Abstract
This Letter reports on adhesive modular proteins recorded by atomic force microscopy on live cells from the extracellular mucilage secreted from, and deposited around, the motile form of the pennate diatom Phaeodactylum tricornutum. This is the first report of modular proteins and their supramolecular assemblies, called adhesive nanofibers (ANFs), to be found on diatoms that use adhesives not only for substratum adhesion, but as a conduit for cell motility. The permanent adhesive pads secreted by Toxarium undulatum, a sessile centric diatom, were previously shown to possess ANFs with a modular protein backbone. Our results reported here suggest that modular proteins may be an important component of diatom adhesives in general, and that diatoms utilize the tensile strength, toughness, and flexibility of ANFs for multiple functions. Significantly, the genome of P. tricornutum has recently been sequenced; this will allow directed searches of the genome to be made for genes with modular protein homologs, and subsequent detailed studies of their molecular structure and function.
Phaeodactylum tricornutum Bohlin (strain CCMP 632) is a polymorphic pennate diatom. Of the three morphotypes, the ovoid form adheres strongly to the substratum and is capable of a gliding motility common in pennate diatoms. Adhesion is facilitated by a slit (raphe) through the silica cell wall of one valve where adhesive, polymeric strands can link the cell cytoplasm to the substratum. Diatom gliding occurs via an actin/myosin motor that applies a driving force to the extracellular strands via connector molecules through the cell membrane, the entire continuum referred to as the adhesion complex (AC) (1). The precise chemical nature of the adhesive strands is not known, though there is compositional evidence that they are glycoproteins (2). Pennate diatoms account for almost all of the world's biofouling diatoms, and are characterized by strong adhesion and active motility.
Atomic force microscopy (AFM) has previously been used to investigate the adhesive mucilage of pennate diatoms. These studies showed that the raphe-derived adhesive mucilage was composed of strands, and that the retraction force curves of strands sometimes had an irregular sawtooth pattern suggesting successive unbinding of modular domains (3).
This Letter is the first report, to our knowledge, of the presence of modular proteins and ANFs in the adhesive mucilage of a motile diatom, where they function in both adhesion and as a conduit for cell movement. The adhesive molecules of P. tricornutum have the nanomechanical properties required of the extracellular, strand component of a functional diatom AC (1).
P. tricornutum ovoid cells were grown in F/2 medium (artificial seawater) on petri dishes, with log phase cells inoculated 1–16 h before use. Stationary cells were probed with AFM (only a small percentage of P. tricornutum cells are motile at any one time) as previously described for the adhesive pads of Toxarium undulatum (4).
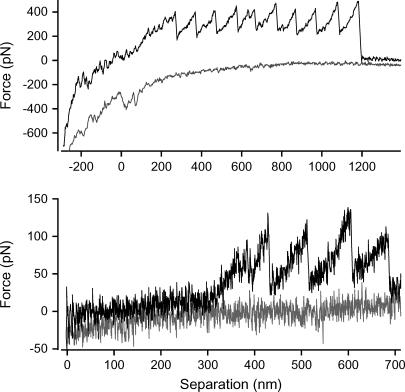
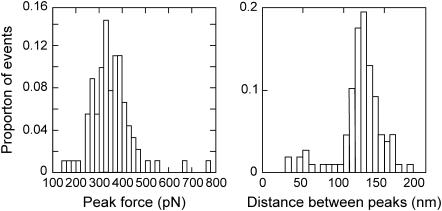
Sawtooth curves were recorded when the AFM tip was retracted from the surface of the cells (Fig. 1). The worm-like chain model of polymer elasticity was fitted to each of the peaks in the curves to determine persistence and contour lengths. The average change in contour length between successive peaks of each of the sawtooth curves was 121 ± 3 (mean ± SE) nm (n = 108; Fig. 2). Sawtooth curves with regular spacing between successive peaks are the characteristic signature of the unfolding of modular proteins (5). Sawtooth curves arise when a molecule is suspended between the tip and substrate: as the separation is increased, the molecule resists extension and a force-induced rearrangement of one of the domains in the molecule occurs. This increases the length of the molecule and allows the cantilever deflection to drop back toward zero. As extension continues, this process is repeated until all of the domains unfold or the molecule detaches from the tip. Detection of this well-defined sawtooth pattern indicates that adhesive modular proteins are present in P. tricornutum extracellular mucilage.
FIGURE 1 .
Force versus separation curves recorded from the surface of stationary ovoid P. tricornutum cells representing ANFs (top) and a single modular protein (bottom).
FIGURE 2 .
Histograms showing the distribution of peak forces (left) and distance between peaks (right) for the sawtooth curves recorded from the extracellular matrix of P. tricornutum. Note there are peaks that have much shorter or greater spacing; these occur within traces where the remainder of the peaks have normal spacing, i.e., ∼120 nm. These can be explained by either modular proteins that are misfolded, or that don't unfold evenly, due to interactions with surrounding fibers and/or diatom extracellular mucilage.
Three further parameters combine to provide the well-defined fingerprint of single-molecule modular proteins as described by Fernandez (5). The peak force values are a measure of the protein's mechanical stability and are typically 100–300 pN for single modular proteins. The regular spacing between peaks measures the length of polypeptide released by the unfolding event. Finally, the persistence length measures the flexibility of the protein polymer and is typically 0.3–1 nm.
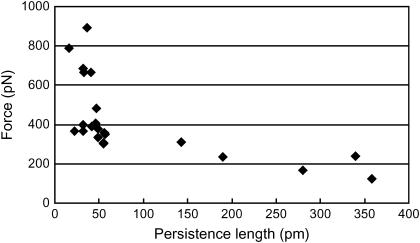
The change in contour length of 121 nm between peaks for P. tricornutum indicates that the unfolded domain contains ∼336 amino acids (assuming an amino acid length of 0.36 nm (6)). These curves were recorded from 22 curves on five cells. The average peak force varied from 123 to 893 pN, whereas the persistence length varied from 21 to 358 pm (Fig. 3). A total of three traces were recorded with single molecule characteristics, i.e., persistence length >250 pm and unfolding force <300 pN; they had between two and four peaks. The number of peaks in the remaining curves, representing unfolding ANFs, is typically greater with an average of 7.2 ± 0.8 (mean ± SE) peaks.
FIGURE 3 .
Average peak force versus average persistence length measured from each of the sawtooth curves recorded from the extracellular matrix of P. tricornutum.
This range of values has previously been recorded in populations of defined polymers. The high force and corresponding low persistence length values are caused by multiple polymers bridged between the tip and substrate and aligned in parallel. The mechanical stability of each of the polymers combines to give the high peak force while the persistence length divides by the number of bridged molecules (7). The true single molecules are therefore represented by the curves with the lowest force and highest persistence length (8,9). This range in the number of parallel molecules has been recorded in another native system of modular proteins, the centric, sessile diatom T. undulatum (4). Centric diatoms do not have raphes and therefore do not posses gliding motility or the standard AC. T. undulatum cells secrete a large, permanent adhesive pad that the cells stand erect in (10,11). We have shown these pads to be a biocomposite composed of a single modular protein arranged into at least five discrete supramolecular assemblies of parallel polymers that we termed ANFs (4). It is therefore likely that the high-force sawtooth curves recorded for P. tricornutum also represent multiple modular proteins aligned in parallel and that the low-force curves represent single modular proteins (Fig. 1). The traces from these ANFs do not overlay precisely because the peak forces differ between peaks. This is most likely explained by the ANFs being composed of a variable number of parallel modular proteins, much like the oligomers recorded for T. undulatum (4). The data presented here for P. tricornutum strongly suggest that the extracellular component of the diatom AC is a molecule with a modular protein backbone, and forms part of the continuum between the surface adhesive at one extreme, and the cellular based motor at the other. The extracellular component of the AC would require a large molecule with great tensile strength and elasticity, both features of modular proteins and ANFs.
The discovery of modular proteins in P. tricornutum is significant because the genome for this species has recently been sequenced (12). Knowledge of nanomechanical signatures derived from AFM studies for native and recombinant modular proteins, e.g., titin (13), will allow directed searches of the genome for genes with modular protein homologs. Once identified, modular proteins could be expressed, adsorbed onto a substratum, and then probed with AFM to sort and identify the molecules responsible for the sawtooth curves and thus the genes responsible for the adhesive modular protein. The AFM approach will thus serve as an analytical screening tool for candidate genes encoding diatom adhesive molecules.
Acknowledgments
The Australian Research Council, our industry partner Akzo Nobel, Gateshead, UK (Industry Linkage Grant No. LP0454982), and the Defense Science and Technology Organization of the Australian Department of Defense provided funding for this study.
References
- 1.Wetherbee, R., J. L. Lind, J. Burke, and R. S. Quatrano. 1998. The first kiss: establishment and control of initial adhesion by raphid diatoms. J. Phycol. 34:9–15. [Google Scholar]
- 2.Chiovitti, A., A. Bacic, J. Burke, and R. Wetherbee. 2003. Heterogeneous xylose-rich glycans are associated with extracellular glycoproteins from the biofouling diatom Craspedostauros australis (Bacillariophyceae). Eur. J. Phycol. 38:351–360. [Google Scholar]
- 3.Higgins, M. J., P. Molino, P. Mulvaney, and R. Wetherbee. 2003. The structure and nanomechanical properties of the adhesive mucilage that mediates diatom—substratum adhesion and motility. J. Phycol. 39:1181–1193. [Google Scholar]
- 4.Dugdale, T. M., R. Dagastine, A. Chiovitti, and R. Wetherbee. 2006. Diatom adhesive mucilage contains distinct supramolecular assemblies of a single modular protein. Biophys. J. 90:2988–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez, J. M. 2005. Fingerprinting single molecules in vivo. Biophys. J. 89:3676–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, H., and J. M. Fernandez. 2003. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J. Mol. Biol. 334:75–86. [DOI] [PubMed] [Google Scholar]
- 7.Bemis, J. E., B. B. Akhremitchev, and G. C. Walker. 1999. Single polymer chain elongation by atomic force microscopy. Langmuir. 15:2799–2805. [Google Scholar]
- 8.Kellermayer, M. S. Z., C. Bustamante, and H. L. Granzier. 2003. Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochim. Biophys. Acta. 1604:105–114. [DOI] [PubMed] [Google Scholar]
- 9.Kellermayer, M. S. Z., S. B. Smith, H. L. Granzier, and C. Bustamante. 1997. Folding-unfolding transitions in single molecules characterized with laser tweezers. Science. 276:1112–1115. [DOI] [PubMed] [Google Scholar]
- 10.Kooistra, W. H. C. F., M. De Stefano, D. G. Mann, N. Salma, and L. K. Medlin. 2003. Phylogenetic position of Toxarium, a pennate-like lineage within centric diatoms (Bacillariophyceae). J. Phycol. 39:185–197. [Google Scholar]
- 11.Dugdale, T. M., R. Dagastine, A. Chiovitti, P. Mulvaney, and R. Wetherbee. 2005. Single adhesive nanofibers from a live diatom have the signature fingerprint of modular proteins. Biophys. J. 89:4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montsant, A., K. Jabbari, U. Maheswari, and C. Bowler. 2005. Comparative genomics of the pennate diatom Phaeodactylum tricornutum. Plant Physiol. 137:500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrion-Vazquez, M., A. F. Oberhauser, T. E. Fisher, P. E. Marszalek, H. Li, and J. M. Fernandez. 2000. Mechanical design of proteins studied by single molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Bio. 74:63–91. [DOI] [PubMed] [Google Scholar]