Short abstract
A report on the Third Annual International Conference on Transposition and Animal Biotechnology, Minneapolis, USA, 23-24 June 2005, and the FASEB Summer Research Conference 'Mammalian Mobile Elements', Tuscon, USA, 4-9 June, 2005.
Abstract
A report on the Third Annual International Conference on Transposition and Animal Biotechnology, Minneapolis, USA, 23-24 June 2005, and the FASEB Summer Research Conference 'Mammalian Mobile Elements', Tuscon, USA, 4-9 June, 2005.
Transposons are mobile genetic elements with the ability to move to new sites in host genomes. This mobility gives them awesome potential as genome-altering tools for somatic and germline mutagenesis, and as gene-delivery tools in the laboratory and for gene therapy. Two meetings on transposons this summer, in Minneapolis and Tucson, revealed the impressive progress in this field, with emphasis on transposons in vertebrates. The following report describes a few of the highlights from these meetings.
Transposable elements are classified as either DNA transposons or retrotransposons on the basis of their mode of transposition. Eukaryotic DNA transposons transpose by a conservative 'cut-and-paste' mechanism; this group includes the Tc1/mariner, hAT, P-element, piggyBac, Mutator, and En/Spm families. Retrotransposons replicate via an RNA intermediate by a 'copy-and-paste' mechanism, and are further subdivided into long terminal repeat (LTR)- and non-LTR types. LTR-retrotransposons are widely distributed among diverse eukaryotes. Phylogenetic analyses based on reverse transcriptase indicate the existence of at least four distinct lineages of LTR-retrotransposons, and five groups of non-LTR retrotransposons. The list is expanding as more organisms are being sequenced and analyzed. Russell Poulter (University of Otago, Dunedin, New Zealand) reported his group's recent identification of an array of transposable elements in fungi and vertebrates, and presented compelling genetic evidence that Zorro-3, a retrotransposon-derived L1 element from Candida albicans, was still transpositionally active.
The Sleeping Beauty transposon
A star of both meetings was the Sleeping Beauty (SB) DNA transposon, a vertebrate member of the Tc1/mariner family that was resurrected from defective ancient elements through site-directed mutagenesis in 1997. SB is typically used as a two-component system: one component is a gutted transposon carrying a reporter gene(s) and/or other molecular bells and whistles, flanked by the inverted repeats containing transposase-binding sites; the second is the SB transposase expressed under the control of a heterologous promoter, which is necessary and sufficient for transposition. The transposition process is not, however, independent of the state of the host cell. Zoltan Ivics (Max Delbruck Center for Molecular Medicine, Berlin-Buch, Germany), who originally revived Sleeping Beauty, reported that SB transposition may be coordinated with cell-cycle control. It is well known that cyclin D1 is a key regulatory factor that promotes cell-cycle progression from G1 to S phase. Interestingly, a reduction of cyclin D1 expression level was observed when SB transposase was overexpressed in human cells, resulting in an extended G1 phase. The molecular mechanism for downregulation of cyclin D1 by SB transposase is being characterized.
The transposition activity of SB has been the focal point of many studies. The element transposes efficiently in a variety of vertebrate cell lines, in mouse somatic tissues, and in the mouse germline, but, unlike retrotransposons, many sites of SB insertion cluster in the vicinity of its chromosome of origin, a phenomenon termed 'local hopping'. To further improve SB transposition activity, SB is being engineered: mutation of the transposase-binding sites and searches for more active versions of the transposase are both being attempted. The stakes for optimization are high, as even a twofold increase in activity could translate into a significant improvement, for example in the efficacy of SB for gene therapy or mutagenesis. This was exemplified by Bradley Fletcher (University of Florida, Gainesville, USA), who reported efforts to develop a more active SB vector system for gene therapy by combining individual improvements discovered by different groups. The new SB system displayed a substantial 16-fold increase in transposition efficiency as compared to the original system in cultured cells, but when it was tested as a non-viral gene-delivery vehicle in mice only a modest twofold increase of transgene expression was achieved.
Cancer gene discovery and germline mutagenesis
In less than a decade, researchers have successfully adapted the SB system to several major applications in vertebrate genomics, summarized by David Largaespada (University of Minnesota, Minneapolis, USA) as germline transgenesis, somatic transgenesis (gene therapy), germline insertional mutagenesis, and somatic cell mutagenesis (Figure 1). Perhaps the most dramatic breakthrough is in somatic cell mutagenesis and its application to the discovery of potential oncogenes, as illustrated in two presentations at the Minneapolis conference. Previously, the limited activity of SB in cultured cells and limited evidence for active somatic transposition in vivo had prevented its use for identifying tumorigenic genes. This barrier has now been broken by the design of more effective mutagenic SB by two collaborating research groups using different approaches. Adam Dupuy (National Cancer Institute, Frederick, USA) has incorporated several proven designs into his system. The transposon itself was first designed to disrupt the expression of an endogenous gene independent of insertion orientation; the new vector also included retroviral enhancer/promoter sequences well known to activate oncogenes, and it had optimized transposase-binding sites and overall size. Second, founder mouse lines with the highest number of unmethylated transposon copies were selected. Finally, a single-copy knock-in line for an improved SB transposase (ROSA-SB11) was constructed, providing ubiquitous and consistent transposase expression. The first sign of success was embryonic lethality in the transposon/transposase double-transgenic lines. By 6 weeks after birth, evidence suggests that the donor copies had virtually all excised from the original integration site and jumped to other genomic locations. The double-transgenic mice were tumor-prone, with high penetrance (the proportion showing a mutant phenotype); by 17 weeks all had succumbed to tumors. On examination, all the tumors contained clonal or subclonal SB insertions. Remarkably, in this study there was little evidence for local hopping, perhaps because the selection for tumors was so strong that rare insertions with strong tumorigenic potential predominated, and/or because transposition rates were so high that there were multiple rounds of transposition in each cell; modeling suggests that multiple rounds of transposition would rapidly reduce the impact of local hopping. It is notable that tumors were induced in a genetic background not predisposed to cancer, demonstrating the feasibility and power of using SB transposon technology in cancer gene discovery. One disappointment was that the tumor spectrum included a preponderance of lymphomas. This may reflect the tropism of the retroviral enhancer/promoter used, or may simply be because hematopoietic stem cells constitute the largest target of self-renewing stem cells in the body. This bias in the tumor spectrum can probably be overcome by tissue-specific expression of SB transposase.
Figure 1.
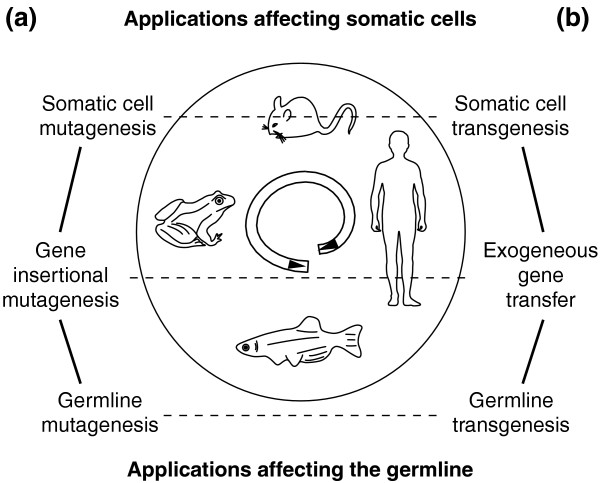
Four major applications of transposon technology in vertebrate functional genomics. The four organisms surrounding the DNA transposon indicate that certain transposons, such as elements belonging to the mariner family, can be used in a broad range of hosts as their movement is largely independent of host functions. (a) Most transposons can disrupt host genes upon insertion. Such insertions can be somatic insertions, which can be used to discover and analyze cancer genes, or germline insertions resulting in heritable mutations that produce phenotypic change in the progeny. (b) Transposons can also be used to deliver exogenous genes into the organism through somatic cell transgenesis (in gene therapy) or germline transgenesis (producing transgenic animals at high efficiency).
Lara Collier (University of Minnesota, Minneapolis, USA) reported the use of a complementary strategy - SB-induced formation of solid tumors in a sensitized p19/Arf mutant mouse line that is predisposed to cancer owing to a deficiency of the Arf tumor suppressor. Retrovirus-mediated mutagenesis screens had previously shown a predisposition to hematopoietic cancers, and to a lesser extent to mammary cancer, but to few other types. Cancer researchers have been waiting for an alternative mutagenesis system to identify genes involved in solid tumor formation. Collier's SB system features a similar transposon vector to Dupuy's, with gene-disrupting elements and the identical retroviral enhancer/promoter, but with a seemingly less aggressive transposase (CAGGS-SB10). In contrast to Dupuy's study, mice doubly transgenic for this transposon/transposase combination did not show increased cancer susceptibility for more than a year after birth. This changed when the system was crossed into the p19/Arf mice; time to morbidity was significantly shortened in the SB animals compared with control p19/Arf-deficient mice. Over 95% of mice from the experimental group in which transposons were mobilizing in the soma succumbed mainly to soft-tissue sarcoma or osteosarcoma in one year compared with around 70% in the controls. One of the common insertion sites is a known oncogene, Braf, which was hit in around 80% of the sarcomas. Analysis of the insertions revealed an approximately twofold higher rate of local hopping compared to Dupuy's work, presumably reflecting the lower frequency of multiple cycles of transposition in Collier's study. So far, no direct comparison has been made between the two systems regarding actual transposition frequencies, transposase expression levels, and mutational patterns. Thus, it remains a mystery as to how much the various components in the two studies contributed to the discrepancies of cancer susceptibility in the wild-type background. Collier's work nevertheless represents a powerful complementary strategy for discovering genes operating in a specific pathway(s) in a sensitized background.
Significant developments are also being made in germline mutagenesis using SB in the mouse. Earlier work suggested that local hopping is most pronounced for SB transposition in the mouse germline. The obvious implication of this is that to achieve unbiased insertion throughout the whole genome, one has to start with a number of independent transgenic lines in which the transposon concatemer is located on different chromosomes. On the other hand, this phenomenon can be exploited for region-specific saturation mutagenesis, as shown in two presentations. Aron Geurts (University of Minnesota, Minneapolis, USA) reported progress on an SB-based forward-genetic screen in the mouse germline. A balancer strain was used to recover recessive lethal mutations and to facilitate a three-generation screening process. More than ten pedigrees with recessive lethal phenotypes and one with a very specific dominant viable polydactyly phenotype were identified. Chikara Kokubu (Osaka University, Osaka, Japan) presented a very elegant exploitation of SB local hopping to engineer a nested series of deletion mutations which he then applied to region-specific mapping of cis-regulatory elements (for example, enhancers and insulators) in the complex Pax1 gene locus of the mouse genome.
The transposon technology toolkit
Although SB is the current bright star, other transposons are also being developed into useful functional genomics tools, such as the fish DNA transposon Tol2, the mouse LTR-retrotransposon IAP and the mammalian non-LTR retrotransposon L1 from human and mouse. Tol2 belongs to the hAT family of DNA transposons, and is so far the only known naturally occurring active DNA transposon in vertebrates. When tested in transgenesis, the germline transmission frequency by the Tol2 system is slightly higher than that by the current SB counterpart. Koichi Kawakami (National Institute of Genetics, Shizuoka, Japan), who first identified a functional Tol2 transposase, discussed the use of Tol2 transposon system in efficient gene and enhancer trapping in zebrafish, and has characterized scores of fish lines with unique expression patterns for the green fluorescent protein (GFP) reporter. Vladimir Korzh (Institute of Molecular and Cell Biology, Singapore) on the other hand elaborated on a range of downstream research applications that may be applicable to existing Tol2-mediated enhancer trap zebrafish lines. Such examples include the potential use of the GFP reporter in some lines as a built-in in vivo histological marker, which can be very useful in tracking single cell fate and/or for morphological studies. In addition, the existing copies of SB may serve as 'launching pads' for further transposition events in somatic tissues if transposase mRNA is supplied by injection.
Kyoji Horie (Osaka University, Osaka, Japan) introduced a new player to the transposon technology field. This is an IAP element initially isolated from a mouse tumor cell line; it shows high transposition activity in cultured cell lines but has not yet been shown to transpose in mice. Currently, the potential mechanism of IAP silencing in transgenic mice is being explored. Eric Ostertag (University of Pennsylvania, Philadelphia, USA) presented an update on the characterization of human L1-mediated insertions in mice, building on his previous demonstration of the active transposition of human L1 in the mouse germline. One of us (J.D.B.) provided initial evidence that a synthetic mouse L1 retrotransposon had high transposition activity in mice. Confirming previous observations from cell-culture-based experiments, neither Ostertag nor ourselves detected any significant bias in integration-site selection by L1 retrotransposons in vivo, which is in sharp contrast to the local hopping by SB, demonstrating the special value of L1 as an additional gadget in the transposon technology toolkit.
In a truly unexpected development that suggests a potential role for natural L1 elements in mammalian biology, Alysson Muotri from Fred Gage's laboratory (The Salk Institute, La Jolla, USA) described the alteration of neuronal gene expression in an L1 transgenic mouse line, and provided a provocative potential connection between L1 retrotransposition and neural somatic mosaicism. Because the notion of 'function benefiting the host' is anathema to the 'selfish' nature of active mobile elements, this interesting study stimulated extensive discussion.
One of the dilemmas for mouse functional genomics researchers is that, for technical and historic reasons, there has been no uniform standard for which mouse strains or genetic background are used in experiments, although C57BL/6 is the obvious candidate, as it has been sequenced. The yeast (Saccharomyces cerevisiae) community found itself in the same predicament a decade ago when the yeast sequence was completed: a sequence of one strain but a multitude of interesting and diverse strains in which to do biology. A concerted community effort was made to construct a uniform mutational resource, the yeast knockout (YKO) collection in the sequenced strain background. The YKO resource has been wildly successful, and is the starting point for virtually every genetic screen done today; it provides the uniform background that can then be embellished by 'genome artists' working in outlying strains of yeast.
The time has come for the mouse functional genomics community to make a similar bold move. Currently, mouse functional genomics is sharply divided between those who use chemical mutagenesis, such as ethylnitrosourea (ENU) mutagenesis, and those who use gene-trapping. Both these approaches are extremely useful, but also have severe limitations: ENU mutations are not easily mapped, and gene-trapping is done in tissue culture, not directly in the mouse. The efforts of groups using these approaches have been piecemeal and disparate, and there has been no effort to provide a mutational resource in a uniform genetic background. The opportunity is here to use the transposon toolbox as part of a community effort to generate a public mouse insertional mutation resource, without the costs of working with embryonic stem cells, but using simple breeding approaches and with the tremendous benefit that each mutation is tagged and can thus easily be mapped. Transposon technology has great potential in vertebrate functional genomics, and as the transposon toolkit is rapidly expanded, we hope to see advances in many important areas, including gene therapy, cancer modeling and gene discovery, in the near future.


