin, which shares 40% sequence identity with FtsH from Escherichia coli, is responsible for an autosomal recessive form of hereditary spastic paraplegia (6). The N terminus of FtsH contains two transmembrane helices followed by an AAA module (ATPases associated with various cellular activities), including the SRH (second region of homology) (3). The C-terminal part of the polypeptide chain bears the HEXXH motif that is characteristic for Zn-dependent metalloproteases, where the two histidines coordinate to the zinc ion and the glutamate serves as a catalytic base. In bacteria, the AAA and protease domains are located on the cytosolic side of the membrane. Of the five ATP-dependent proteases in E. coli, HslVU, Lon, ClpXP, ClpAP, and FtsH, the last is the only one that is essential and universally conserved in bacteria. It degrades membrane proteins like the uncomplexed SecY subunit of translocase (7), the a-subunit of FoF1-ATPase (8), and the photosystem in chloroplasts (9), therefore playing an important role in the quality control of membrane proteins. Further targets comprise regulatory soluble proteins such as σ32 (10, 11) or λ-CII transcriptional activator protein (12). All these substrates are degraded in an ATP-dependent manner where the energy is used for pulling the proteins out of the membrane and/or unfolding and translocation. However, FtsH possesses only a weak unfoldase activity (13) and is not able to degrade stable proteins, contrary to other ATP-proteases like ClpXP (14).
Structural information for FtsH is sparse. As shown by electron microscopic studies, FtsH forms ring-shaped toroidal oligomers like other AAA proteins (12). By comparison with other AAA domains, these rings consist of hexamers; however, no experimental evidence published so far has confirmed this. The transmembrane helices, especially the second one, were reported to be essential for oligomerization and for ATPase and proteolytical activity (15, 16). Two crystal structures of the AAA domain in isolation were reported in the past (17, 18). A hexameric AAA model possessing C6 symmetry was build from these structures essentially in analogy to the crystal structure of N-ethylmaleimide-sensitive factor (NSF), implying a major contribution of the AAA domains for the stabilization of the quaternary structure in addition to the transmembrane part.
FtsH has in recent years been the subject of many biochemical studies. Targeting signals in the substrates were partially identified, and it appears that FtsH recognizes apolar tails (19, 20). The regulation of FtsH activity in E. coli by the periplasmic HflKC complex was examined, and a stable megadalton complex of the composition (FtsH)6(HflKC)6 was proposed to exists in the membrane of E. coli (21). The third zinc ligand other than the two histidines of the HEXXH motif was reported to be a glutamic acid (Glu-476 in E. coli), based on site-directed mutagenesis experiments (22).
To gain a deeper insight into the function and mechanism of FtsH, we undertook the crystal structure determination of a soluble construct of FtsH from Thermotoga maritima lacking both N-terminal transmembrane helices.
Results
T. maritima FtsH Lacking the Transmembrane Domains Is Oligomeric.
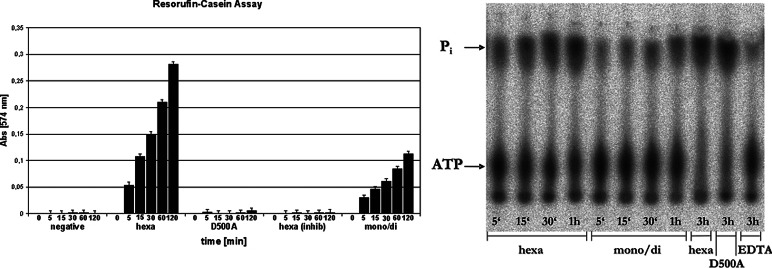
T. maritima FtsH was expressed in E. coli as a construct (residues 147–610) lacking the two transmembrane helices [termed hereafter (Δtm)FtsH]. The expressed protein eluted from a Sephadex-S75 column in the form of two well separated, reproducible, and stable peaks. According to analytical ultracentrifugation (data not shown), the first peak eluting at an apparent molecular mass of ≈600 kDa contains approximately equal amounts of tetramers and hexamers, whereas the second one (≈100 kDa) consists essentially of dimers. ATPase and caseinolytic activity assays showed activity for both peaks, but the larger oligomer possesses significantly higher activity in both assays (Fig. 1). However, caseinolytic activity is not stimulated by ATP in either oligomeric species, and the construct could not be shown to be active in the in vitro degradation of σ32 under the conditions used.
Fig. 1.
Proteolytic and ATPase activity of (Δtm)FtsH. (Left) Resorufin-casein protease assay. Absorbance at 574 nm of the different samples was plotted against time. hexa, samples taken from the “early” peak of the gel-filtration purification step; mono/di, samples from the “late” peak; hexa (inhib), hexa sample incubated with 10 mM ortho-phenanthroline as inhibitor; D500A, mutant of Δtm-FtsH lacking with the true third zinc liganded mutated. The negative control (“negative”) contained no Δtm-FtsH protein. (Right) Radiogram of the [γ-32P]ATPase assay. ATP and free Pi were separated by thin-layer chromatography and are marked at the left side of the picture. Samples were taken at 5, 15, 30, 120, and 180 min. EDTA, sample incubated with 50 mM EDTA to inhibit ATPase activity.
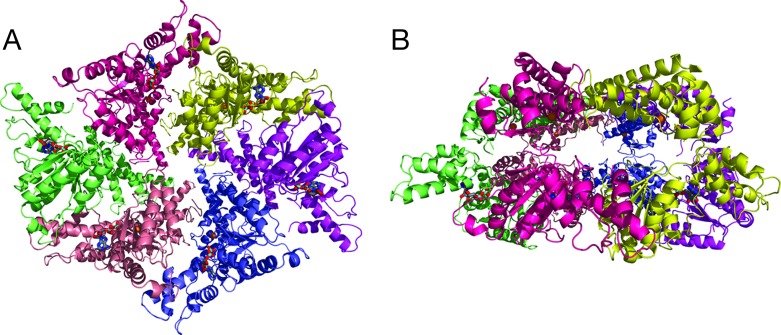
In crystallization experiments, both gel-filtration peaks give rise to the same tetragonal crystal form containing six monomers per asymmetric unit. These belong to two virtually identical half-hexamers; the complete hexameric molecules are generated by a crystallographic twofold axis. The hexamer has a “crown” shape with a diameter and height of ≈100 Å and 65 Å, respectively. It consists of two rings (Fig. 2). The “upper” ring (Fig. 2B) is build by the C-terminal protease domains and possesses virtually exact sixfold symmetry, whereas the lower ring is formed by the AAA domains and follows only the symmetry of the crystallographic twofold. The strongest intersubunit contacts occur between the protease domains of the monomers. Each protease domain interacts with the two neighboring protease domains in the hexameric ring, burying a total of ≈4,000 Å2 of accessible surface, i.e., some 20% of a single protease domain surface. These contacts include hydrophobic and polar interactions including the formation of a small antiparallel β-sheet (residues 466–469 and 492–495 of two subunits).
Fig. 2.
The hexameric structure of FtsH. (A) Top view approximately down the crystallographic twofold axis from the supposed membrane side onto the AAA ring. The colors denote the individual subunits. ADP and active site residues are shown as sticks (gray, carbons; blue, nitrogens; red, oxygens; cyan, phosphorous), and the Zn2+ ions are shown as golden spheres. (B) Side view, the AAA ring is on the bottom, the protease ring on the top.
AAA and Protease Rings Possess Different Symmetries.
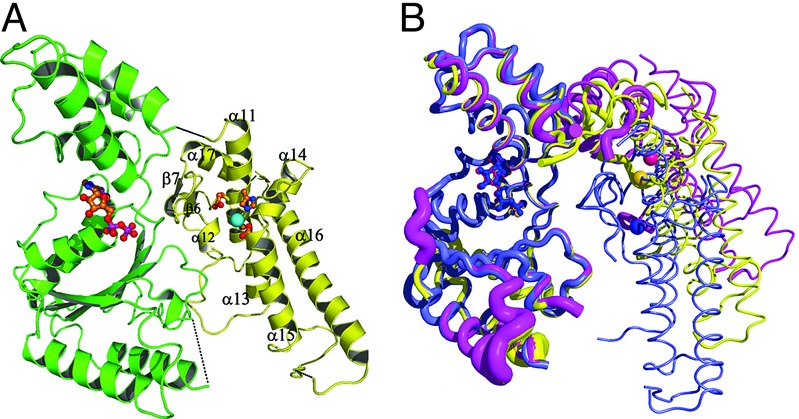
All six independent AAA domains were found to have ADP bound that originates from the cellular pool. Nevertheless, the AAA ring does not form a regular hexagon but shows the crystallographic C2 symmetry only. As a result the contacts of one AAA domain with the two other interacting domains in the ring are much more asymmetric than those observed in the protease domain. Between 1,350 and 2,000 Å2 of accessible surface gets buried for the two contact areas. Two AAA domains show rather weak density, indicating a high flexibility. The average B-factors of AAA domain and protease domain are 95 Å2 and 54 Å2, respectively. This notion of flexibility of the AAA domains is corroborated by an overlay of the three independent monomers of one hexamer. The relative orientation between AAA and protease domain varies significantly between the independent monomers as shown in Fig. 3, which is necessary to switch from the virtually perfect sixfold symmetry in the protease ring to the twofold symmetry in the AAA ring.
Fig. 3.
Conformation of the monomer. (A) Cartoon with secondary structure labeling. The AAA domain is shown in green, the protease domain is shown in yellow, and the zinc ion is shown in cyan. (B) Overlay of the three independent monomers of one hexamer. The AAA domain with bound ADP is at the left, and the protease domain with Zn2+ depicted as spheres is on the right. The tube thickness is proportional to the B-factors. The N-terminal part of the AAA domain was chosen as reference for the overlay. The color coding is the same as in Fig. 2.
The Protease Belongs to a New Asp-Zincin Family.
The protease domain possesses a fold consisting of eight α-helices (α11–α18) and a very short antiparallel β-ribbon (β6 and β7) (Fig. 3 and Fig. 7, which is published as supporting information on the PNAS web site). Database searches using DALI (23) revealed no similar structures. A characteristic feature of this domain is a long, slightly bent α-helix (α16, residues 547–574) that forms the base of the protease domain. This helix and the following helix α17 form the shape of an “L” and are tethered to α11, α13, and α14 by means of a conserved leucine zipper motif (24) consisting of residues 565, 568, 572, and 578. Nonconservative substitutions of these residues abolish activity. The first helix (α11) of the protease domain carries the metalloprotease fingerprint H423EXXH427. Surprisingly, the third zinc ligand is not the reported Glu-486 (Glu-476 in E. coli) (22) but rather the absolutely conserved Asp-500, which is located at the beginning of helix α15 (Fig. 8, which is published as supporting information on the PNAS web site). Glu-486 is in vicinity of the active site and forms hydrogen bonds to Thr-494 and to the first histidine of the HEXXH motif, fixing the imidazole side chain in the proper conformation for zinc ligation. The side chains of the histidines in the HEXXH motif are fixed by hydrogen bonds also in other metalloproteases, usually by carbonyl oxygens. This finding explains the reported significant residual proteolytic activity of ≈10% of the Glu-486Val mutant (22). On the other hand, mutation of Asp-500 to alanine abolishes the proteolytic activity completely in the construct described here (Fig. 1). The crystal structure analysis of this D500A mutant confirmed the loss of the zinc ion.
A striking difference to other HEXXH proteases (“zincins”) is the absence of a significant amount of β structure in the protease domain. In particular, the so-called “edge strand,” which is present in all other HEXXH proteases known so far, is missing. This β-strand that runs just in a parallel direction above the active site helix serves for the orientation of the substrate backbone in an antiparallel extended conformation, which allows efficient cleavage of the target peptide bond. This stretching of the substrate is probably not necessary in the case of unfolded proteins. Instead, substrate orientation is possible in either way, parallel and antiparallel to the HEXXH motif, and hence the observed bidirectional degradation (25) can occur.
Substrate Access Is Granted by a Narrow Cleft Guarded by Aromatic Residues.
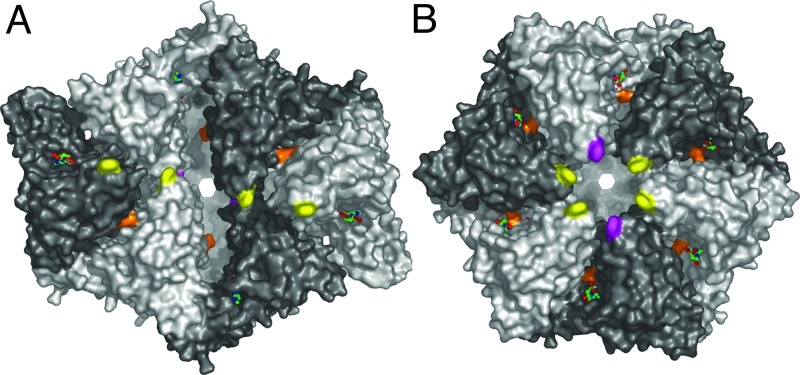
FtsH is a self-compartmentalizing protease (26) with the six active sites located inside the hexameric molecule. The distance between the active centers is ≈34 Å. Access to the active sites is supposed to be mediated by the AAA domains that face the cytosolic leaflet of the membrane. From this side, access to the interior is given by an S-shaped cleft (Fig. 4A) with a width of ≈20 Å. On the side walls of the crown there are six openings with diameters of ≈15 Å, and at the center of the bottom there is a 15-Å-wide circular channel. It is reasonable to assume that the S-shaped cleft facing the membrane is the entry site of the target polypeptide chain while the other channels serve for product release. According to the “pulling model,” after binding to the recognition tag the enzyme begins to translocate this tag through the narrow pore, thereby generating a denaturing force (1). The S-shaped cleft possesses a hydrophobic patch at its center that is mainly built by Phe-234 from the six subunits. The arrangement of these Phe-234 residues is such that one pair of crystallographically related side chains sits on top of the entrance (facing the membrane side of the “crown”) while another pair lies below them (colored magenta in Fig. 4). The two remaining ones are located further away from the S-shaped cleft. Position 234 carries a conserved aromatic residue in all FtsH orthologs from bacteria, chloroplasts, and mitochondria. In E. coli, mutation of the equivalent amino acid (Phe-225) to alanine gives a protein that is not any more able to degrade σ32 but can still hydrolyze unfolded substrates like resorufin-casein, whereas substitution of this phenylalanine by charged residues (e.g., Glu) abolishes activity completely (27).
Fig. 4.
Surface representation. (A) Top view onto AAA ring. Phe-234 residues are colored in yellow and magenta, and Arg-318 is in orange. The orientation is the same as in Fig. 2A. ADP residues are shown as sticks. Subunits are shaded alternately light and dark. (B) Modeled ideal hexameric arrangement of the AAA domains. The protease ring is in the same orientation as in A and Fig. 2A.
Phe-234 is located within a stretch of amino acids strictly conserved in FtsH, but not in HslU or ClpX/B. However, aromatic residues lining the substrate entrance pore were shown to be important for the substrate recognition by ClpB (28), and a pore motif φXG was proposed for HslU, where φ stands for an aromatic residue and X is mostly apolar (29).
Discussion
The crystal structure of FtsH shows some surprising results. Besides the new fold of the protease domain revealing an aspartic acid as the third zinc ligand, the striking breakdown of the expected hexagonal symmetry in the AAA ring requires some discussion. One possibility is a potential requirement for a symmetry mismatch between ATPase and protease moieties during the catalytic cycle. Such symmetry mismatches occur, for example, between hexameric ClpX ATPase and heptameric ClpP protease molecules in the ClpXP complex (30). It is noteworthy that symmetry reduction similar to FtsH was observed in the case of T7 gene 4 ring helicase, where the distortion of the symmetry from C6 to C2 was interpreted in terms of sequential nucleotide hydrolysis and DNA translocation (31). The exact role of such a symmetry mismatch in ATP-dependent proteases is not very clear at the moment and seems not to be a general requirement because HslV and HslU, the two components of the HslUV complex, have sixfold symmetry both (32, 33). FtsH is, like HslUV, a symmetry-matched system in the sense that the numbers of AAA and protease domains are equal. However, the symmetries of ATPase and protease assembly differ much more drastically than in HslUV.
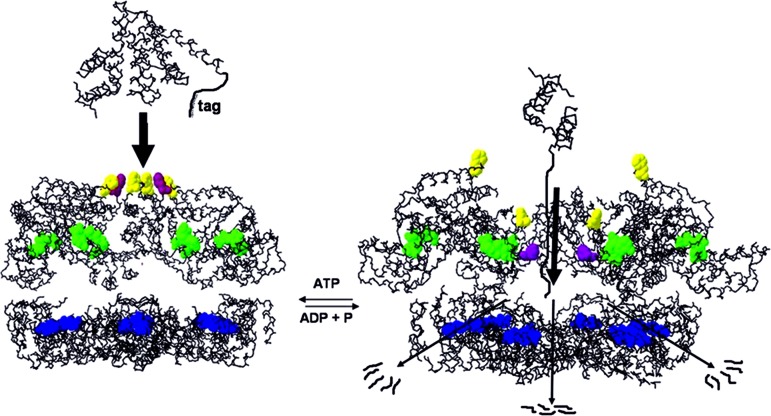
During the catalytic cycle, conformational changes must take place upon ATP hydrolysis to generate a mechanical force, and they can take place within the AAA domain as well as between the AAA and protease or other domains present (34–36). In the structure reported here, we do not observe significant differences between the individual AAA domains (Fig. 3B). In particular, the angle between the N-terminal “wedge” and the C-terminal α-helical part is essentially the same in all six copies of the AAA domains. This finding can be attributed to the fact that all six domains have the same nucleotide (ADP) bound. It is possible that the different symmetries of the two rings represent an intermediate of the translocation/unfolding mechanism and different arrangements of the AAA domains can occur, dependent on catalytic state and nucleotide bound. Arg-321 (and possibly Arg-318 as well) from the SRH is supposed to function as “arginine finger,” i.e., facilitating ATP hydrolysis by positioning the positively charged headgroup close to the γ-phosphate of an ATP bound to a neighboring subunit (3, 33). In the structure presented here, the side chain of Arg-318 is mostly disordered and the charged headgroup can be estimated to be at least some 5–10 Å away from the putative position of the γ-phosphate in a proximal subunit. Arg-321 engages in a salt bridge with the conserved Asp-292 from the same subunit. It is conceivable, however, that the presence of a γ-phosphate in the neighboring subunit can be sensed by at least one of these arginines, leading to a closer contact of the two AAA domains by a rigid-body movement driven by the electrostatic interactions between guanidinium group γ-phosphate and hence to stronger domain–domain interactions in the case of bound ATP. If this occurred to the same extend simultaneously in all subunits, it would lead to a perfect sixfold symmetry in the AAA ring. Modeling an ideal hexameric AAA ring as reported earlier (17) would result in all six Phe-234 residues lining a circular pore located at the center of the former S-shaped cleft (Fig. 4B). One might envisage such an arrangement as another intermediate of the “pulling model.” The central hydrophobic patch could bind to a recognition signal, and subsequent hydrolysis of ATP could then lead to an arrangement similar to the one shown in Fig. 4A. Here, two of the phenylalanines have moved toward the inside of the molecule, and two other have moved further away from the entrance channel. This rearrangement of the AAA domains would lead to an effective movement or “pulling” of the target polypeptide chain attached to the hydrophobic side chains of Phe-234 and also move the target more toward the interior of the hexamer (Fig. 5). This mechanism would still work to some extent even if hydrolysis or ADP–ATP exchange does not occur in all six AAA domains simultaneously but rather in a probabilistic manner (14, 37), as long as there is space for this domain movement. The lack of coupling between ATP hydrolysis and proteolysis in (Δtm)FtsH constructs, and hence the inability in degrading σ32, could be explained by the absence of “elastic springs” formed by the transmembrane helices that would restrain the free movement of the AAA domains and possibly prevent them from being locked into one intermediate conformation on the reaction pathway.
Fig. 5.
Schematic drawing of degradation mechanism. Side view through open-sliced molecule. The AAA ring is on top, and the protease ring is at the bottom. Phe-234 residues are colored as in Fig. 4, nucleotides are shown in green, and active site residues are in blue. After binding of an apolar recognition tag by the hydrophobic patch formed by Phe-324, ATP hydrolysis leads to the inward movement of four Phe residues, translocating the target polypeptide into the interior of the molecule followed by proteolysis.
Experimental evidence for such a model is somewhat contradictory: on the one hand, mutation of Arg-325 in HslU (corresponding to Arg-318 in T. maritima FtsH) or Arg-359/362 in p97/VCP abolishes hexamerization (38, 39), but this does not hold for other AAA proteins (40). Further experiments will be needed to establish the precise mechanism and the nature of the conformational change occurring upon ATP binding.
Materials and Methods
Protein Expression and Purification.
Amino acids 147–610 of FtsH from T. maritima were amplified from genomic DNA and ligated into a pET28a expression vector (Stratagene) adding a C-terminal hexa-His-tag. Expression in Rosetta (DE3) cells (Novagen) was induced by adding 0.5 mM IPTG at an OD600 of 0.5. Heat precipitation of E. coli proteins was carried out at 75°C for 3 min. The supernatant was further purified by Ni-NTA and anion-exchange chromatography. (Δtm)FtsH-containing fractions were again pooled and loaded onto an S75 size-exclusion column (Amersham Pharmacia Biosciences) equilibrated with buffer G (100 mM NaCl/2 mM DTT/20 mM Tris, pH 8.0/0.02% sodium azide). The resulting two well separated peaks (corresponding to molecular masses of ≈600 and 100 kDa) were pooled and concentrated individually.
Point mutations K410L-K415A and D500A were introduced by the QuikChange (Stratagene) mutagenesis protocol, using the pET28a-(Δtm)FtsH vector as a template (mutagenesis sense primers: K410L-K415A, 5′-cccgcaagaaagtcgctgctgatcagccctgcagaaaagcgc-3′; D500A, 5′-cgagtggggcagccaacgcgatcgaaa-gggccacggaaattgcgagg-3′).
Crystallization.
Crystals were obtained at 20°C using the microbatch method by mixing equal volumes of protein (20 mg/ml) and crystallization buffer: 30% (wt/vol) PEG 400, 200 mM CaCl2, 100 mM Hepes (pH 7.5), 15% (wt/vol) xylitol, and 0.1–0.2% (wt/vol) low-melt agarose (FMC BioProduct). Crystals belong to the spacegroup P41212 with cell dimensions of a = 164.7 Å, c = 233.1 Å. To improve reproducibility, especially for the selenomethionine-labeled protein, two lysine residues (Lys-410 and Lys-415) were mutated to amino acids occurring at these positions in bacterial homologs from other sources. This double mutant K410L-K415A gives the same results in ultracentrifugation and activity experiments as the wild type and crystallizes isomorphously. However, diffraction properties and reproducibility were much improved. For cryoprotection, crystals were transferred to crystallization buffer containing 5% (vol/vol) glycerol for 1 min before flash-cooling in liquid nitrogen. Selenomethionine-incorporated protein was produced by the methionine biosynthesis inhibition method (41) and treated as the wild-type protein.
X-Ray Data Collection, Phasing, and Refinement.
X-ray data were collected at 100 K at beamlines BM-14 (European Synchrotron Radiation Facility, Grenoble, France), PX-06 (Swiss Light Source, Villigen), and X11 (Deutsche Elektronen-Synchrotron, European Molecular Biology Laboratory, Hamburg). The data-collection statistics are given in Table 1. Diffraction intensities were processed and scaled with xds (42). Multiple wavelength anomalous diffraction data of selenomethionine-labeled crystals led to the determination of 32 of 48 Se-sites by employing the program shelxd (43). Phasing with sharp (44) followed by density modification and phase extension to 2.8 Å resolution by using resolve (45) resulted in a clearly interpretable electron density map with a figure of merit of 0.60–2.77 Å resolution as computed by sharp. The structure of the published AAA domain from Thermus thermophilus was placed in the electron density with the program essens (46). An initial model was built automatically by using maid (47) and corrected and completed by using o (48).
Table 1.
Data collection, phasing, and refinement statistics
| Crystal 1 native | Crystal 2 Semet1 |
Crystal 3 Semet2 | Crystal 4 D500A | |||
|---|---|---|---|---|---|---|
| Peak | Inflection | Remote | ||||
| Data collection | ||||||
| Wavelength, Å | 1.00531 | 0.9796 | 0.9797 | 0.9184 | 0.9184 | 0.9760 |
| Resolution, Å | 2.77 | 3.20 | 3.20 | 3.20 | 3.20 | 3.00 |
| Rsym, % | 4.4 (28.3) | 9.4 (42.8) | 8.7 (40.6) | 7.1 (29.3) | 7.7 (33.2) | 6.6 (49.6) |
| I/σ(I) | 29.4 (6.6) | 17.3 (4.6) | 15.6 (4.0) | 17.4 (5.0) | 12.5 (4.1) | 20.8 (3.5) |
| Completeness, % | 99.2 (94.2) | 99.8 (99.5) | 99.7 (99.5) | 99.6 (99.2) | 97.9 (97.4) | 99.1 (99.8) |
| Redundancy | 7.1 (7.1) | 7.0 (6.9)* | 5.0 (5.0)* | 5.2 (5.2)* | 5.0 (5.0) | 5.0 (5.0) |
| Refinement | ||||||
| Resolution, Å | 2.77 | 3.00 | ||||
| No. of reflections | 81,127 | 64,723 | ||||
| Rwork/Rfree, % | 25.2/27.6 | 25.7/29.0 | ||||
| No. of atoms | 19,308 | 19,290 | ||||
| Protein | 19,123 | 19,105 | ||||
| Ligand/ion | 168 | 162 | ||||
| Water | 6 | 0 | ||||
| RMS bond lengths, Å | 0.012 | 0.007 | ||||
| RMS bond angles, ° | 1.49 | 1.30 | ||||
Numbers in brackets refer to the highest-resolution shell (2.90–2.77 Å for native, 3.39–3.20 Å for semet, and 3.10–3.00 Å for the mutant D500A data). Semet, selenomethionine.
*Friedel pairs are counted as individual reflections.
Refinement was carried out by using cns (49) and refmac (50) employing experimental phase restraints. Statistics are given in Table 1. A Ramachandran plot according to Kleywegt and Jones (51) shows 3.4% outliers. Waters have not been included so far, other than those coordinating to the Zn2+.
Analytical Ultracentrifugation.
Analytical ultracentrifugation experiments were performed at the Institute of Molecular Biology and Biophysics at the Swiss Federal Institute of Technology by using an Optima XL-1 ultracentrifuge (Beckman). Protein samples were concentrated to 0.5 mg/ml in buffer U (100 mM NaCl/20 mM Tris, pH 8.0). Data were evaluated by using the program sedfit (National Institutes of Health).
Protease Assay.
Tests for proteolytic activity employing resorufin-casein (Roche) were carried out according to the manufacturer’s instructions. Briefly, 0.1 mg (1.9 nmol) of protease in a volume of 50 μl (buffer G) and 50 μg (≈2.5 nmol) of substrate (in 100 μl of water) were mixed in the presence or absence of up to 4 μmol of ATP with 50 μl of incubation buffer (0.2 M Tris, pH 7.8/0.02 M CaCl2). The presence or absence of ATP gave identical results (data not shown). Reactions were stopped by the addition of 480 μl of 5% (wt/vol) trichloroacetic acid. Experiments were performed as recommended by the supplier (Roche) at a temperature of 50°C. The absorbance was measured at various time points at a wavelength of 574 nm (Fig. 1). For inhibition studies, 1.9 nmol of (Δtm)FtsH was incubated with 10 mmol of ortho-phenanthroline (Fluka) for 2 h.
ATPase Assay.
Twenty micrograms (0.38 nmol) of protein in 18 μl of buffer A (100 mM NaCl/20 mM MgCl2/50 mM Tris, pH 8) was mixed with 2 μl of 20 mM ATP and 0.25 μCi/μl (1 Ci = 37 GBq) of [γ-32P]ATP (Amersham Pharmacia Biosciences) and incubated at 37°C. For inhibition of ATPase activity, 50 mM EDTA was added to the samples and incubated for 1 h. Samples were taken at different time points and separated by thin-layer chromatography (PEI-Cellulose F; Merck), run in 0.2 M KH2PO4, 0.33 M HCOOH, and 0.19 M LiCl, and analyzed by using a phosphoimager (FLA-3000, Fujifilm).
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Clemens Schulze-Briese at beamline X06SA of the Swiss Light Source, Paul Scherrer Institute; Max Nanao, Hassan Belrhani, and Martin Walsh at beamline BM14 of the European Synchrotron Radiation Facility; and Brice Kauffmann at Deutsche Elektronen-Synchrotron, European Molecular Biology Laboratory outstation. This work was supported by the Swiss National Science Foundation and the Berner Hochschulstiftung.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2CE7 and 2CEA).
References
- 1.Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., et al. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 3.Ito K., Akiyama Y. Annu. Rev. Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 4.Begg K. J., Tomoyasu T., Donachie W. D., Khattar M., Niki H., Yamanaka K., Hiraga S., Ogura T. J. Bacteriol. 1992;174:2416–2417. doi: 10.1128/jb.174.7.2416-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayasekera M. M., Foltin S. K., Olson E. R., Holler T. P. Arch. Biochem. Biophys. 2000;380:103–107. doi: 10.1006/abbi.2000.1903. [DOI] [PubMed] [Google Scholar]
- 6.Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama Y., Kihara A., Tokuda H., Ito K. J. Biol. Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama Y., Kihara A., Ito K. FEBS Lett. 1996;399:26–28. doi: 10.1016/s0014-5793(96)01283-5. [DOI] [PubMed] [Google Scholar]
- 9.Zaltsman A., Feder A., Adam Z. Plant J. 2005;42:609–617. doi: 10.1111/j.1365-313X.2005.02401.x. [DOI] [PubMed] [Google Scholar]
- 10.Blaszczak A., Georgopoulos C., Liberek K. Mol. Microbiol. 1999;31:157–166. doi: 10.1046/j.1365-2958.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 11.Carmona M., de Lorenzo V. Mol. Microbiol. 1999;31:261–270. doi: 10.1046/j.1365-2958.1999.01169.x. [DOI] [PubMed] [Google Scholar]
- 12.Shotland Y., Shifrin A., Ziv T., Teff D., Koby S., Kobiler O., Oppenheim A. B. J. Bacteriol. 2000;182:3111–3116. doi: 10.1128/jb.182.11.3111-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman C., Prakash S., Lu C. Z., Matouschek A., Gross C. A. Mol. Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 14.Hersch G. L., Burton R. E., Bolon D. N., Baker T. A., Sauer R. T. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Akiyama Y., Ito K. Biochemistry. 2001;40:7687–7693. doi: 10.1021/bi010039w. [DOI] [PubMed] [Google Scholar]
- 16.Makino S., Makino T., Abe K., Hashimoto J., Tatsuta T., Kitagawa M., Mori H., Ogura T., Fujii T., Fushinobu S., et al. FEBS Lett. 1999;460:554–558. doi: 10.1016/s0014-5793(99)01411-8. [DOI] [PubMed] [Google Scholar]
- 17.Niwa H., Tsuchiya D., Makyio H., Yoshida M., Morikawa K. Structure (Cambridge, Mass.) 2002;10:1415–1423. doi: 10.1016/s0969-2126(02)00855-9. [DOI] [PubMed] [Google Scholar]
- 18.Krzywda S., Brzozowski A. M., Verma C., Karata K., Ogura T., Wilkinson A. J. Structure (Cambridge, Mass.) 2002;10:1073–1083. doi: 10.1016/s0969-2126(02)00806-7. [DOI] [PubMed] [Google Scholar]
- 19.Herman C., Thevenet D., Bouloc P., Walker G. C., D’Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobiler O., Koby S., Teff D., Court D., Oppenheim A. B. Proc. Natl. Acad. Sci. USA. 2002;99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saikawa N., Akiyama Y., Ito K. J. Struct. Biol. 2004;146:123–129. doi: 10.1016/j.jsb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Saikawa N., Ito K., Akiyama Y. Biochemistry. 2002;41:1861–1868. doi: 10.1021/bi015748o. [DOI] [PubMed] [Google Scholar]
- 23.Holm L., Sander C. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shotland Y., Teff D., Koby S., Kobiler O., Oppenheim A. B. J. Mol. Biol. 2000;299:953–964. doi: 10.1006/jmbi.2000.3767. [DOI] [PubMed] [Google Scholar]
- 25.Chiba S., Akiyama Y., Ito K. J. Bacteriol. 2002;184:4775–4782. doi: 10.1128/JB.184.17.4775-4782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumeister W., Walz J., Zuhl F., Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 27.Yamada-Inagawa T., Okuno T., Karata K., Yamanaka K., Ogura T. J. Biol. Chem. 2003;278:50182–50187. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- 28.Schlieker C., Weibezahn J., Patzelt H., Tessarz P., Strub C., Zeth K., Erbse A., Schneider-Mergener J., Chin J. W., Schultz P. G., et al. Nat. Struct. Mol. Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Song J. J., Franklin M. C., Kamtekar S., Im Y. J., Rho S. H., Seong I. S., Lee C. S., Chung C. H., Eom S. H. Structure (Cambridge, Mass.) 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 30.Joshi S. A., Hersch G. L., Baker T. A., Sauer R. T. Nat. Struct. Mol. Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- 31.Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 32.Sousa M. C., Trame C. B., Tsuruta H., Wilbanks S. M., Reddy V. S., McKay D. B. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 33.Bochtler M., Hartmann C., Song H. K., Bourenkov G. P., Bartunik H. D., Huber R. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 34.Davies J. M., Tsuruta H., May A. P., Weis W. I. Structure (Cambridge, Mass.) 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 35.DeLaBarre B., Brunger A. T. J. Mol. Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 36.Rouiller I., DeLaBarre B., May A. P., Weis W. I., Brunger A. T., Milligan R. A., Wilson-Kubalek E. M. Nat. Struct. Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 37.Martin A., Baker T. A., Sauer R. T. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 38.Song H. K., Hartmann C., Ramachandran R., Bochtler M., Behrendt R., Moroder L., Huber R. Proc. Natl. Acad. Sci. USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q., Song C., Irizarry L., Dai R., Zhang X., Li C. C. J. Biol. Chem. 2005;280:40515–40523. doi: 10.1074/jbc.M509636200. [DOI] [PubMed] [Google Scholar]
- 40.Karata K., Inagawa T., Wilkinson A. J., Tatsuta T., Ogura T. J. Biol. Chem. 1999;274:26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- 41.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. In: International Tables for Crystallography. Rossmann M. G., Arnold E., editors. Vol. F. The Netherlands: Kluwer Academic, Dordrecht; 2001. pp. 730–734. [Google Scholar]
- 43.Weeks C. M., Adams P. D., Berendzen J., Brunger A. T., Dodson E. J., Grosse-Kunstleve R. W., Schneider T. R., Sheldrick G. M., Terwilliger T. C., Turkenburg M. G., Uson I. Methods Enzymol. 2003;374:37–83. doi: 10.1016/S0076-6879(03)74003-8. [DOI] [PubMed] [Google Scholar]
- 44.Perrakis A., Morris R., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 45.Terwilliger T. C. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleywegt G. J., Jones T. A. Acta Crystallogr. D. 1997;53:179–185. doi: 10.1107/S0907444996012279. [DOI] [PubMed] [Google Scholar]
- 47.Levitt D. G. Acta Crystallogr. D. 2001;57:1013–1019. doi: 10.1107/s0907444901006394. [DOI] [PubMed] [Google Scholar]
- 48.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Kleywegt G. J., Jones T. A. Structure (Cambridge, Mass.) 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.