Abstract
The nanomechanical properties of the coiled-coils of myosin are fundamentally important in understanding muscle assembly and contraction. Force spectra of single molecules of double-headed myosin, single-headed myosin, and coiled-coil tail fragments were acquired with an atomic force microscope and displayed characteristic triphasic force-distance responses to stretch: a rise phase (R) and a plateau phase (P) and an exponential phase (E). The R and P phases arise mainly from the stretching of the coiled-coils, with the hinge region being the main contributor to the rise phase at low force. Only the E phase was analyzable by the worm-like chain model of polymer elasticity. Restrained molecular mechanics simulations on an existing x-ray structure of scallop S2 yielded force spectra with either two or three phases, depending on the mode of stretch. It revealed that coiled-coil chains separate completely near the end of the P phase and the stretching of the unfolded chains gives rise to the E phase. Extensive conformational searching yielded a P phase force near 40 pN that agreed well with the experimental value. We suggest that the flexible and elastic S2 region, particularly the hinge region, may undergo force-induced unfolding and extend reversibly during actomyosin powerstroke.
INTRODUCTION
The simple elegance of the coiled-coil structure belies its diverse functionality in a wide variety of proteins, with myosin being one key example (1). Coiled-coils are two-stranded protein motifs, where each strand is an α-helix with repeated substrings of the form (a-b-c-d-e-f-g). This seven-residue (heptad) repeat generally has apolar residues at the first (a) and fourth (d) position. When the two α-helical strands coil or wrap around one another, the a and d positions are internalized and stabilize the structure. The remaining positions (b, c, e, f, g) are exposed on the surface of the protein, where the side chains are available to interact with other proteins, as well as make intra- and interchain associations that can further stabilize the structure. Most myosins associate into dimers through a coiled-coil interaction along its long tail (termed rod in Fig. 1) and myosin tails further assemble into thick filaments by the interactions between their light meromyosin segments (LMM in Fig. 1). The coiled-coil of muscle myosin exhibits regions of instability that have been documented by proteolytic digestions and microscopy (2–4). The biochemical properties of myosin and its proteolytic fragments: HMM, S1, rod, LMM, and S2 have been studied extensively (5). The myosin rod contains functional diversity along its sequence with coiled-coils, hinges, and polymerizable segments. Two known hinges in S2 are located at the interfaces between S2 and the myosin head (S1) and between S2 and LMM. Such unstable regions contribute to the functional diversity of coiled-coils by conferring additional disorder and compliance. For instance, it has been proposed that some myosins unzip substantial portions of their coiled-coils to enable longer step sizes (6). Several models of muscle contraction suggest that elasticity and flexibility of hinges in S2 may contribute to the ability of myosin heads to orient themselves on actin and produce optimal force generation (7–11).
FIGURE 1.
Coiled-coils in myosin (doubled-headed), single-headed myosin (SHM), and proteolytic fragments rod, S2, and LMM. Hinges and known functional domains along the myosin heavy chain sequence are indicated by the shaded regions. Mutations in β-cardiac myosin (MYH7 gene) that lead to familial hypertrophic cardiomyopathy are indicated as diamonds along the map of the myosin heavy chain in amino acid residues (aa) (13). Many mutations localize to critical functional regions of the myosin head (S1), whereas a substantial number of mutations localize to myosin S2, indicating an important functional role for this domain.
Numerous mutations causing familial hypertrophic cardiomyopathies target the myosin rod (12). Nearly 80% of these mutations affect β-cardiac myosin and myosin binding protein C, which associates with the myosin rod and S2 (13). Most mutation sites in the myosin catalytic domain localize to regions of known functional importance such as the actin-binding site, the nucleotide cleft and the converter. Therefore, the presence of a significant number of mutations in S2 suggests that functionally important structures are being affected (Fig. 1). A different set of mutations in the LMM region of β-cardiac muscle myosin (MYH7 gene) lead to distal myopathy through disruption of the coiled-coil (14). Mutations in the rod domain of nonmuscle myosin IIA (MYH9 gene) can lead to a range of genetic disorders that all exhibit aberrant platelet function, among other symptoms (15,16). Several of these mutations have been shown to disrupt the coiled-coil structure (15).
An elegant crystallographic analysis of scallop muscle myosin S2 near one of the hinges suggests that these coiled-coils are relatively unstable (17). To crystallize this unstable coiled-coil, a leucine zipper was engineered to the normal scallop myosin sequence to enhance its stability. Poorly packed residues were evident toward the interior of the coiled-coil suggesting a propensity for spontaneous unwinding and changes in flexibility. Indeed, such a hinge is thought to enable head separation (8).
The mechanical stability of the coiled-coil domains of myosin and other proteins is clearly important to their function. However, only keratin, being a major component of hair and intermediate filaments (18), has been the subject of extensive mechanical investigation (19). The only single-molecule nanomechanical investigation of a coiled-coil-containing protein has been on skeletal myosin (20). Schwaiger et al. found, by following the force response of a single myosin during stretching and release with an atomic force microscope (AFM), that its coiled-coil is elastic and there was little hysteresis between the extension and relaxing force curves when subjected to a load ∼30 pN (20). Because their pioneering study used full-length myosin, Schwaiger et al. were not able to determine which portions of the myosin coiled-coil were stretched.
To examine the elastic diversity of domains along the myosin rod, we applied the same single-molecule force technique on proteolytic subfragments of myosin (21). Our AFM data indicate a greater degree of compliance in the S2 domain than in the LMM domain of myosin under tensions <40 pN. Molecular mechanics simulations were used to generate force spectra of the available atomic model of S2 and yielded essentially the same characteristic force spectra as from the myosin rod. The data are consistent with a compliant hinge region in S2 that may be easily stretched during the myosin powerstroke.
MATERIALS AND METHODS
Protein purification
Rabbit skeletal myosin was prepared by the method of Godfrey and Harrington (22) and digested with α-chymotrypsin to myosin rod. LMM was prepared by α-chymotryptic digestion of the myosin rod and separation by centrifugation at low ionic strength. Myosin rod and LMM were affinity purified to remove all myosin heads by ultracentrifugation in the presence of excess F-actin at high ionic strength. S2 was prepared by α-chymotryptic digestion of the myosin rod, ethanol precipitation, and ion exchange fast protein liquid chromatography (FPLC) on DEAE sepharose (23,24). Single-headed myosin was prepared from myosin by limited α-chymotryptic digestion, centrifugation, and ion exchange FPLC on DEAE sepharose as previously described (4). Immediately before force microscopy experiments, the myosin and its subfragments were sized by gel filtration chromatography with Superose 6 in high ionic strength buffer (0.5 M KCl, 20 mM imidazole, 1 mM TCEP, pH 7.0) to ensure a monodisperse preparation. Purity and approximate molecular masses were assessed by SDS-PAGE and densitometry.
Protein concentrations were determined spectrophotometrically. The extinction coefficients (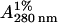 ) and molecular masses were: myosin, 5.5 cm−1, 520,000 Da; SHM, 4.8 cm−1, 390,500 Da; Rod, 2.8 cm−1, 261,000 Da; LMM, 3.0 cm−1, 137,000 Da; S2, 2.6 cm−1, 124,000 Da (25,26).
) and molecular masses were: myosin, 5.5 cm−1, 520,000 Da; SHM, 4.8 cm−1, 390,500 Da; Rod, 2.8 cm−1, 261,000 Da; LMM, 3.0 cm−1, 137,000 Da; S2, 2.6 cm−1, 124,000 Da (25,26).
Single-molecule force spectroscopy
AFM imaging and force measurements were performed on a PicoSPM instrument (Molecular Imaging, Phoenix, AZ) in both contact and noncontact (MAC) mode. Soft cantilevers were obtained from Veeco (Sunnyvale, CA). Cantilevers were 320-μm long (nominal force constant, ∼0.01 N/m; resonance frequency, ∼7 kHz). Cantilevers were cleaned using 0.5 M ethanolic KOH, followed by extensive rinsing with Milli-Q water. Before use, the tips were exposed to oxygen in the presence of high-intensity ultraviolet (UV) to remove any remaining contamination via oxidation from ozone generated by the UV light. Cantilever force constants were measured using the thermal fluctuation method (27) and ranged from 0.012 – 0.015 N/m. Gold surfaces were prepared by epitaxial deposition on heated mica to a thickness of ∼150 nm in vacuum to obtain flat, clean surfaces (Fig. 2 A, inset, bare gold). For adsorbing the myosin on the surface, 190 μl of sterile filtered buffer (0.5 M KCl, 20 mM imidazole, pH 7.0, 1 mM TCEP) was placed on the freshly prepared gold surface and 10 μl of protein solution in the same buffer was carefully injected under the surface and gently mixed. Final protein concentrations were ∼1 μg/ml. The protein was incubated for 30 min at room temperature and the surface was rinsed three times with the same buffer to remove any unattached protein. Buffer containing 0.1 M KCl, 20 mM imidazole, pH 7.0, 1 mM TCEP was then added (550 μl) and force spectroscopy measurements were performed. Several hundred force spectra were obtained for each experiment. The cantilever tip was positioned at different areas on the gold surface, and at least 50 traces were collected at each point. Cantilever speed was ∼1 μm/s and the amplitude setpoint was adjusted so the maximal force of contact was ∼1 nN.
FIGURE 2.
Force microscopy by AFM. (A) Force spectra on Au(111) epitaxially deposited on mica. Insets show the corresponding AFM images of the surface in the absence and presence of protein. On the surface without protein, the tip surface interaction is minimal or absent (black curve “approach”, pink curve “retract”). On the surface with protein, a retracting curve (pink) displaying three distinct phases of stretching of coiled-coil domains is observed. Adsorbed protein on the gold surface is indicated by a roughening of the surface as indicated in the AFM image. (B) Schematics of stretching myosin rod between the Au(111) surface and the tip. The cantilever tip picks up the protein at random positions on the protein along the length, not necessarily end to end.
Force spectra were fit to a worm-like chain model (28):
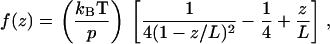 |
(1) |
in which f(z) is the force at a distance of z, p is the persistence length, kB is the Boltzmann's constant, T is the absolute temperature, and L is the contour length of the polymer being stretched. (Note that the contour length is defined as the maximum end-to-end distance of a linear polymer chain and is therefore different from that determined from electron microscopy images of a folded protein). Numerical analysis for this fitting was performed in MathCad (Mathsoft, Cambridge, MA). The spectra were operationally defined by the cantilever movement at the surface, reflecting mainly the stretching of the protein segment between the tip and substrate. However, tip-surface interactions did occur when interfacial surface forces between the cantilever and the protein result in adhesion during contact. To minimize such occurrences, the surfaces were imaged before attachment of protein (Fig. 2 A, bare gold). Gold surfaces were cleaned before use by exposure to high intensity UV to remove any organic contamination. Protein concentration was kept low at ∼1 μg/ml to minimize the possibility of picking up multiple proteins. Force spectra were further analyzed using a scanning probe image processor ((SPIP) Image Metrology, Lyngby, Denmark) before fitting to the worm-like chain (WLC) model. Loess fitting of the curve points using tricube weighting and polynomial regression was used to show the R, P, and E phases (29).
Molecular mechanics simulations of coiled-coil extension
The atomic model derived from x-ray crystallography of scallop muscle myosin subfragment-2 (17) was used as a starting point for the molecular mechanics and conformational searching simulations with Macromodel 8 (Schrödinger, Portland, OR). The leucine zipper residues were removed from the atomic model and hydrogens were added to the remaining intact residues (843–885) of the myosin subfragment-2 coiled-coil (Fig. 3), which is more than 40% identical to vertebrate myosins in this region:
Scallop: EEEMKEQLKQMDKMKEDLAKTERIKKELEEQNVTLLEQKNDLF
Rabbit: EKEMANMKEEFEKTKESLAKAEAKEKELEEKMVALMQEKNDLQ
Chicken: EKEMANMKGEFEKTKEELAKSGAKRKDLEGKMVSLLQEKNDLQ
FIGURE 3.
Molecular model of scallop myosin S2 (17). Each chain of the coiled-coil has the same 43 amino acids shown at the top of the figure and the amino acids numbers run from 843 to 885. The heptads are separated by vertical red bars and the unusual a, d, and g residues are underlined by black bars. The C-chain is shown with the side chains colored as follows: red, acidic; blue, basic; yellow, hydrophobic; green, polar. The D-chain is shown simply as an α-helical coil for simplicity. This orientation is the E0 for the different modes of stretching shown in Figs. 7, B and C, and 8, B and C.
The structure was then energy minimized to convergence using the OPLS-AA force field and a generalized-Born surface area (GBSA) model to simulate water as a continuum at 300 K as done previously (30,31). Four different modes for stretching a coiled-coil were investigated by applying constraints to the amino- and carboxy-termini and then increasing the distance between them at 0.1-nm intervals by successive restrained energy minimizations to convergence or 5000 steps until the polypeptide chains were fully extended (11.7 nm total stretching). A force constant of 16.7 N/m was used in each mode while applying the distance constraints. First, constraints were applied to the amino- and carboxy-terminal atoms of each polypeptide strand allowing the ends of the strands to rotate around one another. Second, the carboxy-termini were fixed and the amino-termini of the two chains were fixed relative to one another so that the chains could not rotate around each other. Third, the constraints and mechanical perturbation were applied to only one chain (D-chain, Fig. 3) and the other chain (C-chain, Fig. 3) was unconstrained. Fourth, one end of each chain is constrained and the carboxyl-terminus of the C-chain and the amino-terminus of the D-chain were separated from one another. In this mode, the distance between the termini was increased to 18 nm to allow for separation of the chains.
To further relax the energy of the first model, every ninth structure (0.9 nm) was entered into a restrained conformational search using a mixed mode Monte Carlo/large-scale, low mode (normal mode) searching algorithm for 1000 steps per structure per cycle, followed by energy minimization to convergence of the lowest energy structures. After two cycles of restrained conformational searching using the standard force constant (16.7 N/m), the force constant for the final cycle of searching was then reduced to 0.0167 N/m to simulate the force constant range used in the force spectroscopy experiments. Forces were calculated by multiplying the force constant by the difference between the target length for the restraint and the actual length of the lowest energy structure following the simulation.
RESULTS
Force spectra of coiled-coils of myosin fragments
An important consideration in analyzing the force spectra of stretched proteins by AFM is distinguishing protein stretching events from nonspecific, tip-surface interactions (sticking), both of which deflect the cantilever tip. Sticking is clearly discernable by the presence of a linear deflection as the cantilever retracts from the surface. As an example, the initial region of the force curve of myosin rod (Fig. 2 A, myosin rod) around ∼20–25 nm of distance shows the presence of such a tip-surface sticking. Typically these regions of force spectra are subtracted or avoided before further analysis. A freshly cleaned gold surface shows little sticking with an AFM tip when measured under buffer. The gold surfaces we used had the expected surface roughness (RMS roughness = 0.3 nm) for this preparation (Fig. 2 A, bare gold). The application of the protein results in an increase in the surface roughness (RMS roughness = 0.68 nm). Occasionally, elongated features of the expected height of a coiled-coil of ∼2 nm were observed (Fig. 2 A, myosin rod). At the protein density examined in these experiments, ∼85% of the total curves either showed only tip-surface sticking (70%) and/or stretching of protein (10–15%). Fig. 2 A (myosin rod) shows a typical spectrum resulting from stretching myosin rod (with the R, P, and E phases; see below) in the presence of tip surface interaction, followed by detachment of the protein at ∼200 pN. About 15% of the total curves showed neither tip-surface interaction nor protein stretching, (cf. retracting curve in Fig. 2 A, bare gold).
Force spectra of single molecules of intact myosin and its proteolytic fragments, including single-headed myosin (SHM), rod, light meromyosin (LMM), and long subfragment-2 (S2), generally displayed three phases: a pronounced rise phase (R) followed by a plateau phase (P) that preceded the final exponential phase (E) in each fragment (Fig. 4 A). After the E phase, there was a sharp drop as the stretched protein detached above 200 pN. Several features are noteworthy and useful for structural interpretation of the force spectra:
The R and P phases of these coiled-coil containing proteins are not typically observed in the force spectra of pure globular proteins, with the exponential phase being the latter's dominant feature (32). This observation suggests that the first two phases arise from stretching coiled-coils. Significantly, these two phases were not analyzable with the WLC model of elasticity.
-
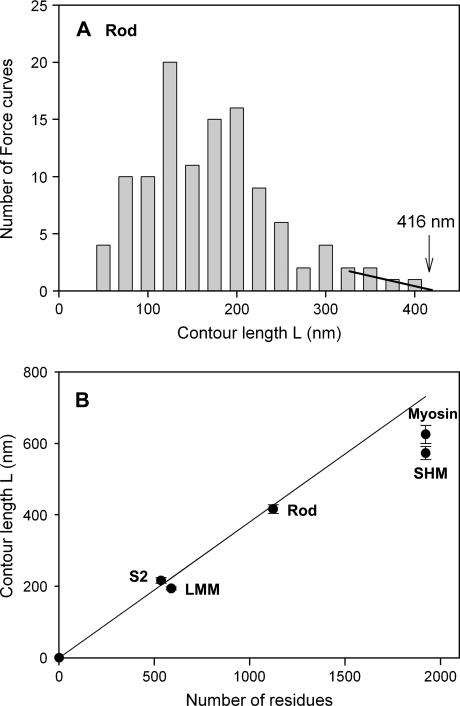
The exponential phase was analyzable with a WLC model of elasticity and yielded two parameters for each curve: the persistence length (p) and the contour length (L). Since the AFM cantilever picked up the single molecule at random sites and stretched the segment between the tip of cantilever and the unspecified anchor sites of the molecule to the gold surface, the resulting force spectra reflect a population of different segment lengths of the polypeptide chains. The average persistence lengths were similar for each of the fragments with mean values ranging from 0.1 to 0.3 nm (Table 1). These values were close to typical values found in mechanical stretching of other proteins (21,33–35). Analysis of the ensemble of contour lengths of extended chains was facilitated by considering the frequency of contour length distribution as if random segments were stretched. Assuming that a chain contained a number of minimal length segments (M) that could be stretched, and the number of potential attachment points was M + 1, then the number of segments with a length equal to a number of contiguous minimal length segments (S) is given by M + 1 − S. If each possible segment were sampled once, then a frequency distribution histogram would begin with M observations of the minimum length segment and decrease linearly with increasing segment length until the maximum segment length was recorded once. In the actual experiments, the shorter length segments were poorly represented due to either steric constraints imposed by the large AFM cantilever or a marginal force signal from a short stretch. Consequently, the frequency distribution will rise to a maximum and then decline. The decline may also deviate from linearity if some segments were preferentially attached or if their mechanical properties varied significantly with composition. Because the composition of the longest segments is expected to be more similar than those among shorter segments, the extrapolation was done only on the upper end of the distribution to determine the longest contour length. Indeed, segments with long contour lengths yielded more uniform persistence lengths than did segments with short contour lengths (data not shown).
The contour length of the extended chain was analyzed by a distribution analysis that suggested the longest observed contour lengths reflect the association of the AFM tip with the longest polypeptide chain in the sample (Fig. 5 A). To test the hypothesis that the upper end of the distribution linearly extrapolates to the contour length of the longest polypeptide chain in the protein sample, the maximum contour lengths from distribution analyses were plotted against the number of amino acids of the protein (Fig. 5 B). The proteins that were primarily coiled-coils showed excellent agreement between the number of residues (of the longest chain in the multichain proteins) and the contour lengths assuming a value for the completely unfolded peptide length of 0.36 nm per amino acid residue (20,36) (line in Fig. 5 A). Double- and single-headed myosins displayed slightly lower than expected maximum contour length values that may reflect a limited capacity of the globular head region to bind to the surface or tip. These results support a direct proportionality between the unfolded coiled-coil peptide length and the contour length L of the WLC model fit to the E phase of the force spectra.
The unique R and P phases of the force spectra leading up to the E phase were analyzed by considering the pattern of variation of these two phases with different protein fragments. We observed that there was a very short R phase length in LMM but longer R phase lengths in proteins containing S2 (Fig. 4 A). The mean R phase lengths were greater for rod than for S2, suggesting that the hinge connecting S2 and LMM contributed to the increased R phase length. In addition, the mean R phase length for myosin was greater than that of rod alone, which is consistent with the idea that the myosin head or the S1/S2 junction hinge also contributes to the R length, although to a somewhat lesser extent than the S2/LMM junction (Fig. 4 B). It thus appears plausible that a prominent R phase results from the stretching of the hinges and the P phase reflects the uncoiling and unfolding of all segments of the coiled-coil.
If this explanation is correct, a plot of the maximum (R + P) lengths versus the estimated coiled-coil length of the protein should yield a slope approximately equal to one. This is expected because the unstretched coiled-coil is ∼0.15 nm/residue and, when stretched, is elongated to a nearly parallel β-strand with ∼0.31 nm/residue. Since (R + P) only accounts for the extension (0.31 nm/residue − 0.15 nm/residue = 0.16 nm/residue), the ratio of 0.16:0.15 is near unity. Maximum (R + P) lengths were estimated by extrapolating linearly the histograms of the (R + P) lengths of each fragment as done for the contour lengths (Fig. 6 A). As shown in Fig. 6 B, the plotted curve indeed supports this interpretation.
FIGURE 4.
Force spectra of double-headed myosin (myosin), single-headed myosin (SHM), and coiled-coil myosin fragments (S2, LMM, and rod). (A) Representative force spectra of LMM, S2, rod, SHM, and DHM (top to bottom). The rise (R) phase begins when the force first reaches 0 and ends when the force reaches the average plateau level after the initial tip-surface interaction, if present. The plateau (P) phase begins after the R phase and has an average force in the range of 15–100 pN (typically 40 pN in these experiments). The exponential (E) phase begins at the end of the P phase with a force level higher than the average plateau force. Solid curves represent a Loess fit of the data points using tricube weighting and polynomial regression to show trend of data. Note that the force spectra of fragments containing S2 have an enhanced R phase that is missing from an otherwise similar force spectrum of LMM. (B) Length distribution of R phase in S2-containing fragments. The mean R lengths from distribution analyses of each fragment analyzed are shown with error bars corresponding to the standard error. Only fragments containing S2 demonstrated substantial R phase lengths. Fragments containing S2 and one or both of its hinges yielded longer R lengths than S2 alone.
TABLE 1.
Mean persistence lengths and plateau forces of myosin fragments
| Fragment (longest chain) | S2 (535 aa) | LMM (588 aa) | Rod (1123 aa) | SHM (1923 aa) | Myosin (1923 aa) |
|---|---|---|---|---|---|
| Persistence length (nm) ± SD | 0.13 ± 0.18 (n = 254) | 0.10 ± 0.08 (n = 24) | 0.29 ± 0.45 (n = 193) | 0.11 ± 0.25 (n = 68) | 0.16 ± 0.38 (n = 73) |
| Plateau force (pN) ± SD | 41 ± 10 (n = 151) | 40 ± 14 (n = 29) | 41 ± 22 (n = 242) | 52 ± 20 (n = 75) | 40 ± 15 (n = 73) |
Means and standard deviations (± SD) for the measured plateau forces and WLC-fitted persistence lengths are calculated as described in the Materials and Method section.
FIGURE 5.
Distribution of contour lengths calculated from WLC fitting of the E phase. (A) The determination of the maximum L of myosin rod by linear regression fitting of the contour length distribution near the maximal values to determine the intercept. (B) The proportionality of the maximum L-values with the longest polypeptide chain of the fragments: LMM, S2, rod, DHM, and myosin. The maximum L-values show a nearly linear correlation with the longest polypeptide chain of the fragments. The line illustrates the expected contour length for a fully extended peptide chain with a 0.36-nm length per residue. The short experimental contour lengths for myosin and SHM may indicate that the globular heads are rarely stretched. Bars represent mean ± SE.
FIGURE 6.
Distribution analyses of (R + P) phase lengths. (A) The determination of the maximum (R + P) lengths by linear regression fitting of the longer values to determine the intercept. (B) The proportionality of maximum (R + P) lengths and the calculated coiled-coil length of the fragments. Bars represent mean ± SE.
Simulation of the mechanical uncoiling and unfolding of a myosin S2 coiled-coil
Further support for the notion that the P region of the force spectra corresponds to the unraveling of the coiled-coil was obtained by performing molecular mechanics simulations on a crystallographic atomic model of scallop S2. Simulated force spectra were calculated by applying constrained energy minimizations to the stretched atomic model of S2 that was held four different ways to mimic possible modes of attachment of the stretched protein segments between a cantilever and the gold surface in the AFM experiments:
Carboxy- and amino-termini of both chains moved away from one another allowing the chains to rotate around one another (Fig. 7, A and B).
Carboxy-termini of both chains fixed and amino-termini moved while maintaining the relative position of the amino termini. This mode of stretching disallows any rotation of the chains around one another (Fig. 7, C and D).
Carboxy- and amino-terminus of the D-chain moved away from one another. The C-chain has no added constraints (Fig. 8, A and B). Only the D-chain is directly stretched.
Carboxy-terminus of the C-chain and amino-terminus of the D-chains moved away from one another. Opposite terminus of each chain is unconstrained (Fig. 8, C and D).
FIGURE 7.
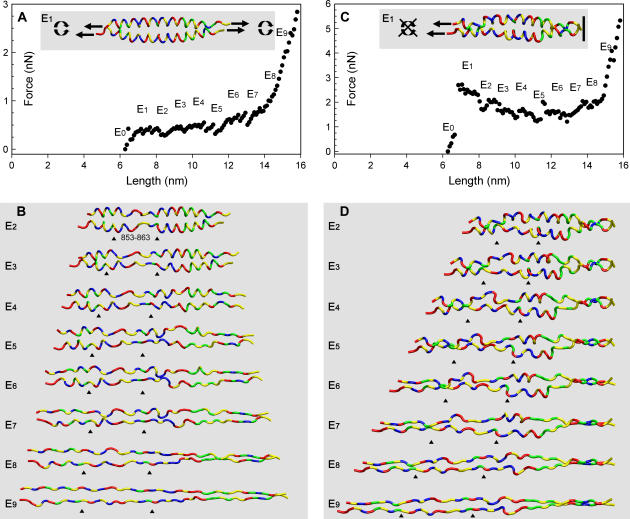
Molecular mechanics simulations of the scallop S2 atomic model allowing rotation (A,B) and disallowing rotation (C,D) of chains relative to one another while stretching both chains. (A,C) The simulated force spectra of incremental extension of the S2 by constrained energy minimizations. Note their similarity to the unique shape of the experimental force spectra from the myosin coiled-coil. E1 configuration showing the stretching mode (inset). (B,D) Images of the S2 atomic model during 1-nm incremental stretching. The coiled-coil unravels at nearly random locations along its length. The ends of the coils unfold first, and then the weakly hydrogen bonded region between residues 853–863 unfolds (E2), followed by the rest of the coils. The position of the 853–863 region at the different extension lengths is marked by black arrowheads. With rotation allowed (A, B), the coiled-coils twists during the extension. With rotation disallowed (C, D), kinks form in the coils. The chains are shown as tubes with color-coded residues: red, acidic; blue, basic; yellow, hydrophobic; green, polar. The E0 configuration is the same as that in Fig. 3. E1–E9 represent structures from 1- to 9-nm extension.
FIGURE 8.
Molecular mechanics simulations of the scallop S2 atomic model with differential stretching of the chains. (A,B) Stretching just the D-chain at both ends. (C,D) Stretching only one end of each chain with the opposite end free. (A,C) The simulated force spectra of incremental extension of the S2 by constrained energy minimizations. E1 configuration showing the stretching mode (inset). Note its similarity to the unique shape of the experimental force spectra from the myosin coiled-coil. Also note that the simulated force spectrum for this mode is different in shape from the other simulated curves and experimental force spectra. (B,D) Images of the S2 atomic model during incremental stretching. The coiled-coil unravels at nearly random locations along its length and twists during the extension. The position of the 853–863 region at the different extension lengths is marked by black arrowheads. In panel B, the weakly hydrogen-bonded region between residues 853 and 863 unfolds in the D-chain at E3 and then in the C-chain at E5. In panel D, the coils in both chains at the end where the restraints are applied unfold first. The coils at the opposite end of the chains remain intact for most of the simulation. The chains are shown as tubes colored as the type of residue: red, acidic; blue, basic; yellow, hydrophobic; green, polar. The E0 configuration is the same as that in Fig. 3. E1–E9 represent structures from 1- to 9-nm extension.
With the exception of Mode 4, all of the simulations showed the characteristic plateau and exponential phases and qualitatively mimicked the experimental data of coiled-coil extension (Figs. 7 and 8 and Supplementary Material). In each mode, the calculated force spectrum for the simulation is shown in the upper panels (A and C), and the modes by which the model is stretched to 1 nm extension (E1) are shown in the insets. Molecular models of the simulation at 2 nm of extension (E2) and every 1-nm increment for 9 nm of extension (E2-E9) are shown in the lower panels (B and D). The initial length of the S2 fragment at zero extension is 6.3 nm and is shown in Fig. 3.
Mode 1
The simulated force spectrum was the most similar in appearance to the experimental data. There was a rapid rise to a bumpy plateau at 0.5 nN and then at the end of the plateau phase the exponential phase started. The length of the plateau region was just over 6 nm, which yielded a total length of the extended molecule of 12.3 nm or roughly a doubling of the S2 fragment initial length of 6.3 nm (Fig. 7 A). Fig. 7 B and the supplemental movie (see Supplementary Material) illustrate that the S2 twisted as it unraveled and the hydrogen bonds along the α-helix popped apart seemingly at random during the stretching to ∼14 nm. The ends of coiled-coil first started to unravel and then the region between residues 853–863 disengaged. Fig. 9 shows the hydrogen bond network in this 11-residue region, the hydrophobic side chains that interact between the coils and two salt bridges between side chains on one of the coils.
FIGURE 9.
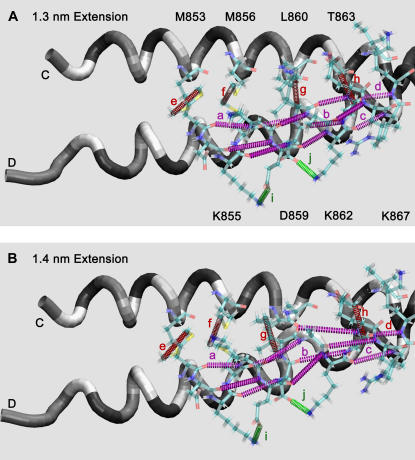
Transformation of hydrogen bonds, hydrophobic interactions, and salt bridges near the N-terminal end of scallop S2 during stretching from (A) 1.3 nm to (B) 1.4 nm of extension. Hydrogen bonds: (a) M853_O:K857_N, (b) D859_O:T863_N, (c) K862_O:I866_N, (d) T863_O:K867_N, and others that are not labeled; intercoil hydrophobic interactions: (e) M853_Cɛ:M853_Cɛ:, (f) M856_Cɛ:M856_Cɛ, (g) L860_Cδ1:L860_Cδ1, (h) T863_Cγ2:T863_Cγ2; salt bridges: (i) K855_Nζ:E858_Oɛ2, (j) D859_Oδ:K862_Nζ. These are shown in the region between residues 853 and 863 as colored, dashed tubes between the interacting atoms. Notice large changes in the α-helical hydrogen bonds between the two panels. See Fig. 10 for details of how these interactions change with extension. The chains are shown as tubes colored as the type of residue: red, acidic; blue, basic; yellow, hydrophobic; green, polar.
Mode 2
The simulated force spectrum rose rapidly to 3 nN then the force dropped over 3 nm of extension to half of this initial rise to ∼1.5 nN (Fig. 7 C). This level of force continued for ∼4 nm then the exponential phase began. Interestingly, the initial increase in force followed by a drop to the plateau level (Fig. 7 C) was similar to some, but not all, of the force spectra for double-headed myosin (Fig. 4 A, myosin). The portion of the myosin coiled-coil that was being stretched in this particular force curve may be a segment where the rotation of the two chains was being inhibited either by the flanking supercoils or the attachment to the tip and substrate.
Mode 3
The simulated force spectrum rose more slowly to a plateau force of ∼0.750 nN over ∼3 nm (Fig. 8 A). The force plateau in this mode was both higher and bumpier than in Mode 1. As the plateau region approached the exponential phase, the variation in force increased. This may be due to the progressive weakening of the stabilizing interactions between the chains as the D-chain was stretched. The C-chain was stretched slightly even though there are no constraints on this chain and only interactions with the D-chain can lead to such an extension (Fig. 8 B). The region of the C-chain that unfolds was the same weak area as on the D-chain, residues 853–863.
Mode 4
The simulated force spectrum increased to ∼1 nN of force over 10 nm of extension without a plateau phase (Fig. 8 C). The force then increased more rapidly to ∼2 nN as if the molecule was in the exponential phase and then the interaction between the chains was disrupted and the force dropped as the two chains separated. The coils at the restrained ends of the two chains were the first to unfold (Fig. 8 D) and then upon further extension, the weak region in the middle of the coils (residues 853–863) unfolded. The coils at the unrestrained ends of the chains remain intact through a greater range of extension than in the other simulations, because only one end of each chain is being stretched. When the chains separated, at >17 nm of extension, the coils were completely disrupted (not shown).
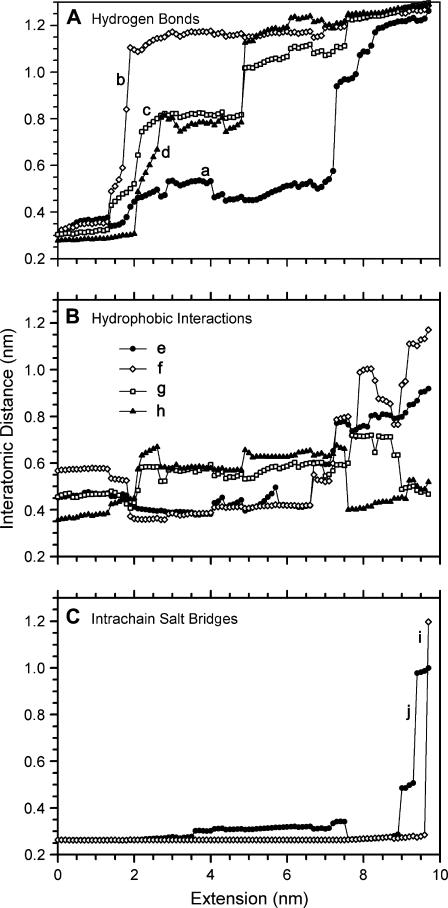
The changes in the hydrogen bond network of the coiled-coil have been analyzed in detail for the force spectrum in Mode 1. As the molecule was stretched, there was an abrupt change in the hydrogen bond network between an extension of 1.3 nm (Fig. 9 A) and 1.4 nm (Fig. 9 B). Hydrogen bonds were assumed to be formed when the O–H distance was <0.26 nm and the O–H-N bond angle was 180 ± 60° (37). The O–N interatomic distance is plotted in Fig. 10 A for several of the (i − i + 4) hydrogen bonded residues. An amide-carbonyl hydrogen bond is considered to be broken at O–N distances >0.4 nm. However, the plot shows that there are several structural transitions that occurred throughout the course of the stretching. The hydrogen bonds breaking at 1.4 and 2.0 nm of extension are seen, as well as jumps in the O–N distance at 4.8 nm (0.78–1.12 nm) and 7.2 nm (0.57–0.94 nm). During these events, the force on the molecule dropped: at 1.4 nm of extension the force dropped 10 pN, at 2.0 nm of extension the force dropped 89 pN, at 4.8 nm of extension the force dropped 25 pN, and at 7.2 nm of extension the force dropped 55 pN.
FIGURE 10.
Interatomic distances of selected residues in the M853–T863 region of one coil during extension. (A) O–N distances of several α-helical hydrogen bonds in the C-coil of the coiled-coil: (a) M853_O:K857_N, (b) D859_O:T863_N, (c) K862_O:I866_N, (d) T863_O:K867_N. Bonded character (O–N distance < 0.4 nm) was mostly absent by 2 nm of extension, although large jumps in the distances indicate the presence of several structural transitions at 4.8 and 7.2 nm of extension. (B) C–C distance for the interchain hydrophobic interactions (heptad a–a and d–d): (e) M853_Cɛ:M853_Cɛ, (f) M856_Cɛ:M856_Cɛ, (g) L860_Cδ1:L860_Cδ1, (h) T863_Cγ2:T863_Cγ2. (C) O–N distances for two i − i + 3 salt bridges: (i) K855_Nζ:E858_Oɛ2, (j) D859_Oδ:K862_Nζ.
The hydrophobic residues that interact between the chains in this region appeared to resist separation until >7 nm of extension (Fig. 10 B). There are four hydrophobic patches that interact between the chains in this region: M853, M856, L860, and T863, which take heptad positions a, d, a, and d, respectively. The distance between these residues varied between 0.35 and 0.65 nm up until ∼8 nm of extension, then the methionine residues separated while the other two came closer together. The two intrachain salt bridges in this region of the coiled-coil were the most stable interactions and they showed little change in the interatomic distance until the extension was >9 nm (Fig. 10 C).
The initial simulation (Fig. 7) yielded a plateau force that was more than 25-fold higher than the values obtained under the experimental conditions, so further simulations were designed to relax the structures to better mimic the slow stretching that occurs under experimental conditions. After lengthy conformational searching using normal mode and Monte Carlo algorithms, the selected structures (every 0.9 nm) relaxed to lower force levels in the experimental range of ∼20–60 pN for the plateau region (Fig. 11). A fit of the WLC model to this simulated data (Fig. 11 A) yielded reasonable values of L = 13.7 nm and p = 0.085 nm, providing support for the validity of the simulations. Before invoking the conformational search for “relaxed” structures (Fig. 7), the persistence length was 36 times lower than the relaxed model, even though the contour length was 14.0 nm. Experimentally, fragments with lengths similar to that of the S2 atomic model were too short to be detected in the force spectroscopy measurements. It is interesting to note, however, that one of the shortest experimentally measured S2 fragments gave a profile similar to the computational model, except with a P region of approximately double the length, which presumably originated from stretching a segment twice as long (Fig. 11 B). The WLC fit to the experimental data from this particular S2 segment yielded a contour length of 30 nm and a persistence length of 0.06 nm, consistent with this segment being approximately twice the length of that in the atomic model. The extension lengths in Fig. 11 were normalized by the WLC contour length to allow for direct comparison of the simulated and experimental force spectra.
FIGURE 11.
Conformational searching simulation of scallop S2 extension and experimental force spectrum. (A) Computational force spectrum of scallop S2 of 43 residues after relaxation by conformational searching. Configurations that were relaxed are those from Fig. 7 taken every 0.9 nm (every 9th configuration). Solid line is a cubic fit through the computed data points. (B) Experimental force spectrum of a short S2 segment of ∼90 residues. Solid line is a cubic fit through the experimental points. Note the presence of R, P, and E phases and that the plateau force levels for both the computed and experimental force spectra are of closely similar magnitude. Extensive computational searching is necessary to relax the structure so that the force levels reach the experimental range. This agreement in force levels has not been attained in molecular dynamics simulations. Extension normalized by the WLC contour length.
Overall, our computational simulations indicated that the coiled-coil chains separated completely from each other at approximately the end of the P region and nearly doubled the initial fragment length. The E phase then began and represents the stretching of the unfolded polypeptide chains. The lack of an enhanced R phase in the computational simulation resulted from the absence of an unfolded coiled-coil/hinge in the atomic model of S2 at the beginning of the simulation.
DISCUSSION
The single-molecule mechanical analysis of myosin fragments containing coiled-coil domains and computational simulations provide a detailed explanation of the unique force spectra of myosin under mechanical load. Our results suggest that the initial R phase is related to hinge regions of the S2 and to a lesser extent the presence of the head in intact or single-headed myosin. The plateau phase is well described by atomic models in which the coiled-coil unfolds first at the weakest intrachain α-helical hydrogen bonds followed by side-chain interactions. Interchain hydrophobic and intrachain ionic interactions remain throughout most of the plateau phase and only become disrupted when in the E phase of the force spectra. The final E phase, and only this phase, can be analyzed with a WLC model to yield L-values that correspond closely to those predicted for the fragment size being analyzed.
Head and tail of the myosin force spectra
The elasticity of single intact myosin has been established recently (20). However, the presence of the myosin heads complicates their interpretation of the force spectra and structural attributes (20). The significant mechanical contribution of the heads to the rapid rise phase is evident in our analysis of a set of myosin fragments. First, the experimental R phase is more extensive in the presence of one or both heads (Fig. 4 A, myosin and SHM) and Schwaiger et al. (20)) than those without heads (Fig. 4 A, LMM, S2, and rod). Second, computational simulations on the atomic model of the coiled-coil yield a force spectrum with the unique P phase and yet lack the extensive R phase found in myosin coiled-coils that contain head and hinge domains (Figs. 7 and 8). Third, the similarity in the results from single-headed versus double-headed myosin indicate that the number of myosin heads or their interactions do not contribute any significant force to the P phase. Rather, the presence of the myosin head domain appears to diminish slightly the estimates of L per amino acid relative to fragments lacking the head domain (Fig. 5), suggesting that the globular head is rarely stretched. Mean persistence length were similar across the different fragments (Table 1).
Experimentally, the R phase is challenging to detect by AFM because the force is low, the noise is high, and the stretch is short. Moreover, the tip/surface interactions often obscure this initial phase of force response. Most AFM force studies delete this large tip/surface attractive force from published force spectra. Careful cleaning of the cantilever tip and a clean gold surface was essential to reproduce this critical phase of the force spectra. Additionally, the simultaneous acquisition of dynamic stiffness and the force spectra of single molecules, as we and others have done recently with a refined AFM (38,39), may reveal additional force bearing structural intermediates as the coiled-coil is being unraveled in this low force region.
Molecular elasticity and modeling by WLC
It is noted that two parameter fits of both experimental and simulated force spectra to the WLC model give persistence lengths that are occasionally shorter than a peptide bond (33–35). Although this may appear puzzling at first sight, we emphasize that a comparison of the model-dependent persistence length with true structural parameters, peptide length and geometry, is unlikely to provide useful insights of the structural basis of molecular elasticity in general. It is useful to keep in mind that the WLC model is a continuum model for describing statistical polymer configurations under the assumptions that there are no excluded volume effects (chain can cross itself), no self-interaction and the chain is inextensible (40,41). Supplementing the model with classical elastic theory (typically applied to bulk materials), showed that a worm-like polymer could be treated as a homogeneous elastic wire with a bending energy density of kBT/p (42). The WLC model cannot be used to make any conclusions about the microscopic structure of the polymer, because it assumes a structureless, homogenous material. The WLC model has been successfully applied to describe the entropic elasticity of the stiff polymer DNA (28,43). Application of Eq. 1 to flexible, self-interacting polymers that phenomenologically appear to behave as WLC may be expected to yield unexpected results as the caveats of the WLC are violated. Elastic persistence lengths shorter than bond lengths is further evidence that the WLC model is not suitable without taking into consideration both entropy and enthalpy in describing the elasticity of coiled-coils. This deficiency of WLC model is even more pronounced in interpreting force spectra of highly charged polyampholytes such as the titin PEVK segments (33). Although the persistence lengths from the WLC are not structurally meaningful, the contour lengths are, as shown by the distribution analyses shown in Fig. 5.
Molecular mechanics simulations of force spectra: uncoiling of superhelix and unfolding of α-helices
Computational simulations provide powerful insights of the force-bearing molecular and atomic events that accompany the mechanical unraveling and unfolding of the coiled-coils of myosin. The recent availability of the atomic model of a segment of S2 from scallop myosin enabled the generation of computational force spectra that simulated the key characteristic phasic features of the experimental data from single-molecule mechanics techniques. Moreover, WLC analysis of experimental and simulated force spectra yielded comparable parameters, providing further validation of the modeling (Fig. 11). We conclude that the unique plateau phase of the force spectrum arises from the uncoiling of the coiled-coil superhelix and the unfolding of the α-helices. The constrained conformational searching method described here enabled precise matching of the absolute force values of the force spectrum to the experimental results. This level of agreement in force levels has not been possible with methods based on molecular dynamics simulations that impose extremely high loading rates that are typically seven orders of magnitude higher than experimentally feasible rates (44). In contrast, conformational searching is not a dynamics method, so the extension rate is closer to the experimental rates. On the other hand, the large extension step size may result in the loss of some molecular details that would be explicitly present in a steered molecular dynamics study. Whether this computational approach is applicable to immunoglobulin and other protein domains remains to be explored.
Stretching coiled-coil α-helices: transformation of helical hydrogen bond networks and salt bridges
Previous simulations and AFM measurements on α-helices agree with these simulations of helix stretching, but lack the contribution from interactions between the two strands (39,45–48). A molecular dynamics study of the GCN4 coiled-coil (49) gave force spectra similar to that in Fig. 7 C. Rohs and colleagues (45) performed a molecular mechanics study of the extension of polyalanine (20-mer) and found that the energetically favorable mechanism was a turn-by-turn unfolding of the helix. This unfolding mechanism leads to a steep rise in force, followed by a plateau in the force similar to that shown in Fig. 11. However, the force versus length curves they report are smooth and do not show the variations in force over the plateau that we have observed in both the simulation of the S2 coiled-coil extension and the force spectroscopy measurements. This difference may result from their method of calculating the force (by differentiating a polynomial fitted to the energy versus length curve) and the homogenous array of hydrogen bonds in this homopolymer.
In our simulations, the ends extend first due to the lack of the i − i + 4 hydrogen bonds, then the next region to uncoil is between residues 853 and 863 as shown in Fig. 9. This region likely ruptures next due to the lack of two α-helical i − i + 4 hydrogen bonds. Of the eleven possible α-helical (i − i + 4) hydrogen bonds in this region, one interaction (D854_O-K857_N) is out of the bonded range and a second (K855_O) is a 310 helical (i − i + 3) hydrogen bond at zero extension. Six of the α-helical hydrogen bonds disappear or convert to 310 helical hydrogen bonds when the extension steps from 1.3 to 1.4 nm. At an extension of 2.0 nm (E2 in Fig. 7), only two hydrogen bonds remain (K855_O-E858_N and T863_O-K867_N). These two hydrogen bonds are lost at an extension of 2.4 nm. Interestingly, at extensions between 3.6 and 7.2 nm, two 310 helical hydrogen bonds form and then break upon further extension of the coiled-coil. The cooperativity of the hydrogen bond network may cause these jumps in the O–N interatomic distance shown in Fig. 10 A as the molecule is stretched and other regions pop apart. Several turns on both chains are also stabilized by i − i + 3 side-chain salt bridges. These salt bridges stabilize the loop to large extension with the salt bridges remaining intact until >9 nm of extension (Fig. 10 C) or 15 nm of total length (Fig. 7 A, E8).
The simultaneous rupturing of several α-helical hydrogen bonds is what leads to the ripples or blunt saw teeth in the plateau region of the force spectrum. When several hydrogen bonds rupture simultaneously there is a drop in the force, which is recovered after a small increase in extension leading to more hydrogen bonds rupturing. This sequential breaking of hydrogen bonds at near constant force is fundamentally different from what happens in the stretching of globular immunoglobulin (Ig) domain. During Ig stretching, force builds rapidly to a high level, then the breakage of an array of β-sheet hydrogen bonds results in the rapid unfolding of a significant portion of the globular domain (44). The force also rapidly drops due to the increased end-to-end distance of the unfolded chain. Experimentally, this rapid buildup and drop in force yields a saw-tooth shape force curve (50). This difference in force extension behavior of coiled-coils and β-barrels is most likely due to the difference in orientation of the hydrogen bonds to the force vector. In the coiled-coil, the hydrogen bonds are co-linear with the force vector and in the β-sheet of Ig, the hydrogen bonds are perpendicular to the force vector. The buildup of strain in the coiled-coil will be mostly along the O–H-N bond, whereas the strain on the β-sheet will result in a change of the O–H-N bond angle.
The residues at the a and d positions in coiled-coil heptad repeat are typically hydrophobic and it is this extended hydrophobic surface at the interface of the two coils that makes the coiled-coil thermodynamically stable. Substitution of a polar or charged side-chain side chain into the interchain interface would be expected to destabilize the coiled-coil structure. However, thermodynamic stability does not necessarily confer resistance to mechanical extension. The scallop S2 has a charged group (K867) at a heptad a position (17) and the separation of this residue on the two chains is similar to those shown in Fig. 10 B. These two polar residues at the hydrophobic interface of the S2 coiled-coil probably significantly reduce the thermodynamic stability of this coiled-coil, but as shown by the simulations, do not detract from its mechanical stability. The mechanical stability or resistance to stretch is dominated by the helical hydrogen bond network and the intrachain salt bridges. Inter- and intrahelical salt bridges contribute to coiled-coil thermodynamic stability (51) and are expected to enhance resistance to mechanical stretch in a nonstereospecific manner.
Protein domains and mechanical attributes
Structural and cytoskeletal proteins have evolved several basic mechanically competent motifs: β-sandwich structures (immunoglobulin and fibronectin type II domains), spectrin repeat (three helical bundle), coiled-coil, and disordered nanogel (titin PEVK) (33). Only two β-sandwich type proteins have been studied extensively, titin I27 and tenascin TNfn3 domains. These two globular proteins unfold at very different force levels and the molecular mechanisms for stability are also different (52). In titin I27, resistance to force-induced unfolding is dominated by hydrogen bonds in a limited region of the protein, where as in TNfn3, resistance to unfolding arises from both hydrogen bonds and packing rearrangements over a larger portion of the protein. Spectrin repeats also unfold in an all or nothing manner (53) where one helix unfolds followed by a simultaneous unraveling of the other two helices. PEVK unwraps smoothly with no bursts in the force response. The forced unfolding behavior of coiled-coil domains lies between the two extremes of all-or-nothing and smoothly unfolding/unwrapping. Interestingly, coiled-coils unfold progressively under nearly constant force, which starts to rise again only when it is almost completely unfolded. Coiled-coils also refold rapidly without hysteresis (20). These distinct elastic behaviors of the three types of fundamental protein domains allow complex multidomain proteins to generate a broad spectrum of mechanical attributes.
Muscle contraction and S2 coiled-coil
Optimal force generation in muscle requires that the myosin heads bind to the actin thin filaments in the proper orientation. However, it is unlikely that the myosin head can always bind efficiently to actin if the attachment to the thick filaments is through a stiff rod. Several models have been proposed that allow for hinges in the S2 region as well as extension of S2 to facilitate binding of the S1 head to actin (7,54). A number of studies on the properties of myosin and S2 have given strong evidence for conformational flexibility or elasticity in the S2 region of myosin. Proteolytic studies identified a major hinge at the S2/LMM junction (2). Thermal stability studies have verified that the S2 has regions of low melting temperature (23). S2 has been shown to have pronounced torsional flexibility, which is consistent with unwinding of the coiled-coil (4).
The nanomechanical properties of S2 suggest that the flexible S2, particularly the hinge region, might undergo force-induced unfolding and extend reversibly during muscle contraction. The amount of force that S2 may experience in muscle under physiological conditions can be estimated from single-molecule studies. The upper limit of this force would be the actomyosin unbinding force that has been measured to range from 9 to 35 pN depending upon how the force was applied to the actomyosin interface (55,56). The power stroke from an individual myosin head has been estimated to generate 1–10 pN of force over the 10-nm motion of the myosin head (57). Taking into consideration that the S2 coiled coils unfold and unwind below ∼40 pN, and even at lower force during the R phase, the possible contribution of S2 or hinge elasticity toward muscle contraction is intriguing indeed. Our molecular mechanical simulation studies also suggest that the manner of attachment of the two-headed myosin to actin may also alter the mechanical response of the S2 region. For example, simultaneous attachments of both heads would constrain the coiled-coil of the S2 as in the Mode 2 simulation (Fig. 7, C and D), whereas a one-head attachment scheme would be simulated by Mode 1, yielding different force spectra. Whether the mode of myosin head attachment to the thin filament affects force spectra and torsional flexibility of the S2 (4) remain to be established experimentally.
The mutations in the coiled-coil domain of myosins in certain muscle diseases vary in their position in the heptad repeat. Some are at positions a and d and may alter the stability of the coiled-coil as well as the force generating/transmission capability of the mutated myosins (12,14). Other mutations switch charges, which in some cases disrupt salt bridges and in others affect the interaction with other sarcomeric proteins (15,16). Some of these mutations could also affect the helical hydrogen bond network as seen here for the scallop S2; however, high-resolution structural information on the coiled-coil are still lacking for meaningful modeling studies.
Single-molecule mechanical studies of coiled-coil proteins with disease-causing mutations provide the possibility of determining whether these mutations affect the mechanical competency of the coiled-coil. These types of nanomechanical measurements are complementary to thermodynamic measurements of coiled-coil stability and may give an indication of the molecular mechanism of some of these disease-causing mutations, even in the absence of detailed structural information. Investigation of the mechanical properties of other coiled-coil proteins and the effects of disease-causing mutations are likely to yield valuable information on how the proteins perform their mechanical and structural roles in muscle and other tissues.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Health and Human Services (AR44737 to D.D.R.) and by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Health and Human Services (K.W.).
Douglas D. Root, Vamsi K. Yadavalli, and Jeffrey G. Forbes contributed equally to the work.
Abbreviations used: LMM, light meromyosin; aa, amino acids; AFM, atomic force microscopy; DTT, dithiothreitol; E, exponential phase; FPLC, kB, Boltzmann's constant; L, contour length; M, molecular mass; p, persistence length; P, plateau phase; R, rise phase; SHM, single-headed myosin; S1, myosin subfragment-1; S2, myosin long subfragment-2; TCEP, tri-(2-carboxyethyl)phosphine hydrochloride.
References
- 1.Rose, A., and I. Meier. 2004. Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins. Cell. Mol. Life Sci. 61:1996–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mihalyi, E., and W. F. Harrington. 1959. Studies on the tryptic digestion of myosin. Biochim. Biophys. Acta. 36:447–466. [Google Scholar]
- 3.Walker, M., P. Knight, and J. Trinick. 1985. Negative staining of myosin molecules. J. Mol. Biol. 184:535–542. [DOI] [PubMed] [Google Scholar]
- 4.Gundapaneni, D., J. Xu, and D. D. Root. 2005. High flexibility of the actomyosin crossbridge resides in skeletal muscle myosin subfragment-2 as demonstrated by a new single molecule assay. J. Struct. Biol. 149:117–126. [DOI] [PubMed] [Google Scholar]
- 5.Sellers, J. R. 1999. Myosins. Oxford University Press, New York.
- 6.Rock, R. S., S. E. Rice, A. L. Wells, T. J. Purcell, J. A. Spudich, and H. L. Sweeney. 2001. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 98:13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huxley, A. F., and R. M. Simmons. 1971. Proposed mechanism of force generation in striated muscle. Nature. 233:533–538. [DOI] [PubMed] [Google Scholar]
- 8.Malnasi-Csizmadia, A., E. Shimony, G. Hegyi, A. G. Szent-Gyorgyi, and L. Nyitray. 1998. Dimerization of the head-rod junction of scallop myosin. Biochem. Biophys. Res. Commun. 252:595–601. [DOI] [PubMed] [Google Scholar]
- 9.Holmes, K. C., and M. A. Geeves. 2000. The structural basis of muscle contraction. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Root, D. D. 2002. The dance of actin and myosin: a structural and spectroscopic perspective. Cell Biochem. Biophys. 37:111–139. [DOI] [PubMed] [Google Scholar]
- 11.Levitsky, D. I. 2004. Actomyosin systems of biological motility. Biochemistry (Mosc.). 69:1177–1189. [DOI] [PubMed] [Google Scholar]
- 12.Blair, E., C. Redwood, M. D. Oliveira, J. C. Moolman-Smook, P. Brink, V. A. Corfield, I. Ostman-Smith, and H. Watkins. 2002. Mutations of the light meromyosin domain of the beta-myosin heavy chain rod in hypertrophic cardiomyopathy. Circ. Res. 90:263–269. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann, J., S. Daehmlow, S. Wischke, M. Senyuva, U. Werner, J. Raible, N. Tanis, S. Dyachenko, M. Hummel, R. Hetzer, and V. Regitz. 2003. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin. Genet. 64:339–349. [DOI] [PubMed] [Google Scholar]
- 14.Meredith, C., R. Herrmann, C. Parry, K. Liyanage, D. E. Dye, H. J. Durling, R. M. Duff, K. Beckman, M. de Visser, M. M. van der Graaff, P. Hedera, J. K. Fink, et al. 2004. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1). Am. J. Hum. Genet. 75:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke, J. D., F. Dong, W. L. Rickoll, M. J. Kelley, and D. P. Kiehart. 2005. Rod mutations associated with MYH9-related disorders disrupt nonmuscle myosin-IIA assembly. Blood. 105:161–169. [DOI] [PubMed] [Google Scholar]
- 16.Heath, K. E., A. Campos-Barros, A. Toren, G. Rozenfeld-Granot, L. E. Carlsson, J. Savige, J. C. Denison, M. C. Gregory, J. G. White, D. F. Barker, A. Greinacher, C. J. Epstein, et al. 2001. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am. J. Hum. Genet. 69:1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y., J. H. Brown, L. Reshetnikova, A. Blazsek, L. Farkas, L. Nyitray, and C. Cohen. 2003. Visualization of an unstable coiled coil from the scallop myosin rod. Nature. 424:341–345. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, E. 1995. Keratins and the skin. Annu. Rev. Cell Dev. Biol. 11:123–153. [DOI] [PubMed] [Google Scholar]
- 19.Hearle, J. W. S. 2000. A critical review of the structural mechanics of wool and hair fibres. Int. J. Biol. Macromol. 27:123–138. [DOI] [PubMed] [Google Scholar]
- 20.Schwaiger, I., C. Sattler, D. R. Hostetter, and M. Rief. 2002. The myosin coiled-coil is a truly elastic protein structure. Nat. Mater. 1:232–235. [DOI] [PubMed] [Google Scholar]
- 21.Wang, K., J. G. Forbes, and A. J. Jin. 2001. Single molecule measurements of titin elasticity. Prog. Biophys. Mol. Biol. 77:1–44. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey, J., E., and W. Harrington, F. 1970. Self-association in the myosin system at high ionic strength. I. Sensitivity of the interaction to pH and ionic environment. Biochemistry 9:886–893. [DOI] [PubMed] [Google Scholar]
- 23.Rodgers, M. E., and W. F. Harrington. 1987. Hinging of rabbit myosin rod. Biochemistry. 26:8697–8703. [DOI] [PubMed] [Google Scholar]
- 24.Borras-Cuesta, F., and E. Truche. 1984. The purification of myosin long subfragment-2 by DEAE-Sepharose Cl-6b ion-exchange chromatography. Anal. Biochem. 142:84–87. [DOI] [PubMed] [Google Scholar]
- 25.Weeds, A. G., and B. Pope. 1977. Studies on chymotryptic digestion of myosin: effects of divalent-cations on proteolytic susceptibility. J. Mol. Biol. 111:129–157. [DOI] [PubMed] [Google Scholar]
- 26.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326. [DOI] [PubMed] [Google Scholar]
- 27.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 28.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 29.Cleveland, W. S. 1993. Visualizing Data. Hobart Press, Summit, NJ.
- 30.Root, D. D., S. G. Xin, X. Jin, and M. A. McAllister. 1999. Determination of fluorescent probe orientations on biomolecules by conformational searching: algorithm testing and applications to the atomic model of myosin. J. Struct. Biol. 127:22–34. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Z. L., J. P. Jin, and D. D. Root. 2004. Binding of calcium ions to an avian flight muscle troponin T. Biochemistry. 43:2645–2655. [DOI] [PubMed] [Google Scholar]
- 32.Craig, A., and E. M. Terentjev. 2005. Stretching globular polymers. I. Single chains. J. Chem. Phys. 122:194901. [DOI] [PubMed] [Google Scholar]
- 33.Forbes, J. G., A. J. Jin, G. Gutierrez-Cruz, W. L. Tsai, and K. Wang. 2005. Charge-driven single molecule elasticity of the open and flexible titin PEVK polyampholyte. J. Muscle Res. Cell Motil. In press. [DOI] [PMC free article] [PubMed]
- 34.Labeit, D., K. Watanabe, C. Witt, H. Fujita, Y. M. Wu, S. Lahmers, T. Funck, S. Labeit, and H. Granzier. 2003. Calcium-dependent molecular spring elements in the giant protein titin. Proc. Natl. Acad. Sci. USA. 100:13716–13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar, A., S. Caamano, and J. M. Fernandez. 2005. The elasticity of individual titin PEVK exons measured by single molecule atomic force microscopy. J. Biol. Chem. 280:6261–6264. [DOI] [PubMed] [Google Scholar]
- 36.Lantz, M. A., S. P. Jarvis, H. Tokumoto, T. Martynski, T. Kusumi, C. Nakamura, and J. Miyake. 1999. Stretching the alpha-helix: a direct measure of the hydrogen- bond energy of a single-peptide molecule. Chem. Phys. Lett. 315:61–68. [Google Scholar]
- 37.Baker, E. N., and R. E. Hubbard. 1984. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 44:97–179. [DOI] [PubMed] [Google Scholar]
- 38.Forbes, J. G., and K. Wang. 2004. Simultaneous dynamic stiffness and extension profiles of single titin molecules: nanomechanical evidence for unfolding intermediates. J. Vac. Sci. Technol. A. 22:1439–1443. [Google Scholar]
- 39.Kageshima, M., M. A. Lantz, S. P. Jarvis, H. Tokumoto, S. Takeda, A. Ptak, C. Nakamura, and J. Miyake. 2001. Insight into conformational changes of a single alpha-helix peptide molecule through stiffness measurements. Chem. Phys. Lett. 343:77–82. [Google Scholar]
- 40.Kratky, O., and G. Porod. 1949. Rontgenuntersuchung Geloster Fadenmolekule. [in German.] Recl. Trav. Chim. Pay. B. 68:1106–1122. [Google Scholar]
- 41.Yamakawa, H. 1997. Helical wormlike chains in polymer solutions. Springer-Verlag, Berlin, Germany.
- 42.Landau, L. D., and E. M. Lifshitz. 1986. Theory of Elasticity. J. B. Sykes and W. H. Reid, translator. Butterworth-Heinemann, Oxford, UK.
- 43.Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- 44.Lu, H., B. Isralewitz, A. Krammer, V. Vogel, and K. Schulten. 1998. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohs, R., C. Etchebest, and R. Lavery. 1999. Unraveling proteins: a molecular mechanics study. Biophys. J. 76:2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Idiris, A., M. T. Alam, and A. Ikai. 2000. Spring mechanics of alpha-helical polypeptide. Protein Eng. 13:763–770. [DOI] [PubMed] [Google Scholar]
- 47.Ptak, A., S. Takeda, C. Nakamura, J. Miyake, M. Kageshima, S. P. Jarvis, and H. Tokumoto. 2001. Modified atomic force microscope applied to the measurement of elastic modulus for a single peptide molecule. J. Appl. Phys. 90:3095–3099. [Google Scholar]
- 48.Masugata, K., A. Ikai, and S. Okazaki. 2002. Molecular dynamics study of mechanical extension of polyalanine by AFM cantilever. Appl. Surf. Sci. 188:372–376. [Google Scholar]
- 49.Akkermans, R. L. C., and P. B. Warren. 2004. Multiscale modelling of human hair. Philos. Transact. A. Math. Phys. Eng. Sci. 362:1783–1793. [DOI] [PubMed] [Google Scholar]
- 50.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 51.Burkhard, P., R. A. Kammerer, M. O. Steinmetz, G. P. Bourenkov, and U. Aebi. 2000. The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure. 8:223–230. [DOI] [PubMed] [Google Scholar]
- 52.Ng, S. P., R. W. S. Rounsevell, A. Steward, C. D. Geierhaas, P. M. Williams, E. Paci, and J. Clarke. 2005. Mechanical unfolding of TNfn3: the unfolding pathway of a fnIII domain probed by protein engineering, AFM and MD simulation. J. Mol. Biol. 350:776–789. [DOI] [PubMed] [Google Scholar]
- 53.Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. 1999. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561. [DOI] [PubMed] [Google Scholar]
- 54.Kad, N. M., A. S. Rovner, P. M. Fagnant, P. B. Joel, G. G. Kennedy, J. B. Patlak, D. M. Warshaw, and K. M. Trybus. 2003. A mutant heterodimeric myosin with one inactive head generates maximal displacement. J. Cell Biol. 162:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishizaka, T., H. Miyata, H. Yoshikawa, S. Ishiwata, and K. Kinosita. 1995. Unbinding force of a single motor molecule of muscle measured using optical tweezers. Nature. 377:251–254. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima, H., Y. Kunioka, K. Nakano, K. Shimizu, M. Seto, and T. Ando. 1997. Scanning force microscopy of the interaction events between a single molecule of heavy meromyosin and actin. Biochem. Biophys. Res. Commun. 234:178–182. [DOI] [PubMed] [Google Scholar]
- 57.Tyska, M. J., and D. M. Warshaw. 2002. The myosin power stroke. Cell Motil. Cytoskeleton. 51:1–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.