Short abstract
Expansins are plant proteins that loosen cell walls via a nonenzymatic mechanism; they are involved in cell expansion and other developmental events during which cell-wall modification occurs.
Abstract
The expansin superfamily of plant proteins is made up of four families, designated α-expansin, β-expansin, expansin-like A and expansin-like B. α-Expansin and β-expansin proteins are known to have cell-wall loosening activity and to be involved in cell expansion and other developmental events during which cell-wall modification occurs. Proteins in these two families bind tightly to the cell wall and their activity is typically assayed by their stimulation of cell-wall extension and stress relaxation; no bona fide enzymatic activity has been detected for these proteins. α-Expansin proteins and some, but not all, β-expansin proteins are implicated as catalysts of 'acid growth', the enlargement of plant cells stimulated by low extracellular pH. A divergent group of β-expansin genes are expressed at high levels in the pollen of grasses but not of other plant groups. They probably function to loosen maternal cell walls during growth of the pollen tube towards the ovary. All expansins consist of two domains; domain 1 is homologous to the catalytic domain of proteins in the glycoside hydrolase family 45 (GH45); expansin domain 2 is homologous to group-2 grass pollen allergens, which are of unknown biological function. Experimental evidence suggests that expansins loosen cell walls via a nonenzymatic mechanism that induces slippage of cellulose microfibrils in the plant cell wall.
Gene organization and evolutionary history
Expansins are plant cell-wall loosening proteins involved in cell enlargement and in a variety of other developmental processes in which cell-wall modification occurs [1]. They are typically 250-275 amino acids long and are made up of two domains (domain 1 and domain 2) preceded by a signal peptide (Figure 1). On the basis of phylogenetic sequence analysis (Figure 2), four families of expansins are currently recognized in plants [2]. From the largest family to the smallest they are designated α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA) and expansin-like B (EXLB). α-Expansin and β-expansin proteins have been demonstrated experimentally to cause cell-wall loosening [3,4], whereas expansin-like A and expansin-like B proteins are known only from their gene sequences.
Figure 1.
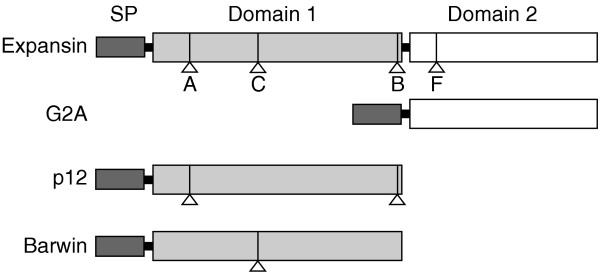
The domain structure of expansins and a comparison with that of distantly related single-domain plant proteins (G2A, p12 and barwin). The expansin signal peptide (SP) is 20-30 amino acids long, domain 1 is 120-135 amino acids, and domain 2 is 90-120 amino acids. Some barwin proteins have an additional chitin-binding domain after the signal peptide (not shown). The positions of the introns that are present in more than one expansin family are indicated by lettered triangles; homologous introns are present in p12 and barwin proteins. Intron letters are as in [7]. The position of intron B suggests that it could have participated in exon shuffling.
Figure 2.
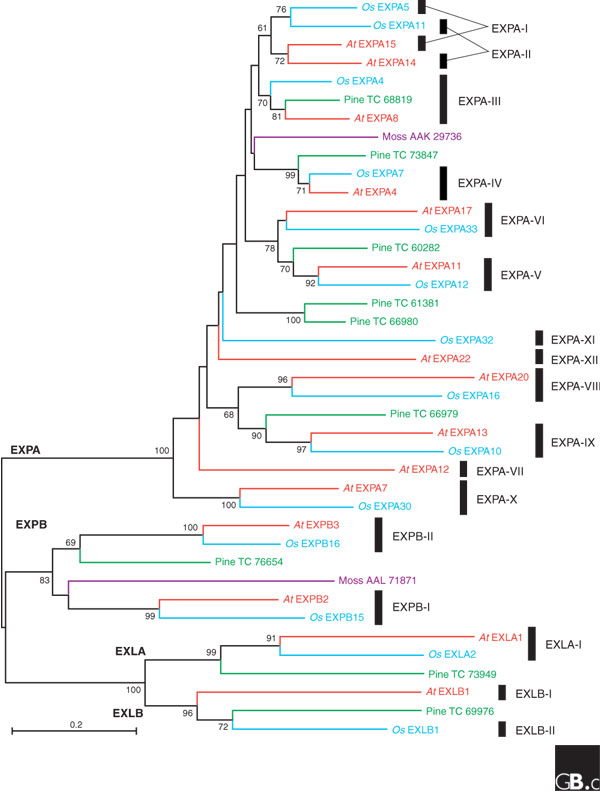
A phylogenetic tree of the expansin superfamily, including protein sequences from Arabidopsis thaliana (At), Oryza sativa (Os), Pinus species (pine) and Physcomitrella patens (moss). These sequences were selected to showcase expansin diversity. They were aligned with CLUSTALW (see Additional data file 1) and a neighbor-joining tree was constructed with MEGA 3. Bootstrap values above 60 are indicated next to the relevant node, and the four families are labeled at their roots. Clades, defined as all the descendants of the same ancestral gene in the last common ancestor of monocots and eudicots, are indicated by black bars to the right and given Roman numbers as in [7]. This tree does not correctly resolve clades EXPA-I and EXPA-II, possibly because of changes in amino-acid usage between Arabidopsis and rice expansins [7]. The numbers for pine sequences are from TIGR Pinus Gene Index [70]; GenBank accession numbers are shown for moss sequences. EXPA, α-expansin; EXPB, β-expansin; EXLA, expansin-like A; EXLB, expansin-like B.
It has not been established when expansins first appeared in evolution, but the α-expansin and β-expansin families already existed by the time the vascular plants and mosses diverged (Figure 2) [5,6]. So far, the expansin-like A and expansin-like B families can be traced back only to the last ancestor of angiosperms and gymnosperms (Figure 2). More recently the expansin families have continued to grow and diversify in different plant lineages. Table 1 shows the number of genes for each family found in the available angiosperm genomes, as well as the numbers of genes estimated for the last common ancestor of eudicots (including Arabidopsis) and monocots (including rice). On the basis of this reconstruction, we have recently proposed a subdivision of the four expansin families of angiosperms into 17 clades (Figure 2) [7]. As shown in Table 1, the number of genes has doubled in the Arabidopsis lineage and more than tripled in rice since these two species diverged, approximately 150 million years ago. The main reason for this difference is the larger number of tandem duplications present in the rice genome (Figure 3). The growth of the β-expansin family in grasses is particularly impressive, with 18 genes in rice compared with 6 in Arabidopsis.
Table 1.
Sizes of the four expansin families in different plants
| Species | EXPA | EXPB | EXLA | EXLB |
| Last common ancestor | 12 | 2 | 1 | 2 |
| Arabidopsis | 26 | 6 | 3 | 1 |
| Poplar | 27 | 2 | 2 | 4 |
| Rice | 34 | 19 | 4 | 1 |
The number of genes in each family is listed for the three plant species whose genomes have been sequenced. The number of genes in the last common ancestor of monocots and eudicots was estimated from an analysis of the rice and Arabidopsis genomes [7]. Numbers for poplar do not include partial gene fragments and should be taken as minimum estimates given that its genome is incompletely sequenced. EXPA, α-expansin; EXPB, β-expansin; EXLA, expansin-like A; EXLB, expansin-like B.
Figure 3.
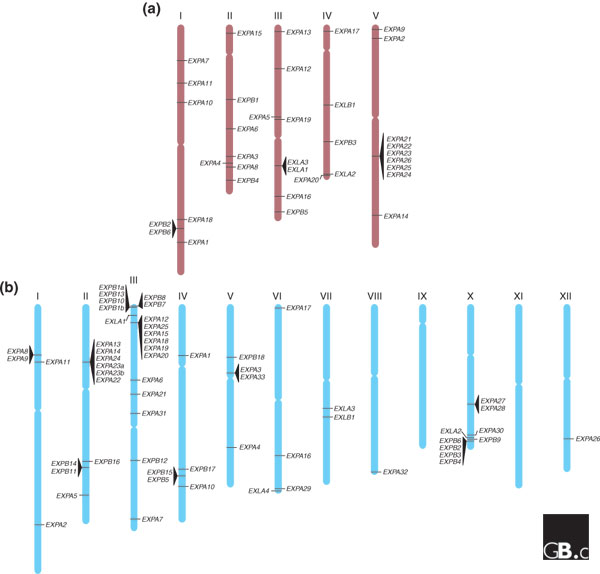
Genomic locations of expansin genes. (a) Arabidopsis; (b) rice. Genes in tandem are indicated by triangles and chromosome numbers are shown with Roman numerals. EXPA, α-expansin; EXPB, β-expansin; EXLA, expansin-like A; EXLB, expansin-like B.
Curiously, grasses (but only grasses) also have an additional group of secreted proteins homologous only to expansin domain 2; these are known in the immunological literature as grass group-2 pollen allergens (G2As). They seem to have evolved from a truncated copy of a β-expansin gene and they share about 35-45% protein identity with their closest β-expansin relatives; their native biological function is uncertain. Although G2As evolved from a β-expansin ancestor, because of the loss of domain 1 they are considered a separate family and not part of the expansin superfamily.
Two other families of plant proteins show distant homology to expansin domain 1, but as they lack domain 2 they are not considered part of the superfamily. The closest (approximately 25-35% identity) has been variously called p12 and plant natriuretic peptide (PNP). These proteins become abundant in the xylem of blighted citrus trees [8], and they have been ascribed a signaling function [9,10]. No wall-loosening activity has been found in extracts containing p12 (D.J.C. and T. Ceccardi, unpublished observations). More distantly related (about 20-30% identity) is the barwin-like domain that defines the PR4 family of antifungal proteins [11]. Both these protein families were already present in the last ancestor of mosses and vascular plants.
Turning to non-plant organisms, various proteins with distant homology to full-length expansins or exclusively to domain 1 are found from bacteria to nematodes and mollusks [12-15]. Many of these are probably involved in the digestion of plant cell-wall material. A family of expansin-like proteins has been found in the slime mold Dictyostelium discoideum, where they could help to maintain the fluidity of the cellulosic cell walls in the stalk structure [16]. Recent nomenclature rules [2] recommend that only proteins with homology to both expansin domains should be designated expansins. The polyphyletic group of non-plant expansins, such as those in Dictyostelium, can be referred to as expansin-like family X (EXLX). The relationship of the various groups of expansin-like X proteins with the plant expansins is unclear at the moment. Their divergence could predate the origin of land plants, or they could have been acquired later through horizontal transfer of a gene from one of the plant expansin families. The same applies to proteins with homology only to domain 1, both in plants and other organisms, in that it is possible that some of them originally evolved from an expansin protein with both domains.
Characteristic structural features
Expansin proteins from different families share only 20-40% identity with each other. The degree of conservation is highest in domain 1, as shown in Figure 4. Expansin domain 1 has a distant homology to glycoside hydrolase family 45 (GH45) proteins [17], most of which are fungal β-1,4-D-endoglucanases. Proteins from this family have been crystallized and their mechanism of action determined [18]: they form a six-stranded β-barrel with a groove for substrate binding (Figure 5a). Barwin also has a similar β-barrel structure [19]. On the basis of hydrophobic cluster analysis, we expect this structural motif also to be present in expansins (Figure 6). Furthermore, expansin domain 1 shares with GH45 a number of conserved cysteines that form disulfide bridges in the fungal enzymes. It is interesting that several residues that make up the catalytic site of GH45 endoglucanases are also conserved in expansin (see Figures 4, 5). Despite the presence of these conserved GH45 motifs, no hydrolytic activity has been detected for either α-expansin or β-expansin proteins.
Figure 4.
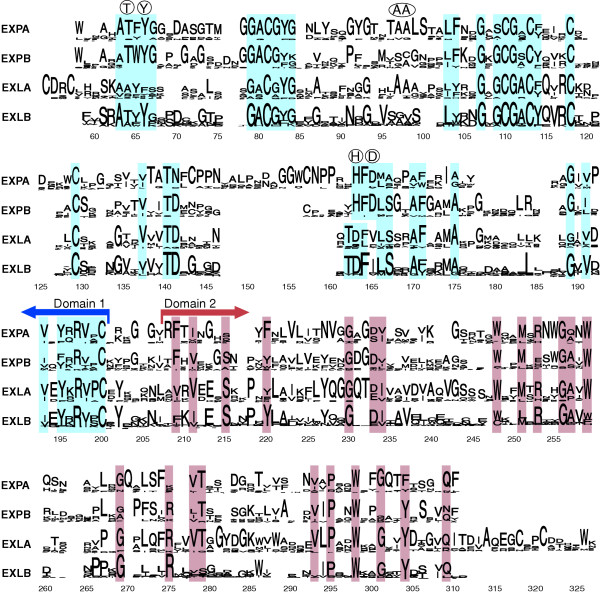
Sequence conservation in the expansin superfamily. Sequence logos for the four expansin families were generated with WebLogo [71] and manually aligned. The signal peptide and the poorly conserved amino terminus of the mature proteins have been removed from the alignment; because some expansins have exceptionally large signal peptides and amino-terminal extensions the alignment starts around position 60. In these sequence logos the height of the stack of amino-acid symbols at each position indicates the degree of sequence conservation, and the height of each letter within the stack indicates the frequency of the corresponding amino acid. Residues conserved between families are shaded, and the boundary between the two domains is indicated by arrows. Key residues that are part of the catalytic site of GH45 proteins and that are conserved in domain 1 of some expansin families are shown in circles above the logos. EXPA, α-expansin; EXPB, β-expansin; EXLA, expansin-like A; EXLB, expansin-like B.
Figure 5.
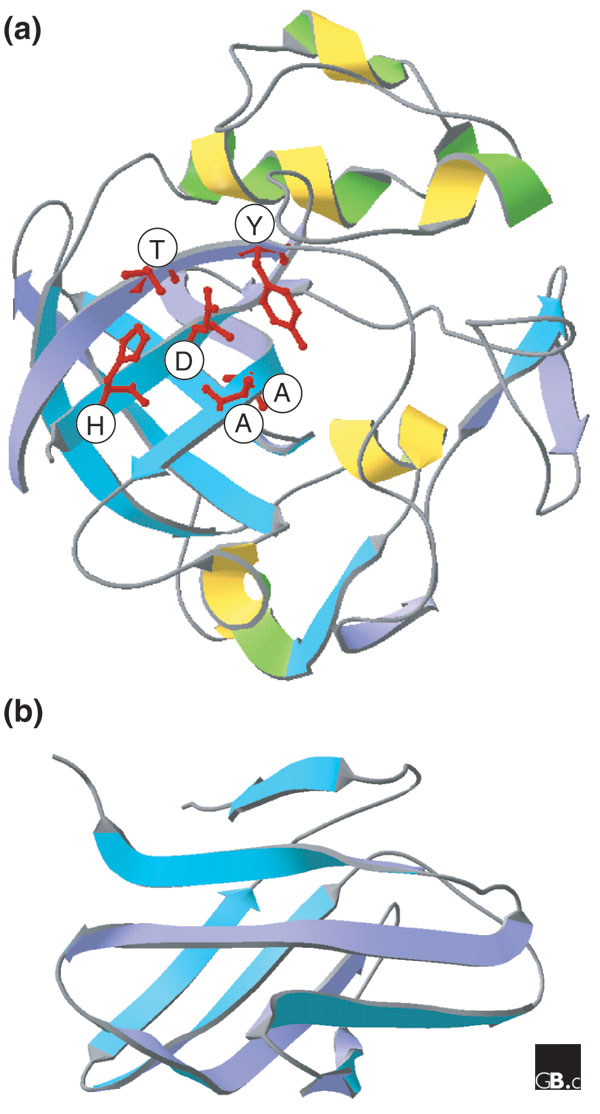
Structure of proteins homologous to expansin domains. (a) Expansin domain 1 (the catalytic domain of a GH45 endoglucanase from Humicola insolens; Protein Data Bank (PDB) code 2ENG). (b) Expansin domain 2 (a G2A protein from Phleum pratense; PDB 1WHO). In (a), the domain forms a β barrel; amino-acid residues that are conserved in expansins are indicated in the single-letter amino-acid code.
Figure 6.
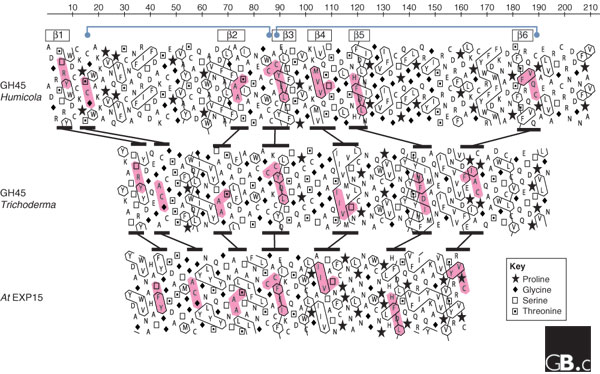
Aligned hydrophobic cluster analysis (HCA) plots of the catalytic domains of two GH45 proteins and domain 1 of an α-expansin protein, Arabidopsis EXPA15, with additional annotation based on the crystal structure of Humicola GH45. The GH45 sequences are from Humicola insolens (GenBank accession number P43316) and Trichoderma reesei (AAQ21385). HCA plots were constructed with DrawHCA [72]. In these plots, the amino-acid sequence of each protein is written out in duplicate in a helical representation that puts together amino-acid residues that would be next to each other in an α helix. The six β sheets that form a barrel in the GH45 from Humicola (see Figure 5a) are indicated by boxes above the plot. Cytosine residues involved in intramolecular bridges and conserved in expansins and GH45 proteins are shown by blue dots connected by blue lines, also above the plot. Selected conserved motifs are highlighted in pink and the differences in their relative positions between proteins are indicated by black lines between the plots. The interpretation of HCA plots is summarized in [73]. HCA uses the standard one-letter amino acid abbreviations except for four amino acids, as shown in the key. Hydrophobic residues are outlined. Clusters of hydrophobic residues are usually associated with regular secondary structures (α helices or β sheets). Zigzagging vertical lines of hydrophobic residues indicate alternating hydrophobic and non-hydrophobic residues, typical of exposed β sheets (for example, β2, β3, β5 and β6). Continuous hydrophobic clusters are more common in internal β sheets (for example, β4). Conservation of clusters and sequence motifs suggests that the core β-barrel structure with stabilizing cysteine bridges is conserved in the three proteins and that the differences are mostly in the size of the intervening loops. In Humicola GH45, the loops between β1 and β2 and between β5 and β6 have expanded considerably, while the other loops appear reduced in comparison with Trichoderma GH45. The latter appears more similar to expansin domain 1, which has an even more compact structure.
α-Expansin proteins can be distinguished from other expansins by the presence of a large insertion and a nearby deletion in domain 1; these are at either side of a conserved motif that is part of the conserved GH45 active site (HFD in the single-letter amino-acid code; Figure 4). Expansin-like A and expansin-like B proteins lack the HFD motif, which suggests that their action may differ from that of other expansins. Furthermore, expansin-like A proteins have a unique conserved motif (CDRC) at the amino terminus of domain 1, and their domain 2 has an extension of approximately 17 amino acids that is not found in other expansin families (Figure 4). The functional implications of these differences among families are currently unknown.
No proteins homologous to expansin domain 2 have yet been identified except for the G2A family. The structure of a G2A protein consists of two stacked β sheets with an immunoglobulin-like fold (Figure 5b) [20]. On the basis of this structure, some highly conserved aromatic residues present in expansin domain 2 have been hypothesized to form a binding strip for cell-wall polysaccharides [1,21], but this speculative idea has yet to be tested experimentally.
Localization and function
Expansins were first identified as wall-loosening proteins in studies of 'acid-induced growth' [3,22-24]. It was known for years that low extracellular pH (< 5.5) causes cell-wall loosening in land plants as well as in a subset of green algae that have walls of similar structure [25]. The process is mediated in large part by wall-bound expansins with an acidic pH optimum [3]. Wall pH is normally determined by the activity of the plasma membrane H+ ATPase, which pumps protons to the cell wall; the pH of the wall is typically about 5.5 but may go below 4.5 in some circumstances [25,26]. Acid-induced growth and expansin action are implicated in the growth responses of plants to hormones and to external stimuli such as light, drought, salt stress and submergence (anoxia) and in morphogenetic processes such as root-hair formation [27-31].
Expansin activity is usually assayed as the ability of a protein sample to induce extension of isolated cell walls (Figure 7). It may also be measured in stress-relaxation assays, in which the decay in wall stress is measured after the wall is rapidly extended and then held to a constant dimension [22]. Plant cell walls extend or relax by a process of molecular 'creep', in which the cellulose microfibrils and associated matrix polysaccharides separate from one another [32]. The energy needed to overcome the viscous resistance and entanglement of wall polymers comes from cell-wall stress, which in living plants arises from the turgor pressure within cells. Such molecular creep occurs only when the cell wall is loosened by expansins or by other factors (Figure 8); otherwise, the cellulose microfibrils are firmly held in place by matrix polysaccharides [27]. Artificial cell walls made of bacterial cellulose and xyloglucan have also been used as materials to investigate expansin action [33].
Figure 7.
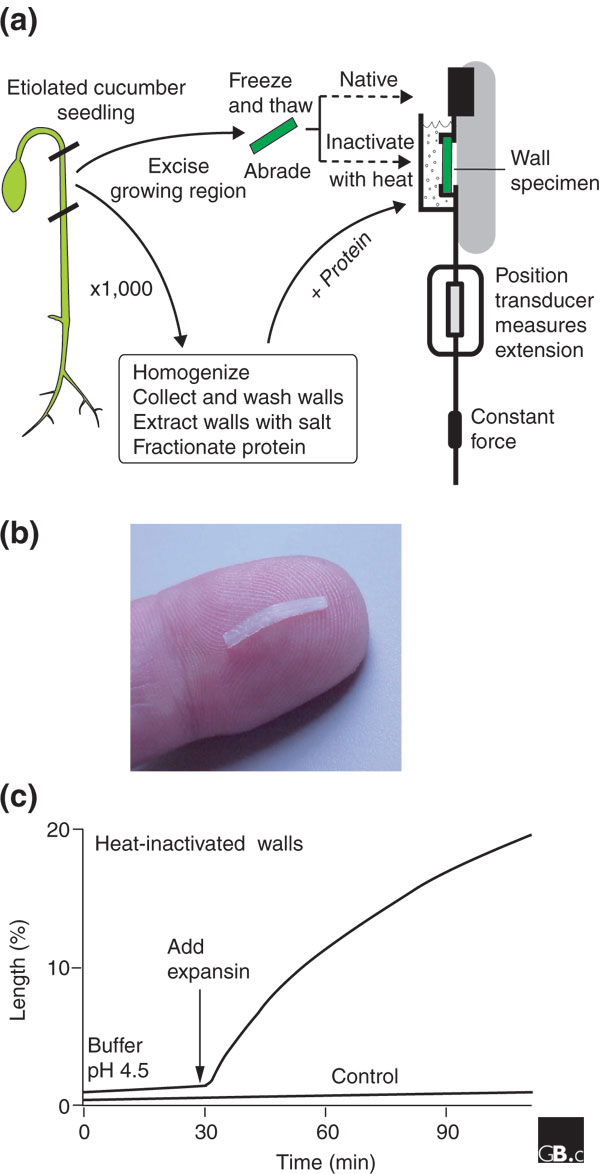
A common method for measuring the cell-wall extension activity of expansins. (a) Cell-wall specimens are excised from the growing region of a young seedling that has been grown in the dark (etiolated). The specimens are frozen and thawed in order to destroy the cells but leave the cell walls intact (the cuticle is abraded to facilitate penetration of proteins). The specimens are heat-treated to inactivate endogenous expansins and then clamped under constant tension in an extensometer. The extensometer measures the change in length of the sample, with or without the addition of exogenous expansins. Walls may be collected in parallel from other seedlings and extracted to obtain fractions with expansin activity, assayed as an increase in cell-wall length. (b) Photograph of a typical cell wall sample, placed on an index finger for scale, prior to clamping in the extensometer. (c) Time course for irreversible wall extension (creep) of heat-treated walls with and without the addition of expansin.
Figure 8.
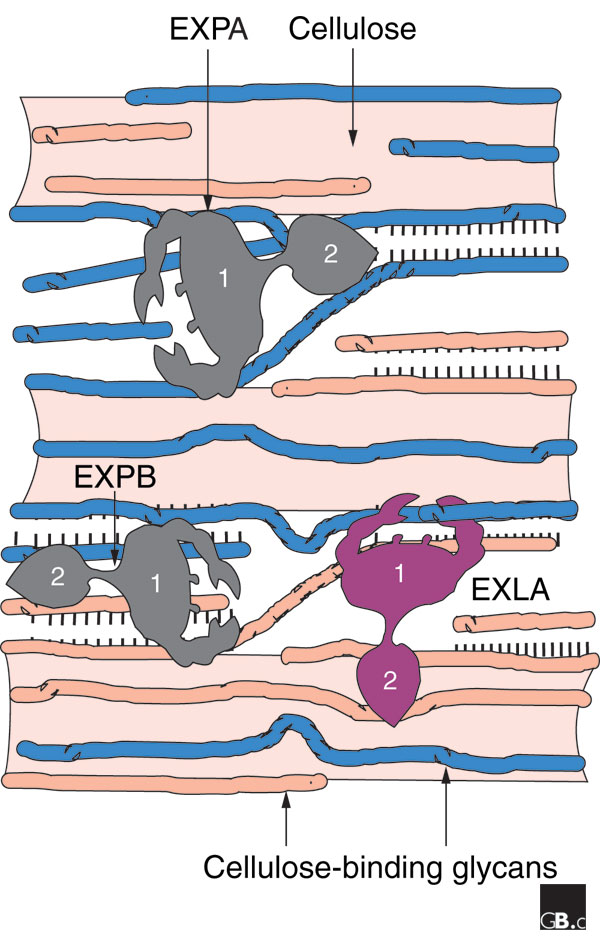
A simplified model of the plant cell wall and its loosening by expansins. The cell wall consists of a scaffold of cellulose microfibrils (shaded areas) to which are bound various glycans such as xyloglucan or xylan (thin strands); together these polysaccharides form a strong, flexible, load-bearing network based on hydrogen bonds (indicated by rows of short lines). Extension of the cell wall entails movement and separation of the cellulose microfibrils by a process of molecular creep. α-Expansins (EXPA) may promote such movement by inducing local dissociation and slippage of xyloglucans on the surface of the cellulose, whereas β-expansins (EXPB) work on a different glycan, perhaps xylan, for similar effect. Expansin-like A (EXLA) and expansin-like B (EXLB) proteins are predicted to be secreted to the cell wall, but their activity has not yet been established.
Expansin activity is most often associated with cell-wall loosening in growing cells [34]; this connection has been confirmed and extended by experiments in which expansin gene expression is manipulated in transgenic plants [35-38]. In most cases, silencing of expansin genes leads to inhibition of growth, whereas excessive ectopic expression leads to faster or abnormal growth. Localized expression of expansins is associated with the meristems and growth zones of the root and stem, as well as the formation of leaf primordia on shoot apical meristems [39] and the outgrowth of the epidermal cell walls during root-hair formation [40]. Additionally, expansins are implicated in other developmental processes during which wall loosening occurs, such as fruit softening [41-46], xylem formation [47], abscission (leaf shedding) [48], seed germination [49], penetration of pollen tubes through the stigma and style [4,50], formation of mycorrhizal associations with symbiotic fungi in root tissues [51], development of nitrogen-fixing nodules in legumes [52], development of parasitic plants [53,54], and rehydration of 'resurrection' plants, which curl up tightly when dry and expand when wet [55]. Some plants that are adapted to an aquatic environment respond to submergence with a pronounced elongation. This depends on wall acidification [56] and is correlated with activation of expansin gene expression [57-59].
In cell-fractionation studies, expansins are found bound to the cell wall, as expected from their activity [23,60,61]. With immunolocalization by light and electron microscopy, expansin proteins were localized to the cell wall [51,61,62], where they were found to be distributed throughout the thickness of the walls rather than concentrated in specific strata. There is at least one report that expansin mRNA can be found specifically at the polar ends of developing xylem cells [63]; transcript localization may be a means for ensuring that protein production and secretion is directed to the ends of these cells. It is not clear whether this mRNA targeting is unique to expansins in xylem or whether it is a more general phenomenon. Finally, grass pollen produces and secretes specialized β-expansin proteins in copious amounts (they are known as grass pollen group-1 allergens) [4,64], but this is an unusual situation: expansins in other tissues have been found only at low concentrations.
Mechanism and regulation
All the α-expansin proteins that have been characterized so far have a pH optimum for cell-wall extension of about 4 [3,23,60]. This situation permits the cell to regulate α-expansin activity rapidly by modulating wall pH. The pH optimum of only one class of β-expansin proteins has been characterized, namely the group-1 grass pollen allergens (such as EXPB1 from maize), and it has a broad pH optimum centered at about 5.5 [65]. These pollen proteins are probably not involved in acid growth but rather in the wall loosening that is associated with invasion of maternal tissues by pollen tubes. It is expected that β-expansin proteins in somatic tissues have a pH dependence more similar to that of α-expansins, but so far β-expansin proteins in an active state have not been extracted from somatic tissues, so this expectation remains to be tested experimentally. Also, both α-expansin and β-expansin proteins are activated by reducing agents [3,4,65]; this could be biologically significant, as the cell-wall redox potential can be modulated by electron transport across the plasma membrane.
Expansins do not have hydrolytic activity or any of the other enzymatic activities yet assayed [64,66,67]. A report that they are proteases was later refuted [64]. Expansins also act very quickly - they induce rapid extension within seconds of addition to wall specimens, but they do not affect the plasticity or elasticity of the cell wall [68]. In contrast, cell-wall creep caused by an endoglucanase has a long lag time and is accompanied by large increases in wall plasticity and elasticity, indicative of major structural changes in the cell wall (cutting of cross-links) [68]. Thus, expansin's effects on cell walls are distinct from those expected of hydrolytic enzymes.
A nonenzymatic mechanism has been proposed for expansin action, in which expansin disrupts noncovalent bonds that tether matrix polysaccharides to the surface of cellulose microfibrils or to each other [1,66,69]. In this model, the expansin is thought to act like a zipper that enables microfibrils to move apart from each other by ungluing the chains that stick them together. This idea is also supported by experiments in which an expansin is applied to artificial composites made of bacterial cellulose and xyloglucan [33]. Whatever their biochemical mechanism of action, expansins act in catalytic amounts to stimulate wall polymer creep without causing major covalent alterations of the cell wall [66].
Frontiers
In the published literature on expansins, gene expression has drawn the greatest amount of attention, but given the large size of the superfamily, the expression and presumptive role of many expansin genes remains unexplored. Although expression of specific expansin genes has been shown to be induced by hormones, by submergence, by drought stress, or by other stimuli, the signaling pathway has not been worked out in detail in even a single case. Major biochemical questions also remain regarding the specific wall polysaccharides on which expansins act, the differences between the action of α-expansins and β-expansins, and the molecular mechanisms underlying wall loosening. Answering these questions will require a much deeper understanding of cell-wall structure and in particular of how the cell wall is able to expand in a controlled fashion. Finally, it remains to be established whether expansin-like A and expansin-like B proteins have cell-wall loosening activity or not.
Additional data files
An alignment of the sequences used to make the phylogenetic tree in Figure 2 is available as Additional data file 1.
Supplementary Material
An alignment of the sequences used to make the phylogenetic tree in Figure 2
Acknowledgments
Acknowledgements
This work has been supported by grants to D.J.C. from the US National Science Foundation, the Department of Energy, and the National Institutes of Health.
References
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. A review of cell-wall structure and the action of expansins. [DOI] [PubMed] [Google Scholar]
- Kende H, Bradford K, Brummell D, Cho HT, Cosgrove DJ, Fleming A, Gehring C, Lee Y, Queen-Mason S, Rose J, Voesenek LA. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol. 2004;55:311–314. doi: 10.1007/s11103-004-0158-6. This article by the expansin research community defines expansins and recommends naming conventions. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. The purification of two related proteins that cause pH-dependent wall extension, later named expansins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. Shows that group-1 grass pollen allergens have expansin activity and defines the second family of expansins (β-expansins) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128:854–864. doi: 10.1104/pp.010658. Phylogenetic analysis of the expansin superfamily in Arabidopsis and identification of more distantly related sequences; also proposes a nomenclature system superseded by [2]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper O, Schaefer D, Reski R, Fleming A. Expansins in the bryophyte Physcomitrella patens. Plant Mol Biol. 2002;50:789–802. doi: 10.1023/A:1019907207433. Identification of expansin family members in a moss and analysis of gene expression. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Lee Y, Carey RE, dePamphilis CW, Cosgrove DJ. Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J. 2005;44:409–419. doi: 10.1111/j.1365-313X.2005.02540.x. Traces the evolution of the expansin superfamily in Arabidopsis and rice since their last common ancestor. [DOI] [PubMed] [Google Scholar]
- Ceccardi TL, Barthe GA, Derrick KS. A novel protein associated with citrus blight has sequence similarities to expansin. Plant Mol Biol. 1998;38:775–783. doi: 10.1023/A:1006039016393. Identification of a 12 kDa protein (p12) in xylem of blighted citrus trees that is distantly related to expansin domain 1. [DOI] [PubMed] [Google Scholar]
- Rafudeen S, Gxaba G, Makgoke G, Bradley G, Pironcheva G, Raitt L, Irving H, Gehring C. A role for plant natriuretic peptide immuno-analogues in NaCl- and drought-stress responses. Physiol Plant. 2003;119:554–562. doi: 10.1046/j.1399-3054.2003.00201.x. Implicates a 12 kDa protein, with distant sequence similarity to expansin domain 1, in plant water stress responses. [DOI] [Google Scholar]
- Gehring CA, Irving HR. Natriuretic peptides - a class of heterologous molecules in plants. Int J Biochem Cell Biol. 2003;35:1318–1322. doi: 10.1016/S1357-2725(03)00032-3. Reviews the properties of PNPs, which are homologous to expansin domain 1. [DOI] [PubMed] [Google Scholar]
- Friedrich L, Moyer M, Ward E, Ryals J. Pathogenesis-related protein 4 is structurally homologous to the carboxy-terminal domains of hevein, Win-1 and Win-2. Mol Gen Genet. 1991;230:113–119. doi: 10.1007/BF00290658. Characterizes the PR-4 family, whose barwin-like domain is distantly related to expansin domain 1. [DOI] [PubMed] [Google Scholar]
- Laine M, Haapalainen M, Wahlroos T, Kankare K, Nissinen R, Kassuwi S, Metzler MC. The cellulase encoded by the native plasmid of Clavibacter michiganensis subsp. sepedonicus plays a role in virulence and contains an expansin-like domain. Physiol Mol Plant Pathol. 2001;57:221–233. doi: 10.1006/pmpp.2000.0301. A Clavibacter virulence factor contains a domain distantly related to expansin domain 1. [DOI] [Google Scholar]
- Xu B, Janson JC, Sellos D. Cloning and sequencing of a molluscan endo-beta-1,4-glucanase gene from the blue mussel, Mytilus edulis. Eur J Biochem. 2001;268:3718–3727. doi: 10.1046/j.1432-1327.2001.02280.x. Cloning and sequence analysis of the first family-45 endoglucanase from a mollusk. [DOI] [PubMed] [Google Scholar]
- Kudla U, Qin L, Milac A, Kielak A, Maissen C, Overmars H, Popeijus H, Roze E, Petrescu A, Smant G, et al. Origin, distribution and 3D-modeling of Gr-EXPB1, an expansin from the potato cyst nematode Globodera rostochiensis. FEBS Lett. 2005;579:2451–2457. doi: 10.1016/j.febslet.2005.03.047. Phylogenetic analysis and molecular modeling of a nematode protein with homology to expansin domain 1. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. Identification of a fungal protein with distant sequence similarity to expansin and which disrupts cotton fibers with hydrolytic action. [DOI] [PubMed] [Google Scholar]
- Darley CP, Li Y, Schaap P, McQueen-Mason SJ. Expression of a family of expansin-like proteins during the development of Dictyostelium discoideum. FEBS Lett. 2003;546:416–418. doi: 10.1016/S0014-5793(03)00598-2. Identifies a small group of Dictyostelium genes related to expansins and characterizes their expression during development. [DOI] [PubMed] [Google Scholar]
- Carbohydrate-Active enZYmes http://afmb.cnrs-mrs.fr/CAZY/ A database that describes the families of structurally related catalytic enzymes that degrade, modify, or create glycosidic bonds.
- Davies GJ, Tolley SP, Henrissat B, Hjort C, Schulein M. Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 Å resolution. Biochemistry. 1995;34:16210–16220. doi: 10.1021/bi00049a037. A structural analysis of the Humicola GH45 enzyme complexed with cello-oligosaccharide identifies key catalytic residues as well as a large conformational change in the enzyme upon substrate binding. [DOI] [PubMed] [Google Scholar]
- Ludvigsen S, Poulsen FM. Three-dimensional structure in solution of barwin, a protein from barley seed. Biochemistry. 1992;31:8783–8789. doi: 10.1021/bi00152a014. The barwin structure includes a β barrel somewhat resembling the catalytic domain of GH45 proteins. [DOI] [PubMed] [Google Scholar]
- Fedorov AA, Ball T, Valenta R, Almo SC. X-ray crystal structures of birch pollen profilin and Phl p 2. Int Arch Allergy Immunol. 1997;113:109–113. doi: 10.1159/000237520. Structural analysis of the group-2 grass pollen allergen, Phl p 2, which is homologous to expansin domain 2. [DOI] [PubMed] [Google Scholar]
- Barre A, Rouge P. Homology modeling of the cellulose-binding domain of a pollen allergen from rye grass: structural basis for the cellulose recognition and associated allergenic properties. Biochem Biophys Res Commun. 2002;296:1346–1351. doi: 10.1016/S0006-291X(02)02091-0. A homology model of domain 2 from EXPB with identification of a groove and extended strip of aromatic and polar residues that might function in carbohydrate binding. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta. 1989;177:121–130. doi: 10.1007/BF00392162. An analysis of acid growth of isolated cell walls, hypothesizing a wall-loosening enzyme with unusual properties, later purified and named expansin. [DOI] [PubMed] [Google Scholar]
- Li Z-C, Durachko DM, Cosgrove DJ. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta. 1993;191:349–356. doi: 10.1007/BF00195692. The first use of the name expansin; this article identifies a wall-loosening protein from oat seedlings with many similarities to cucumber expansins. [DOI] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins - a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. The cloning of α-expansin shows that it belongs to a multigene family and lacks sequence similarity to any enzymes known at the time of publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. Reviews the evidence that cell-wall acidification is part of the mechanism of auxin-induced cell elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Jacob T, Dahse I, Gilroy S. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development. 1998;125:2925–2934. doi: 10.1242/dev.125.15.2925. Shows that outgrowth of the outer epidermal cell wall during root-hair initiation starts with local wall acidification (to pH 4.5). [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. This review summarizes wall structure and recent progress in understanding the biosynthesis and expansion of the plant cell wall. [DOI] [PubMed] [Google Scholar]
- Bogoslavsky L, Neumann PM. Rapid regulation by acid pH of cell wall adjustment and leaf growth in maize plants responding to reversal of water stress. Plant Physiol. 1998;118:701–709. doi: 10.1104/pp.118.2.701. Provides evidence that water availability affects leaf-cell elongation by changes in cell-wall acidification and consequent changes in wall extensibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanova IB, Sabirzhanov BE, Chemeris AV, Veselov DS, Kudoyarova GR. Fast changes in expression of expansin gene and leaf extensibility in osmotically stressed maize plants. Plant Physiol Biochem. 2005;43:419–422. doi: 10.1016/j.plaphy.2005.01.021. Maize seedlings respond to osmotic stress by an increase in wall extensibility that is correlated with an increase in EXPA transcript levels. [DOI] [PubMed] [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and alpha-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. Shows that cells in suspension culture elongate faster in response to fusicoccin (which induces wall acidification) and express expansins at the mRNA and protein level. Cells also grow faster upon treatment with exogenous expansin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Steinbaker CR, Crowell DN. Expression and processing of a hormonally regulated beta-expansin from soybean. Plant Physiol. 2001;126:244–252. doi: 10.1104/pp.126.1.244. Auxin and cytokinin synergistically enhance the accumulation of EXPB protein (CIM1) in soybean cell cultures. Evidence for stepwise proteolysis of the extracellular protein is also reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marga F, Grandbois M, Cosgrove DJ, Baskin TI. Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J. 2005;43:181–190. doi: 10.1111/j.1365-313X.2005.02447.x. Shows that cell-wall extension does not entail passive reorientation of cellulose microfibrils, implying a specific loosening mechanism that results in lateral separation of microfibrils. [DOI] [PubMed] [Google Scholar]
- Whitney SE, Gidley MJ, McQueen-Mason SJ. Probing expansin action using cellulose/hemicellulose composites. Plant J. 2000;22:327–334. doi: 10.1046/j.1365-313x.2000.00742.x. EXPA induces rapid, transient extension of artificial cellulosic composites (derived from Acetobacter pellicles) made with xyloglucan, but not those made with glucomannan or galactomannan. [DOI] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/S1369-5266(00)00211-9. A review of expansin gene expression and gene structure and their implications for expansin function and evolution. [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. Transgenic experiments to increase or reduce expansin gene expression support the role of expansin in cell growth and in abscission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Lee Y, Cho HT, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. Plants expressing an antisense EXPA RNA were shorter than controls, whereas plants overexpressing the EXPA gene had more leaves but a mixed phenotype, some taller and some shorter than controls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. The authors used transient local micro-induction of EXPA genes on the shoot apical meristem and the flanks of leaf primordia to demonstrate that EXPA overexpression can markedly stimulate plant cell growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Ahn JH, Song SK, Choi YD, Lee JS. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol. 2003;131:985–997. doi: 10.1104/pp.009902. This study shows that GmEXPA1 expression is maximal in the elongation zone of the soybean root, and ectopic expression of this gene in tobacco roots leads to faster root growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. Shows that localized expression of an EXPA gene on the flanks of the shoot apical meristem precedes emergence of the leaf primordium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002;14:3237–3253. doi: 10.1105/tpc.006437. Two expansin genes (AtEXPA7 and AtEXP18) are turned on specifically at the place and time of root-hair formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell. 1999;11:2203–2216. doi: 10.1105/tpc.11.11.2203. Transgenic tomato fruits with higher levels of LeEXPA1 gene expression were much softer than controls, whereas fruits with reduced LeEXPA1 expression were firmer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. Identifies a tomato EXPA gene (LeEXPA1) that is expressed during fruit ripening and proposes a role for expansins in cell-wall disassembly in nongrowing tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamaki MS, Powell AL, Struijs K, Labavitch JM, Reid DS, Bennett AB. Transgenic overexpression of expansin influences particle size distribution and improves viscosity of tomato juice and paste. J Agric Food Chem. 2003;51:7465–7471. doi: 10.1021/jf0341666. Overexpression of EXPA1 in tomato fruit leads to higher-viscosity tomato paste and juice with larger particle size, perhaps as a result of increased cell-wall hydration. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Gao ZF, Cantini C, Loescher WH, van Nocker S. Fruit ripening in sour cherry: changes in expression of genes encoding expansins and other cell-wall-modifying enzymes. J Am Soc Hortic Sci. 2003;128:16–22. Expression of four EXPA genes is strongly upregulated upon onset of fruit ripening in the sour cherry. [Google Scholar]
- Harrison EP, McQueen-Mason SJ, Manning K. Expression of six expansin genes in relation to extension activity in developing strawberry fruit. J Exp Bot. 2001;52:1437–1446. doi: 10.1093/jexbot/52.360.1437. Developmental analysis of expression of six EXPA genes in strawberry fruit indicates distinctive roles for the different genes. [DOI] [PubMed] [Google Scholar]
- Obenland DM, Crisosto CH, Rose JKC. Expansin protein levels decline with the development of mealiness in peaches. Postharvest Biol Technol. 2003;29:11–18. doi: 10.1016/S0925-5214(02)00245-4. Downregulation of EXPA gene expression in pear fruit is associated with onset of mealiness. [DOI] [Google Scholar]
- Gray-Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzell A, Sterky F, Blomqvist K, McQueen-Mason S, Teeri TT, Sundberg B. Expansins abundant in secondary xylem belong to subgroup a of the alpha-expansin gene family. Plant Physiol. 2004;135:1552–1564. doi: 10.1104/pp.104.039321. Analysis of expansin gene expression indicates that certain EXPA genes are involved in secondary wall formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason SJ. Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot. 2005;56:817–823. doi: 10.1093/jxb/eri076. Transcripts of two EXPA genes accumulate specifically in leaf abscission zone during ethylene-stimulated leaf abscission. [DOI] [PubMed] [Google Scholar]
- Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000;124:1265–1274. doi: 10.1104/pp.124.3.1265. An EXPA gene is expressed specifically in the endosperm region that is selectively loosened to allow emergence of the root from the seed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzotti M, Feron R, Mariani C. Pollination modulates expression of the PPAL gene, a pistil-specific beta-expansin. Plant Mol Biol. 2002;49:187–197. doi: 10.1023/A:1014962923278. Documents expression of an EXPB mRNA in the secretory zone of the stigma and in the epidermal layer of the placenta. [DOI] [PubMed] [Google Scholar]
- Balestrini R, Cosgrove DJ, Bonfante P. Differential location of alpha-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta. 2005;220:889–899. doi: 10.1007/s00425-004-1431-2. Evidence by immunolocalization experiments that α-expansins are involved in cell-wall remodeling during formation of the mycorrhizal infection. [DOI] [PubMed] [Google Scholar]
- Giordano W, Hirsch AM. The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol Plant Microbe Interact. 2004;17:613–622. doi: 10.1094/MPMI.2004.17.6.613. Identified an EXPA gene in a legume (sweetclover) that is upregulated during Sinorhizobium-induced nodule formation. [DOI] [PubMed] [Google Scholar]
- O'Malley RC, Lynn DG. Expansin message regulation in parasitic angiosperms: marking time in development. Plant Cell. 2000;12:1455–1465. doi: 10.1105/tpc.12.8.1455. When Striga switches to its parasitic mode, three EXPA genes are activated and are involved in developmental changes that include formation of the root-like parasitic organ (haustorium). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel RL, Yoder JI. Differential RNA expression of alpha-expansin gene family members in the parasitic angiosperm Triphysaria versicolor (Scrophulariaceae). Gene. 2001;266:85–93. doi: 10.1016/S0378-1119(01)00376-6. The authors identified EXPA genes that are upregulated in the parasitic plant Triphysaria by cytokinin application; they are probably not involved in haustorium development. [DOI] [PubMed] [Google Scholar]
- Jones L, McQueen-Mason S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett. 2004;559:61–65. doi: 10.1016/S0014-5793(04)00023-7. Wall extensibility and expression of an EXPA gene are markedly increased during dehydration and rehydration of this resurrection plant. [DOI] [PubMed] [Google Scholar]
- Ridge I, Osborne DJ. Wall extensibility, wall pH and tissue osmolality: significance for auxin and ethylene-enhanced petiole growth in semi-aquatic plants. Plant Cell Environ. 1989;12:393. Submergence-induced and ethylene-induced elongation of aquatic plants depends on cell-wall acidification. [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;9:1661–1671. doi: 10.1105/tpc.9.9.1661. Expression of certain EXPA genes in rice is increased upon submergence-induced internode elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeburg RA, Benschop JJ, Peeters AJ, Colmer TD, Ammerlaan AH, Staal M, Elzenga TM, Staals RH, Darley CP, McQueen-Mason SJ, Voesenek LA. Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J. 2005;43:597–610. doi: 10.1111/j.1365-313X.2005.02477.x. In Rumex, a leafy herb adapted to wet conditions, submergence increases ethylene concentration which promotes RpEXPA1 expression and apoplastic acidification, which combine to stimulate growth. [DOI] [PubMed] [Google Scholar]
- Colmer TD, Peeters AJ, Wagemaker CA, Vriezen WH, Ammerlaan A, Voesenek LA. Expression of alpha-expansin genes during root acclimations to O2 deficiency in Rumex palustris. Plant Mol Biol. 2004;56:423–437. doi: 10.1007/s11103-004-3844-5. An analysis of EXPA gene expression, showing selective expression of certain genes in root tips, cambium and other cells involved in plant growth. [DOI] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expansins in deepwater rice internodes. Plant Physiol. 1997;113:1137–1143. doi: 10.1104/pp.113.4.1137. Purification and partial characterization of two EXPA proteins from rice internodes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant Cell Physiol. 2002;43:1436–1444. doi: 10.1093/pcp/pcf180. A review of expansin gene expression and its actions on the plant cell wall. [DOI] [PubMed] [Google Scholar]
- Zhang N, Hasenstein KH. Distribution of expansins in gravi-responding maize roots. Plant Cell Physiol. 2000;41:1305–1312. doi: 10.1093/pcp/pcd064. By fluorescence immunolocalization, EXPA signals in the cell wall were shown to become stronger on the upper side of roots bending downwards during gravitropism. [DOI] [PubMed] [Google Scholar]
- Im KH, Cosgrove DJ, Jones AM. Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 2000;123:463–470. doi: 10.1104/pp.123.2.463. EXPA mRNAs show polar localization to the apical or basipetal ends of xylem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Cosgrove DJ. Grass group I pollen allergens (beta-expansins) lack proteinase activity and do not cause wall loosening via proteolysis. Eur J Biochem. 2001;268:4217–4226. doi: 10.1046/j.1432-1327.2001.02336.x. Using five protease assays, this study tests the idea that grass pollen EXPBs (group-1 allergens) loosen cell walls via proteolytic action; the results refute this idea. [DOI] [PubMed] [Google Scholar]
- Li LC, Bedinger PA, Volk C, Jones AD, Cosgrove DJ. Purification and characterization of four beta-expansins (Zea m 1 isoforms) from maize pollen. Plant Physiol. 2003;132:2073–2085. doi: 10.1104/pp.103.020024. A detailed biochemical characterization of the physical properties and action of maize group-1 pollen allergens (EXPBs). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. A detailed biochemical and biophysical analysis of how α-expansins loosen plant cell walls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Fry SC, Durachko DM, Cosgrove DJ. The relationship between xyloglucan endotransglycosylase and in vitro cell wall extension in cucumber hypocotyls. Planta. 1993;190:327–331. doi: 10.1007/BF00196961. Presents evidence that expansins do not have xyloglucan endotransglycosylase (XET) activity and that XET lacks wall extension activity, either by itself or in the presence of xyloglucan oligosaccharides. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wu Y, Cosgrove DJ. A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 2001;127:324–333. doi: 10.1104/pp.127.1.324. The first demonstration that plant cell walls extend as a result of hydrolysis by an endoglucanase (fungal not plant); the characteristics of this extension are very different from that caused by expansin action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. This report shows that α-expansin loosens paper made of pure cellulose, without hydrolysis, and proposes a model for expansin action involving local dissolution of the adhesion of matrix polysaccharides to the surface of cellulose microfibrils. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Institute for Genomic Research http://www.tigr.org The Institute for Genomic Research (TIGR) website.
- Weblogo http://weblogo.berkeley.edu WebLogo is a web-based application for the generation of sequence logos (graphical representations of amino acid or nucleic acid multiple sequence alignments).
- Salle de Modelisation et d'Imagerie http://smi.snv.jussieu.fr The DrawHCA program available at this site allows the construction of HCA plots.
- Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon JP. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. A summary of the interpretation of HCA plots. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An alignment of the sequences used to make the phylogenetic tree in Figure 2


