Abstract
Polymerization dynamics of single actin filaments is investigated theoretically using a stochastic model that takes into account the hydrolysis of ATP-actin subunits, the geometry of actin filament tips, and the lateral interactions between the monomers as well as the processes at both ends of the polymer. Exact analytical expressions are obtained for the mean growth velocity, for the dispersion in the length fluctuations, and the nucleotide composition of the actin filaments. It is found that the ATP hydrolysis has a strong effect on dynamic properties of single actin filaments. At high concentrations of free actin monomers, the mean size of the unhydrolyzed ATP-cap is very large, and the dynamics is governed by association/dissociation of ATP-actin subunits. However, at low concentrations the size of the cap becomes finite, and the dissociation of ADP-actin subunits makes a significant contribution to overall dynamics. Actin filament length fluctuations reach a sharp maximum at the boundary between two dynamic regimes, and this boundary is always larger than the critical concentration for the actin filament's growth at the barbed end, assuming the sequential release of phosphate. Random and sequential mechanisms of hydrolysis are compared, and it is found that they predict qualitatively similar dynamic properties at low and high concentrations of free actin monomers with some deviations near the critical concentration. The possibility of attachment and detachment of oligomers in actin filament's growth is also discussed. Our theoretical approach is successfully applied to analyze the latest experiments on the growth and length fluctuations of individual actin filaments.
INTRODUCTION
Actin filaments are major component of cytoskeleton in eukaryotic cells, and they play important roles in many biological processes, including the organization of cell structures, transport of organelles and vesicles, cell motility, reproduction, and endocytosis (1–3). Biological functions of actin filaments are mostly determined by the dynamic processes that take place during the growth or shrinking of these biopolymers. However, our understanding of mechanisms of assembly and disassembly of these filaments is still very limited.
In recent years, the number of experimental investigations of the growth dynamics of rigid cytoskeleton filaments, such as actin filaments and microtubules, at a single-molecule level has increased significantly (4–12). Dynamic behavior of individual microtubules has been characterized by a variety of experimental techniques such as video and electron microscopy, fluorescence spectroscopy, and optical trap spectrometry (4–9), while the studies of the single actin filaments have just started (10–12). The assembly dynamics of individual actin filaments revealed a treadmilling phenomenon, i.e., the polymer molecule tends to grow at the barbed end and to depolymerize at the pointed end (11). Similar picture has been observed earlier for microtubules (13,14). Although the conventional actin filaments do not exhibit the dynamic instability as observed in microtubules (13,15), it was shown recently that the DNA-segregating prokaryotic actin homolog ParM displays two phases of polymer elongation and shortening (16). ATP hydrolysis is not required for actin assembly (17), but it is known to play an important role in the actin polymerization dynamics. Experimental observations suggest that the nucleotide bound to the actin filament acts as a timer to control the filament turnover during the cell motility (18). Hydrolysis of ATP and release of inorganic phosphate are assumed to promote the dissociation of the filament branches and the disassembly of ADP-actin filaments (19).
Recent experimental studies of the single actin filament growth (11,12) have revealed unexpected properties of actin polymerization dynamics. A large discrepancy in the kinetic rate constants for actin assembly estimated by average length change in the initial polymerization phase and determined from the analysis of length fluctuations in the steady-state phase (by a factor of 40) has been observed. Several possible explanations of this intriguing observation have been proposed (11,12,20,21). First, the actin polymerization dynamics might involve the assembly and disassembly of large oligomeric actin subunits. However, this point of view contradicts the widely accepted picture of single-monomer polymerization kinetics (1,21). In addition, as we argue below, it would require the association/dissociation of actin oligomers with 30–40 monomers, but the annealing of such large segments has not been observed in experiments or it has been excluded from the analysis (11,12). Second, the stochastic pauses due to filament-surface attachments could increase the apparent dispersion in the length of the single actin-filaments, although it seems that the effect is not significant (12). Third, the errors in the experimental measurements could contribute into the observation of large apparent diffusion constants (12). Another possible reason for the discrepancy is the use of the oversimplified theoretical model in the analysis that neglects the polymer structure and the lateral interaction in the actin filament. However, the detailed theoretical investigation of the growth of single actin filaments (20) indicates that large length fluctuations still cannot be explained by correctly describing the structure of the filament's tip and the lateral interactions between the monomers.
The fact that hydrolysis of ATP bound to the actin monomer is important stimulated a different model to describe the actin polymerization dynamics (21). According to this approach, the ATP-actin monomer in the filament can be irreversibly hydrolyzed and transformed into the ADP-actin subunit. The polymer growth is a process of adding single ATP-actin monomers and deleting hydrolyzed or unhydrolyzed subunits, and the actin filament consists of two parts—a hydrolyzed core in the middle and unhydrolyzed caps at the polymer ends. Large length fluctuations are predicted near the critical concentration for the barbed end. Although this model provides a reasonable description of the filament growth rates for different ATP-actin concentrations, the position of the peak in dispersion is below the critical concentration for the barbed end, while in the experiments (11,12) large dispersion is observed at, or slightly above, the critical concentration. In addition, the proposed analytical method (without approximations) (21) cannot calculate analytically cap sizes and dispersion.
The goal of this work is to develop a theoretical model of polymerization of single actin filaments that incorporates the ATP hydrolysis in the polymer, the structure of the filament tips, lateral interactions between the monomers, and the dynamics at both ends. Our theoretical method is based on the stochastic models developed for describing the growth dynamics of rigid multifilament biopolymers (20,22), and it allows us to calculate explicitly all dynamic parameters of the actin filament's growth. Different mechanisms of ATP hydrolysis in actin filaments are compared. The possibility of adding or deleting oligomeric actin subunits is also discussed. Finally, we analyze the latest experiments on the growth dynamics of single actin filaments (11,12).
MODEL OF ACTIN FILAMENT ASSEMBLY
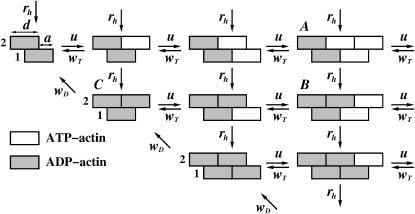
Let us consider an actin filament as a two-stranded polymer, as shown in Fig. 1. It consists of two linear protofilaments. The size of the monomer subunit in this polymer is equal to d = 5.4 nm, and two protofilaments are shifted with respect to each other by a distance a = d/2 = 2.7 nm (1,2). Each monomer in the actin filament lattice carries a nucleotide molecule: it can be either ATP, or ADP (see Fig. 1). Shortly after actin monomers assemble into filaments, the ATP is hydrolyzed to ADP. For simplicity, we do not differentiate between the initial ATP-actin and intermediate ADP-Pi-actin states since the off-rates for these states are assumed to be very close, and the transition between states is relatively rapid (21). Thus, we only consider two states of actin monomers in the filament—hydrolyzed and unhydrolyzed. It is argued that only the dynamics of capped (unhydrolyzed) and uncapped (hydrolyzed) states effect the length fluctuations in the actin filaments (21). The hydrolyzed nucleotide remains bound to the polymer, and at the physiological conditions it is not exchangeable with free ATP molecules from the solution. The dynamical and biochemical properties of ATP-bound (T-state) and ADP-bound (D-state) monomers are known to be different (23). The dissociation rate of ADP-actin subunits from the actin filaments is estimated to be 2–5 times larger than the rate for the ATP-actin subunits, whereas the association rate is considerably slower (by a factor of 10) than that for the nonhydrolyzed analog (12,23).
FIGURE 1.
Schematic picture of polymer configurations and possible transitions in the vectorial model of the single actin filament's growth. The size of the monomer subunit is d, while a is a shift between the parallel protofilaments equal to one-half of the monomer size. Two protofilaments are labeled 1 and 2. The transition rates and labels to some of the configurations are explained in the text.
ATP hydrolysis plays an important role for the overall actin filament assembly dynamics; however, the details of this process are not clear. Several mechanisms of ATP-actin hydrolysis in the filament have been proposed. In a random mechanism (24–26) any ATP-actin subunit can hydrolyze in a stochastic manner independently of the states of the neighboring monomers. The rate of hydrolysis in this case is proportional to the amount of nontransformed nucleotide in the polymer. A different approach is a sequential, or vectorial, mechanism (27–29), that assumes a high degree of cooperation during the hydrolysis. According to this mechanism, the recently assembled actin monomer hydrolyzes its ATP only if it touches the more-interior, already hydrolyzed subunits. In this mechanism, there is a sharp boundary between the unhydrolyzed cap and the hydrolyzed core of the filament, whereas, in the random mechanisms, there are many interfaces between ATP-actin and ADP-actin subunits. Also, there are experimental evidences for the intermediate ADP-Pi-actin state in the nucleotide transformation. The phosphate release was found to be slow (30,31), and ADP-Pi and ATP-actins are practically indistinguishable (21). A little is known about the mechanism of Pi dissociation, although most people assume that it follows a random mechanism (30,31). Finally, it is also possible that a mixed mechanism, which combines the properties of random and vectorial approaches, describes the hydrolysis in actin filaments. The available experimental data cannot clearly distinguish between these mechanisms. In our model we view the hydrolysis of actin filaments as a slow one-step sequential nucleotide transformation without considering the intermediate states. One can associate the phosphate release with this process since ATP-Pi-actin subunits are practically indistinguishable from the ATP-actin monomers, and the dissociation of the phosphate is the rate-limiting stage of the hydrolysis. However, as we show below, the exact details of hydrolysis do not much influence the dynamic properties of the actin filament's growth.
There is an infinite number of possible polymer configurations depending on the nucleotide state of each monomer and the geometry of polymer ends (20): see Fig. 1. However, we assume that only the so-called one-layer configurations, where the distance between two edge monomers at parallel protofilaments is less than d, are relevant for actin polymerization dynamics. This is based on the previous theoretical studies (20,22), which showed that the one-layer approach is an excellent approximation to a full dynamic description of growth of two-stranded polymers with large lateral interactions between the subunits. It is also known that for actin filaments the lateral interaction energy is larger than 5 kBT per monomer (20,29), and it strongly supports the one-layer approximation.
Each configuration we label with two pairs of integers, (l1, k1;l2, k2), where li is the total number of monomers (hydrolyzed and unhydrolyzed) in the ith protofilament, while ki specifies the number of ATP-actin subunits in the same protofilament. For example, the configuration A from Fig. 1 is labeled as 2,1:3,2 and the configuration B is described as 2,1:3,1. The polymerization dynamics at both ends of the actin filament is considered independently from each other.
As shown in Fig. 1, at each end free ATP-actin molecules from the solution can attach to the actin filament with the rate u = kTc, where kT is the ATP-actin polymerization rate constant and c is the concentration of free ATP-actin species in the solution. Because of the excess of free ATP molecules in the solution only ATP-actin monomers are added to the filament (27–29). We also assume that the dissociation rates of actin monomers depend on their nucleotide state, and only the leading subunits dissociate from the filament. Specifically, ATP-actin monomer may detach with the rate wT, while the hydrolyzed subunit dissociates with the rate wD (see Fig. 1). In addition, the sequential mechanism of hydrolysis is assumed, i.e., ATP-actin monomer can transform into ADP-actin state with the rate rh if it touches two already hydrolyzed subunits.
Although our theoretical picture is similar to the model proposed in Vavylonis et al. (21), there are many differences between two approaches. Vavylonis et al. (21) investigated the dynamics of single chain polymers at barbed ends by explicitly taking into account the intermediate ADP-Pi-actin state and by assuming random mechanisms of ATP hydrolysis and Pi dissociation. In our model, the actin filament is viewed as two growing interacting protofilaments, the dynamics at both ends is considered explicitly, the existence of the intermediate states is ignored, and the vectorial mechanism of hydrolysis is utilized.
The growth dynamics of single actin filaments can be determined by solving a set of master equations for all possible polymer configurations. The mathematical derivations and all details of calculations are given in Appendix. Here we only present the explicit expressions for the dynamics properties of actin filament growth at stationary state. Specifically, the mean growth velocity is equal to
 |
(1) |
and dispersion is given by
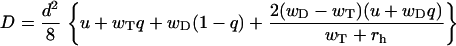 |
(2) |
for 0 ≤ q ≤ 1, where
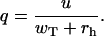 |
(3) |
Note also that expression similar to Eq. 1 has been derived earlier (29), and Eq. 2 in the limit of rh → 0 reduces to an expression derived by Vavylonis et al. (21) using the scaling arguments.
The parameter q plays a critical role for understanding mechanisms of actin growth dynamics. It has a meaning of probability that the system is in a capped state with Ncap ≥ 1 ATP-actin monomers, i.e., it is a fraction of time that the actin filament can be found in any configuration with at least one unhydrolyzed subunit. For example, in Fig. 1 the configurations A and B are capped (with Ncap = 3 and 2, correspondingly), while the configuration C is uncapped with Ncap = 0. The parameter q increases linearly with the concentration of free ATP-actin monomers because of the relation u = kTc. However, it cannot be larger than 1, and this observation leads to existence of a special transition point with the concentration
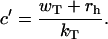 |
(4) |
Above the transition point we have q(c ≥ c′) = 1, and the probability to have a polymer configuration with Ncap = 0 is zero and the unhydrolyzed ATP cap grows steadily with time. At large times, for c ≥ c′ the average length of ATP cap is essentially infinite, whereas below the transition point (c < c′) this length is always finite. Note that our theoretical approach accounts for fluctuations in the ATP cap; however, for c ≥ c′, the probability of the fluctuation that completely removes all unhydrolyzed monomers is an exponentially decreasing function of time and cap size—leading to zero probability to find a hydrolyzed monomer at the end of the filament in the stationary-state limit.
At the critical concentration for each end of the filament, by definition, the mean growth velocity for this end vanishes. Using Eqs. 1 and 3 it can be shown that
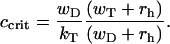 |
(5) |
An important observation is the fact the critical concentration is always below the transition point,
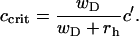 |
(6) |
Because at concentrations larger than the transition point the dissociation events of hydrolyzed actin monomers are absent, the explicit expressions for the mean growth velocity and dispersion in this case are given by
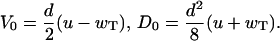 |
(7) |
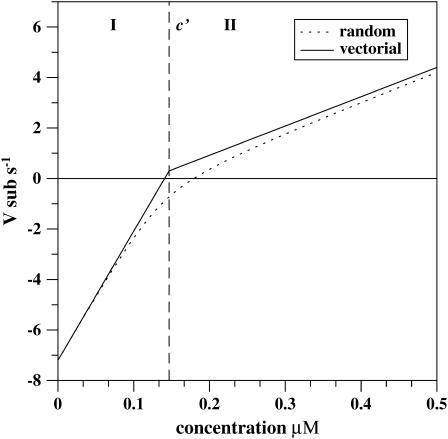
The calculated mean growth velocity for the barbed end of the actin filament is shown in Fig. 2 for parameters specified in Table 1. It can be seen that the velocity depends linearly on the concentration of free ATP-actin particles in the solution, although the slope changes at the transition point. This is in agreement with experimental observations on actin filament's growth (32). However, the behavior of dispersion is very different; see Fig. 3. It also grows linearly with concentration in both regimes, but there is a discontinuity in dispersion at the transition point. From Eqs. 2 and 7 we obtain that the size of the jump is equal to
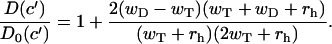 |
(8) |
The origin of this phenomenon is the fact that ADP-actin subunits dissociations contribute to overall growth dynamics only below the transition point c′. Note, as shown in Fig. 3, this contribution can increase the length fluctuations when wD > wT (the barbed end of the filament), or dispersion can be reduced for wD < wT (the pointed end of the filament). The jump disappears when wD = wT. For the barbed end of the actin filament we calculate, using the parameters from Table 1, that D(c′)/D0(c′) ≃ 20.6 and it approaches to 26.5 when rh → 0. This result agrees quite well with the experimentally observed apparent difference in the kinetic rate constants (35–40 times) (11,12).
FIGURE 2.
Comparison of the growth velocities for the barbed end of the single actin filament as a function of free monomeric actin concentration for the random and the vectorial ATP hydrolysis mechanisms. A vertical dashed line indicates the transition point c′ (in the vectorial hydrolysis). It separates the dynamic regime I (low concentrations) from regime II (high concentrations). The kinetic rate constants used for calculations of the velocities are taken from Table 1.
TABLE 1.
Summary of rate constants
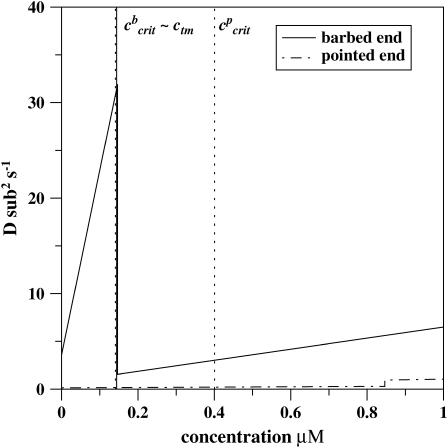
FIGURE 3.
Dispersion of the length of the single actin filament as a function of free monomer concentration for the barbed end and for the pointed end (the vectorial mechanisms). The kinetic rate constants are taken from Table 1. (Vertical dotted lines indicate the critical concentrations ccrit for the barbed and the pointed ends; thin solid line corresponds to the concentration of the treadmilling, which is also almost the same as the transition point for the barbed end.) The transition point for the pointed end is at 0.85 μM. Total dispersion is a sum of the independent contributions for each end of the filament.
To compare our theoretical predictions with experimental observations the dynamics at both ends should be accounted for. However, as we showed earlier (20), the total velocity of growth and the overall dispersion are the sums of the corresponding contributions for each end of the filament. The parameters we use in the calculations are shown in Table 1.
The process of ATP hydrolysis at the ATP-actin monomer consists of two steps: the relatively rapid chemical cleavage of ATP into ADP and Pi; and the slow rate-limiting release of the phosphate (12,30,33). Experimental measurements of Pi dissociation suggest that the phosphate release rate is rather small ≈0.003 s−1 (30). However, this value is obtained assuming the random mechanism for ATP hydrolysis and Pi release, in which the transformation can take place at any unhydrolyzed subunit. In our calculations we assume the sequential mechanism, where only one subunit can be hydrolyzed at any time. Thus it can be concluded that rh(sequential)q(sequential) = rh(random)Ncap(random). The number of ATP-actin subunits in the actin filament depends on the concentration of free ATP-actin monomers, but in the region around the transition point (q ≈ 1) it can be estimated that Ncap < 100 (21), and we took rh = 0.3 s−1 as an upper bound for the hydrolysis rate in our calculations for both ends of the actin filaments. Note, however, that the specific value of rh does not strongly influence our calculations as long as it is small in comparison with the association/dissociation rates (see Eq. 8).
More controversial is the value of ADP-actin dissociation rate constant wD. Most experiments indicate that wD is relatively large, ranging from 4.3 s−1 (32) to 11.5 s−1 (34); however, the latest measurements performed using the TIRF method (12) estimated that the dissociation rate is lower, wD = 1.3 s−1. We choose for wD the value of 7.2 s−1 as better describing the majority of experimental work.
For the actin filament system with the parameters given in Table 1 we can calculate from Eq. 5 that the critical concentration for the barbed end is ccrit ≃ 0.141 μM, while for the pointed end it is equal to ccrit ≃ 0.401 μM. However, the contribution of the pointed end processes to the overall growth dynamics is very small. As a result, the treadmilling concentration, when the overall growth rate vanishes, is estimated as ctm ≃ 0.144 μM, and it is only slightly above the critical concentration for the barbed end (see Fig. 3). The treadmilling concentration also almost coincides with the transition point for the barbed end, as can be calculated from Eq. 4, c′ ≃ 0.147 μM. According to Eq. 2, the dispersion at treadmilling concentration at stationary-state conditions is equal to D(ctm) ≃ 31.6 sub2 × s−1, with the contribution from the pointed end equal to 0.5%. From experiments, the values 29 sub2 × s−1 (11) and 31 sub2 × s−1 (12) are reported for the filaments grown from Mg-ATP-actin monomers, and 25 sub2 × s−1 (11) is the dispersion for Ca-ATP-actin filaments. The agreement between theoretical predictions and experimental values is very good. It is also important to note that, in contrast to the previous theoretical description (21), our model predicts large length fluctuations slightly above the ccrit for the barbed end of the filaments, exactly as was observed in the experiments (11,12).
The presented theoretical model allows us to calculate explicitly not only the dynamic properties of actin growth but also the nucleotide composition of the filaments. As shown in Appendix, the mean size of the cap of ATP-actin monomers is given by
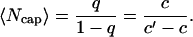 |
(9) |
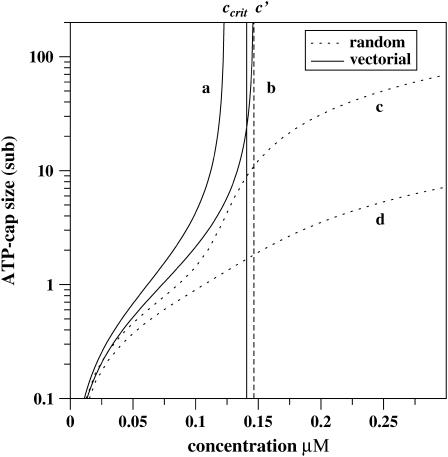
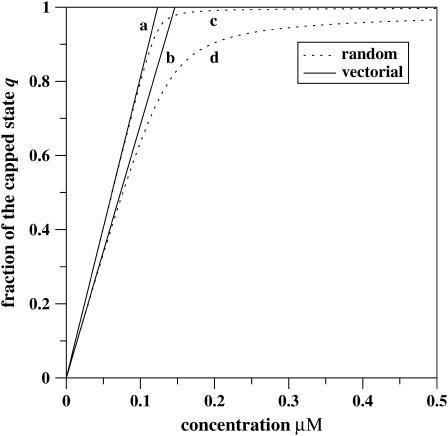
Then at the critical concentration for the barbed end, ccrit ≃ 0.141 μM, the cap size at the barbed end is Ncap ≃ 24, while the cap size at the pointed end at this concentration (with the transition point c′ ≃ 0.846 μM) is <1 monomer. These results agree with the Monte Carlo computer simulations of actin polymerization dynamics (21,35). A large ATP-cap appears at the barbed end of actin filament and a smaller cap is found at the pointed end (35). This is quite reasonable since the transition point for the barbed end is much smaller than the corresponding one for the pointed end. The overall dependence of Ncap on the concentration of free actin monomers is shown in Fig. 4.
FIGURE 4.
The size of ATP cap as a function of free monomer concentration for the barbed end of the single actin filament within the vectorial (a and b) and the random (c and d) mechanisms of ATP hydrolysis. (Thick solid lines describe the vectorial mechanism, while dotted lines correspond to the random mechanism.) The kinetic parameters for constructing curves b and d are taken from Table 1. For the curves a and c, the kinetic rate constants are also taken from Table 1 with the exception of the smaller hydrolysis rate rh = 0.03 s−1. (Vertical dashed line and thin solid line indicate the transition point c′ and the critical concentration ccrit, respectively, for curve b.)
RANDOM VERSUS VECTORIAL ATP HYDROLYSIS IN ACTIN FILAMENTS
An important issue for understanding the actin polymerization dynamics is the nature of the ATP hydrolysis mechanism. In our theoretical model the vectorial mechanism is utilized. To understand which features of actin dynamics are independent of the details of hydrolysis, it is necessary to compare the random and the vectorial mechanisms for this process. In our model of the polymer's growth with a vectorial mechanism, the actin filament consists of two parallel linear chains shifted by the distance a = d/2 from each other. According to our dynamic rules, the addition (removal) of one actin subunit increases (decreases) the overall length of the filament by the distance a. Then the growth dynamics of two-stranded polymers can be effectively mapped into the polymerization of single-stranded chains with an effective monomer's size deff = d/2.
A single-stranded model of the actin filament's growth that assumes association of ATP-actin monomers and dissociation of ATP-actin and ADP-actin subunits along with the random hydrolysis has been developed earlier (33,36). In this model the parameter q is also introduced, and it has a meaning of the probability to find the leading subunit of the polymer in the unhydrolyzed state. However, the parameter q in the random hydrolysis model has a more complicated dependence on the concentration than in the vectorial model. It can be found as a root of the cubic equation,
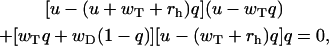 |
(10) |
with the obvious restriction that 0 ≤ q ≤ 1. Note that this equation is a result of the approximate description of the process.
Using the parameters given in Table 1, the fraction of the capped configurations for two different mechanisms of hydrolysis is shown in Fig. 5. The predictions for both mechanisms are close at very low and very high concentrations, but deviate near the critical concentration. It can be understood if we analyze the special case wT = wD, although all arguments are still valid in the general case of wD ≠ wT. From Eq. 10 we obtain
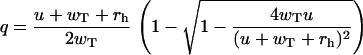 |
(11) |
FIGURE 5.
The fractions of the capped configurations q for the barbed end of the single actin filament as a function of free monomer concentration. The results for the vectorial (a and b) and the random (c and d) mechanisms of hydrolysis are presented. (Thick solid lines describe the vectorial mechanism, while dotted lines correspond to the random mechanism.) The kinetic parameters for constructing curves b and d are taken from Table 1. For curves a and c the kinetic rate constants are also taken from Table 1 with the exception of the smaller hydrolysis rate rh = 0.03 s−1.
In the limit of very low concentrations, the fraction q approaches
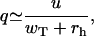 |
(12) |
whereas for c ≫ 1 it can be described as
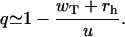 |
(13) |
Generally, in the limit of very low hydrolysis rates the random and the vectorial mechanisms should predict the same dynamics, as expected. It can be seen by taking the limit of rh → 0 in Eq. 10, which gives q = u/wT for u < wT and q = 1 for u > wT. These results are illustrated in Fig. 5.
To calculate the size of the ATP-cap in the actin filament for the random mechanism we introduce a function Pn defined as a probability to find in the ATP-state the monomer positioned n subunits away from the leading one. Then it can be shown that this probability is an exponentially decreasing function of n (35),
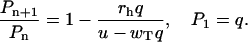 |
(14) |
The size of the unhydrolyzed cap in the polymer is associated with the total number of ATP-actin monomers (21),
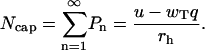 |
(15) |
The results of the different mechanisms for Ncap are plotted in Fig. 4. The random and vectorial mechanism agree at low concentrations, but the predictions differ for large concentrations.
The mean growth velocity in the model with the random mechanism is given by
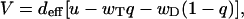 |
(16) |
which is exactly the same as in the vectorial mechanism (see Eq. 1), although the fraction of ATP-cap configurations q have a different behavior in two models. Mean growth velocities for different mechanisms are compared in Fig. 2. Again, the predictions for different mechanisms of hydrolysis converge for very low and very high concentrations of free actin, but differences arise near the critical concentration.
In comparing two mechanisms of hydrolysis it was assumed that the hydrolysis rate constants are the same, which is correct only in the limit of low concentrations of free ATP-actin monomers. In the sequential model, only one ATP-subunit can hydrolyze, while in the random model, the hydrolysis can take place at any of Ncap subunits. As was discussed earlier, the general relation between the hydrolysis rates for the random and sequential models is given by
 |
(17) |
This means that the sequential model with given rh should be compared with the random model with the smaller hydrolysis rate. However, as we showed above, in this case the agreement in the prediction of the dynamic properties of growing actin filaments between two different models of hydrolysis will be even better.
In the model with the random mechanism of hydrolysis (21,36) the analytical expressions for dispersion have not been found. However, from the comparison of the fraction of ATP-capped configurations q, the size of the ATP-cap Ncap, and the mean growth velocity V, it can be concluded that both mechanisms predict qualitatively and quantitatively similar pictures for the dynamic behavior of the single actin filaments. Thus, it can be expected that, similarly to the vectorial mechanism, there is a sharp peak in the dispersion near the critical concentration in the model with the random hydrolysis, in agreement with the latest Monte Carlo computer simulations results (21). However, the biggest remaining problem is the position of this peak with respect to the critical concentration. Monte Carlo computer simulations (21) indicate that the peak of fluctuations in the random mechanism is below ccrit.
ASSEMBLY/DISASSEMBLY OF OLIGOMERS IN ACTIN FILAMENT DYNAMICS
The association and dissociation of large oligomers of actin monomers has been suggested as a possible reason for large fluctuations during the elongation of single actin filaments (11,12). Let us consider this possibility more carefully. Suppose that the oligomeric particles that contain n ATP-actin monomers can attach to or detach from the filament. Then the mean growth velocity can be written as
 |
(18) |
where u(n) and wT(n) are the assembly and disassembly rates of oligomeric subunits. Similarly, the expression for dispersion is given by
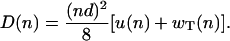 |
(19) |
At the same time, in the analysis of the experimental data (11,12) the addition or removal of single subunits has been assumed. This suggests that the rates has been measured using the expression
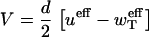 |
(20) |
Comparing this equation with Eq. 18, it yields the relation between the effective rates ueff and 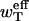 per monomer and the actual rates u(n) and wT(n) per oligomer,
per monomer and the actual rates u(n) and wT(n) per oligomer,
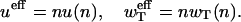 |
(21) |
The substitution of these effective rates into the expression for dispersion (19) with n = 1 produces
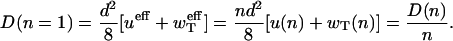 |
(22) |
This means that dispersion calculated by assuming monomer association/dissociation underestimates the true dispersion by a factor of n rather than by a factor of n2 as suggested previously (11,12).
The experimental results (11,12) suggest that only the addition or dissociation of oligomers with n = 35–40 can explain the large length fluctuation in the single actin filaments if one accepts the association/dissociation of oligomers. These particles are quite large by size (≃100 nm), and, if present in the system, they would be easily observed in the experiments. However, no detectable amounts of large oligomers have been found in studies of kinetics of the actin polymerization. It has been reported (37) that only small oligomers (up to n = 4 – 8) may coexist with the monomers and the polymerized actin under the special, not physiological, solution conditions. Therefore, it is very unlikely that the presence of very small (if any) amounts of such oligomers might influence the dynamics and large length fluctuations in the actin growth.
SUMMARY AND CONCLUSIONS
The growth dynamics of single actin filaments is investigated theoretically using the stochastic model that takes into account the dynamics at both ends of filament, the structure of the polymer's tip, lateral interactions between the protofilaments, the hydrolysis of ATP bounded to the actin subunit, and assembly and disassembly of hydrolyzed and unhydrolyzed actin monomers. It is assumed that sequential (vectorial) mechanism of hydrolysis controls the transformation of ATP-actin subunits. Using the analytical approach, exact expressions for the mean growth velocity, the length dispersion, and the mean size of fluctuating ATP-cap are obtained in terms of the kinetic rate constants that describe the assembly and disassembly events, and the hydrolysis of nucleotides. It is shown that there are two regimes of single actin filament's growth. At high concentrations the size of the ATP-cap is very large and the fully hydrolyzed core is never exposed at filament's tip. As a result, the disassembly of ADP-actin subunits does not contribute to the overall dynamics. The situation is different at low concentrations, where the size of ATP-cap is always finite. Here the dissociation of both hydrolyzed and unhydrolyzed actin monomers is critical for the growth dynamics of filaments. The boundary between two regimes is defined by the transition point, which depends on the association/dissociation rate constants for the ATP-actin monomers and on the hydrolysis rate. For any nonzero rate of hydrolysis the transition point is always above the critical concentration where the mean growth velocity becomes equal to zero. The most remarkable result of our theoretical analysis is the nonmonotonic behavior of dispersion as the function of concentration and large length fluctuations near the critical concentration. These large fluctuations are explained by alternation between the relatively slow assembly/disassembly of ATP-actin subunits and the rapid dissociation of ADP-actin monomers.
For the experimentally determined kinetic rates our theoretical analysis suggests that the contribution of the dynamics at the pointed end of the actin filament is very small. The treadmilling concentration for the system, as well as the transition point for the barbed end, are only slightly above the critical concentration for the barbed end. At this transition point the dispersion of length reaches a maximal value, and it is smaller for larger concentrations. Our theoretical predictions are in excellent agreement with all available experimental observations on the dynamics of single actin filaments. However, more measurements of dispersion and other dynamic properties at different monomeric concentrations are needed to check the validity of the presented theoretical method. The predictions of our model can be checked by measuring the dynamic properties of single actin filaments at different monomer concentrations.
Since the exact mechanism of ATP hydrolysis in actin is not known, we discussed and compared the random and the vectorial mechanisms for the simplified effective single-stranded model of actin growth. It was shown that the mean growth velocity, the fraction of the configurations with unhydrolyzed cap and the size of ATP-cap are qualitatively and quantitatively similar at low and at high concentrations, although there are deviations near the transition point. It was suggested then that the length fluctuations in the random mechanism, like in the vectorial mechanism, might also exhibit a peak near the critical concentration. The fact that our model is able to explain the peak in the length fluctuations and its position is mainly due to the assumption of the vectorial mechanism of hydrolysis that has not been accounted for in the previous theoretical models. This strongly suggests a significant contribution from the vectorial mechanism in the overall hydrolysis process. Future experimental measurements of dynamic properties at different concentrations might help to distinguish between the hydrolysis mechanisms in the actin growth.
The possibility of attachment and detachment of large oligomers in the single actin filaments has been also discussed. It was argued that the experimental observations of large length fluctuations can only be explained by addition of oligomers consisting of 35–40 monomers. However, these oligomers have a large size and such events have not been observed or have been excluded from the analysis of the experiments. Thus the effect of the oligomer assembly/disassembly on the single actin filament's growth is probably negligible.
Although the effect of ATP hydrolysis on polymerization dynamics of actin filaments has been studied before (21,36,38,39), to the best of our knowledge, the present work is the first that provides rigorous calculations of the mean growth velocity, dispersion, the size of ATP-cap, and the fraction of capped configurations simultaneously. It is reasonable to suggest that this method might be used to investigate the dynamic instability in microtubules because the polymer can be viewed as growing in two dynamic phases. In one phase the ATP-cap is always present at the end of the filament, while in the second phase it is absent. A similar approach to investigate the dynamic phase changes has been proposed earlier (38). The model can also be improved by considering the intermediate states of hydrolysis and the release of inorganic phosphate (3,21), and the possible exchange of nucleotide at the terminal subunit of the barbed end of the actin filaments (34).
Acknowledgments
The authors are grateful to M.E. Fisher for valuable comments and discussions.
The authors acknowledge support from the Welch Foundation (grant No. C-1559), the Alfred P. Sloan foundation (grant No. BR-4418), and the U.S. National Science Foundation (grant No. CHE-0237105).
APPENDIX: ONE-LAYER POLYMERIZATION MODEL WITH SEQUENTIAL ATP HYDROLYSIS FOR TWO-STRANDED POLYMERS
Let us define a function P(l1, k1;l2, k2;t) as the probability of finding the two-stranded polymer in the configuration (l1, k1;l2, k2). Here li, ki = 0, 1, … (ki ≤ li, i = 1 or 2) are two independent parameters that count the total number of subunits (li) and the number of unhydrolyzed subunits (ki) in the ith protofilament. We assume that the polymerization and hydrolysis in the actin filament can be described by one-layer approach (20,22). It means that l2 = l1 or l2 = l1 + 1, and k2 = k1 or k2 = k1 ± 1 (see Fig. 1). Then the probabilities can be described by a set of master equations. For configurations with l1 = l2 = l and 1 ≤ k < l we have
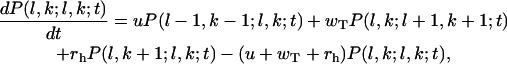 |
(A1) |
and
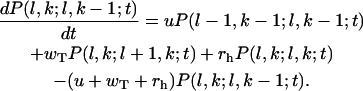 |
(A2) |
Similarly for the configurations with l1 = l2 – 1 = l and 1 ≤ k < l + 1 the master equations are
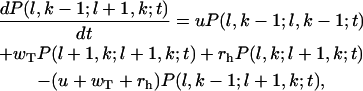 |
(A3) |
and
 |
(A4) |
Then the polymer configurations without ATP-actin monomers (k = 0) can be described by
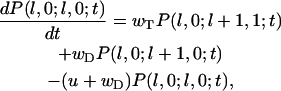 |
(A5) |
and
 |
(A6) |
Finally, for the configurations consisting of only unhydrolyzed subunits we have
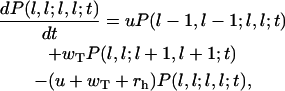 |
(A7) |
and
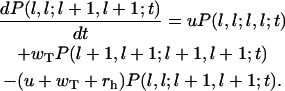 |
(A8) |
The conservation of probability leads to
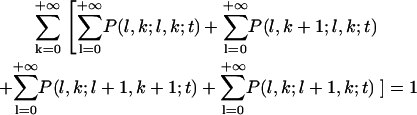 |
(A9) |
at all times.
Following the method of Derrida (40), we define two sets of auxiliary functions (k = 0, 1, …),
 |
(A10) |
and
 |
(A11) |
Note that the conservation of probability gives us
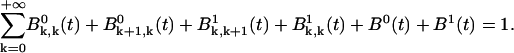 |
(A12) |
Then, from the master equations (Eqs. A1–A4), we derive for k ≥ 1
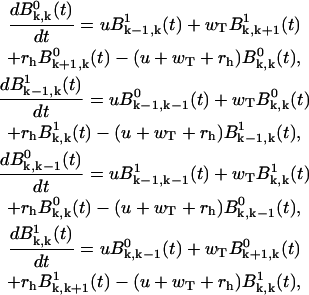 |
(A13) |
while the master equations (Eqs. A5 and A6) for k = 0 yield
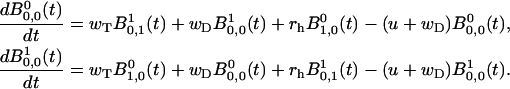 |
(A14) |
Finally, Eqs. A7 and A8 lead to
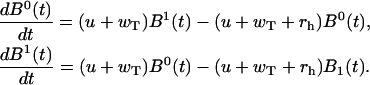 |
(A15) |
Similar arguments can be used to describe the functions  and
and 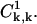 Specifically, for k ≥ 1 we obtain
Specifically, for k ≥ 1 we obtain
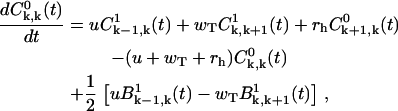 |
(A16) |
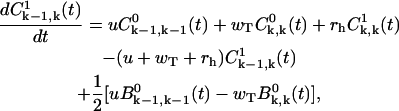 |
(A17) |
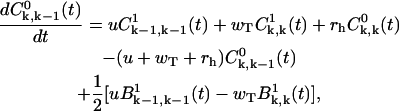 |
(A18) |
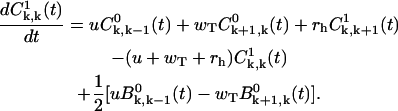 |
(A19) |
For k = 0, the expressions are
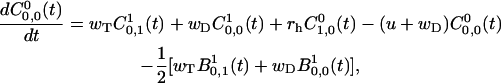 |
(A20) |
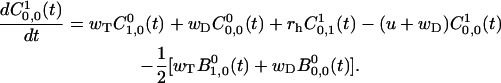 |
(A21) |
Again following the Derrida's approach (40) we introduce an ansatz that should be valid at large times t, namely,
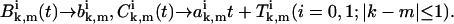 |
(A22) |
At steady-state 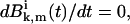 and Eqs. A13 and A14 yield for k ≥ 1,
and Eqs. A13 and A14 yield for k ≥ 1,
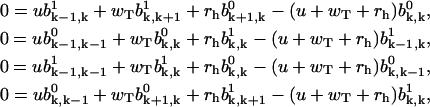 |
(A23) |
while for k = 0 we obtain
 |
(A24) |
Finally, from Eq. A15 we have
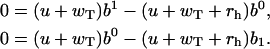 |
(A25) |
Due to the symmetry of the system we can conclude that the probabilities 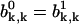 and
and 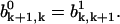 Then the solutions of Eqs. A23 and A24 can be written in the form
Then the solutions of Eqs. A23 and A24 can be written in the form
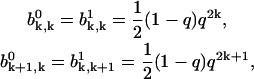 |
(A26) |
where k = 0, 1, .., and
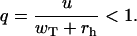 |
(A27) |
In addition, the expressions in Eq. A25 have only a trivial solution b0 = b1 = 0. Recall that b0 and b1 give the stationary-state probabilities of the polymer configurations with all subunits in ATP state, i.e., it corresponds to the case of very large k. The solution agrees with the results for 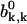 and
and 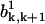 at k → ∞ (see the expressions in Eq. A26).
at k → ∞ (see the expressions in Eq. A26).
For q > 1, the systems of equations in Eqs. A23 and A24 have the trivial solutions, with all 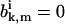 for finite k and m. It means that at the stationary conditions the polymer can only exist in the configurations with very large number of unhydrolyzed subunits and the size of ATP-cap is infinite, whereas for q < 1 the size of ATP-cap is always finite. The case q = 1 is a boundary between two regimes. At this condition there is a qualitative change in the dynamic properties of the system.
for finite k and m. It means that at the stationary conditions the polymer can only exist in the configurations with very large number of unhydrolyzed subunits and the size of ATP-cap is infinite, whereas for q < 1 the size of ATP-cap is always finite. The case q = 1 is a boundary between two regimes. At this condition there is a qualitative change in the dynamic properties of the system.
To determine the coefficients 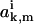 and
and 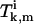 from Eq. A22, the ansatz for the functions
from Eq. A22, the ansatz for the functions 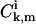 is substituted into the asymptotic expressions (Eqs. A16–A21), yielding for k ≥ 1,
is substituted into the asymptotic expressions (Eqs. A16–A21), yielding for k ≥ 1,
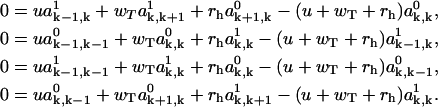 |
(A28) |
At the same time, for k = 0 we obtain
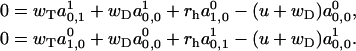 |
(A29) |
The coefficients 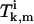 satisfy the following equations (for k ≥ 1),
satisfy the following equations (for k ≥ 1),
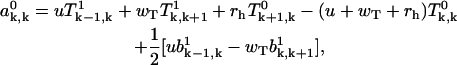 |
(A30) |
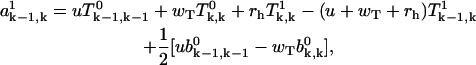 |
(A31) |
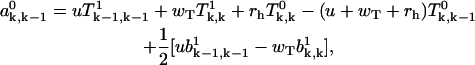 |
(A32) |
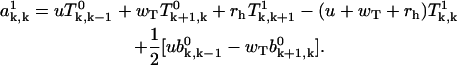 |
(A33) |
For k = 0 the expressions are given by
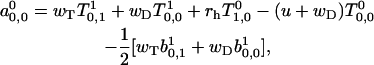 |
(A34) |
 |
(A35) |
Comparing Eqs. A23 and A24 with Eqs. A28 and A29, we conclude that
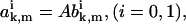 |
(A36) |
with the constant A. This constant can be calculated by summing over the left and right sides in Eq. A36 and recalling the normalization condition (Eq. A12). The summation over all ak, i in Eqs. A28 and A29 produces
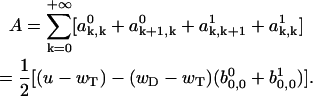 |
(A37) |
To determine the coefficients 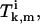 we need to solve Eqs. A30–A35. Again, due to the symmetry, we have
we need to solve Eqs. A30–A35. Again, due to the symmetry, we have 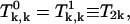 and
and 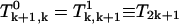 for all k. The solutions for these equations are given by
for all k. The solutions for these equations are given by
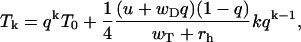 |
(A38) |
where k = 0, 1, … and T0 is an arbitrary constant.
It is now possible to calculate explicitly the mean growth velocity, V, and dispersion, D, at steady-state conditions. The average length of the polymer is given by
 |
(A39) |
Then, using Eq. A36, we obtain for the velocity
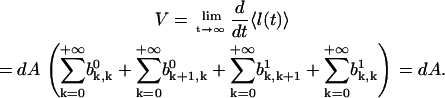 |
(A40) |
A similar approach can be used to derive the expression for dispersion. We start from
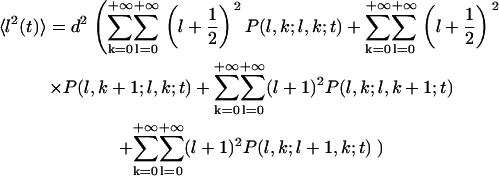 |
(A41) |
Then, using the master equations (Eqs. A1–A6), it can be shown that
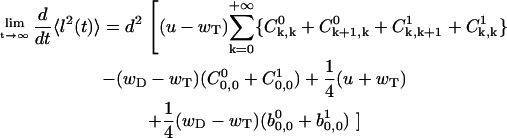 |
(A42) |
Also, the following equation can be derived using Eq. A39,
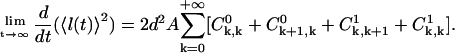 |
(A43) |
The formal expression for dispersion is given by
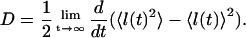 |
(A44) |
Then, substituting into this expression Eqs. A42 and A43, we obtain
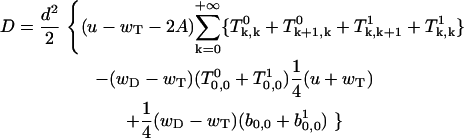 |
(A45) |
Note that 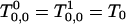 and for sum of all Tk, m we have from Eq. 60
and for sum of all Tk, m we have from Eq. 60
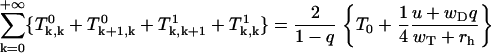 |
(A46) |
Finally, after some algebraic transformations of Eqs. A37 and A45, we derive the final expression for the growth velocity, V and dispersion, D, which are given in Eqs. 1 and 2 in Model of Actin Filament Assembly. Note that the constant T0 cancels out in the final equation.
The mean size of ATP-cap can be calculated as
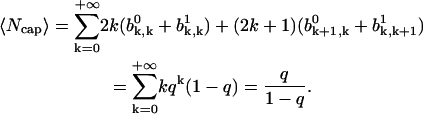 |
(A47) |
The average relative fluctuation in the size of the ATP-cap, by definition, is given
 |
(A48) |
where
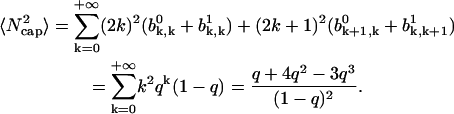 |
(A49) |
Then from Eqs. A49 and A47 we have
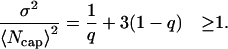 |
(A50) |
References
- 1.Howard, J. 2001. Mechanics of Motor Proteins and Cytoskeleton. Sinauer Associates, Sunderland, MA.
- 2.Bray, D. 2001. Cell Movements. From Molecules to Motility. Garland Publishing, New York.
- 3.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 4.Erickson, H. P., and E. T. O'Brien. 1992. Microtubule dynamic instability and GTP hydrolysis. Annu. Rev. Biophys. Biomol. Struct. 21:145–166. [DOI] [PubMed] [Google Scholar]
- 5.Desai, A., and T. J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- 6.Dogterom, M., and B. Yurke. 1997. Measurement of the force-velocity relation for growing microtubules. Science. 278:856–860. [DOI] [PubMed] [Google Scholar]
- 7.Shaw, S. L., R. Kamyar, and D. W. Ehrhardt. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 300:1715–1718. [DOI] [PubMed] [Google Scholar]
- 8.Kerssemakers, J. W. L., M. E. Janson, A. van der Horst, and M. Dogterom. 2003. Optical trap setup for measuring microtubule pushing forces. Appl. Phys. Lett. 83:4441–4443. [Google Scholar]
- 9.Janson, M. E., and M. Dogterom. 2004. Scaling of microtubule force-velocity curves obtained at different tubulin concentrations. Phys. Rev. Lett. 92:248101. [DOI] [PubMed] [Google Scholar]
- 10.Lehto, T., M. Miaczynska, M. Zerial, D. J. Muller, and F. Severin. 2003. Observing the growth of individual actin filaments in cell extracts by time-lapse atomic force microscopy. FEBS Lett. 551:25–28. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara, I., S. Takahashi, H. Tadakuma, T. Funatsu, and S. Ishiwata. 2002. Microscopic analysis of polymerization dynamics with individual actin filaments. Nat. Cell Biol. 4:666–673. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn, J. R., and T. D. Pollard. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88:1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotani, H., and T. Horio. 1988. Dynamics of microtubules visualized by dark-field microscopy: treadmilling and dynamic instability. Cell Motil. Cyroskel. 10:229–236. [DOI] [PubMed] [Google Scholar]
- 14.Grego, S., V. Cantillana, and E. D. Salmon. 2001. Microtubule treadmilling in vitro investigated by fluorescence speckle and confocal microscopy. Biophys. J. 81:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchison, T. J. 1992. Compare and contrast actin-filaments and microtubules. J. Mol. Biol. Cell. 3:1309–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, E. C., C. S. Campbell, and R. D. Mullins. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 306:1021–1025. [DOI] [PubMed] [Google Scholar]
- 17.De La Cruz, E. M., A. Mandinova, M. O. Steinmetz, D. Stoffler, U. Aebi, and T. D. Pollard. 2000. Polymerization and structure of nucleotide-free actin filaments. J. Mol. Biol. 295:517–526. [DOI] [PubMed] [Google Scholar]
- 18.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in non-muscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- 19.Blanchoin, L. A., T. D. Pollard, and R. D. Mullins. 2000. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10:1273–1282. [DOI] [PubMed] [Google Scholar]
- 20.Stukalin, E. B., and A. B. Kolomeisky. 2005. Polymerization dynamics of double-stranded biopolymers: chemical kinetic approach. J. Chem. Phys. 122:104903. [DOI] [PubMed] [Google Scholar]
- 21.Vavylonis, D., Q. Yang, and B. O'Shaughnessy. 2005. Actin polymerization kinetics, cap structure and fluctuations. Proc. Natl. Acad. Sci. USA. 102:8543–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stukalin, E. B., and A. B. Kolomeisky. 2004. Simple growth models of rigid multifilament biopolymers. J. Chem. Phys. 121:1097–1104. [DOI] [PubMed] [Google Scholar]
- 23.Pollard, T. D. 1986. Rate constants for the reaction of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohm, T., and A. Wegner. 1994. Mechanism of ATP hydrolysis by polymeric actin. Biochim. Biophys. Acta. 1208:8–14. [DOI] [PubMed] [Google Scholar]
- 25.Pieper, U., and A. Wegner. 1996. The end of a polymerizing actin filament contains numerous ATP-subunit segments that are disconnected by ADP-subunits resulting from ATP hydrolysis. Biochemistry. 35:4396–4402. [DOI] [PubMed] [Google Scholar]
- 26.Blanchoin, L., and T. D. Pollard. 2002. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 41:597–602. [DOI] [PubMed] [Google Scholar]
- 27.Carlier, M. F., D. Pantaloni, and E. D. Korn. 1987. The mechanism of ATP hydrolysis accompanying the polymerization of Mg-actin and Ca-actin. J. Biol. Chem. 262:3052–3059. [PubMed] [Google Scholar]
- 28.Korn, E. D., M. F. Carlie, and D. Pantaloni. 1987. Actin polymerization and ATP hydrolysis. Science. 238:638–644. [DOI] [PubMed] [Google Scholar]
- 29.Pantaloni, D., T. L. Hill, M.-F. Carlier, and E. D. Korn. 1985. A model for actin polymerization and the kinetic effects of ATP hydrolysis. Proc. Natl. Acad. Sci. USA. 82:7207–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melki, R., S. Fievez, and M. F. Carlier. 1996. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry. 35:12038–12045. [DOI] [PubMed] [Google Scholar]
- 31.Carlier, M. F., and D. Pantaloni. 1986. Direct evidence fro ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 25:7789–7792. [DOI] [PubMed] [Google Scholar]
- 32.Carlier, M. F., D. Pantaloni, and E. D. Korn. 1986. The effects of Mg2+ at the high-affinity and low-affinity sites on the polymerization of actin and associated ATP hydrolysis. J. Biol. Chem. 261:10785–10792. [PubMed] [Google Scholar]
- 33.Bindschadler, M., E. A. Osborn, C. F. Dewey, Jr., and J. L. McGrath. 2004. A mechanistic model of the actin cycle. Biophys. J. 86:2720–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teubner, A., and A. Wegner. 1998. Kinetic evidence for a readily exchangeable nucleotide at the terminal subunit of the barbed ends of actin filaments. Biochemistry. 37:7532–7538. [DOI] [PubMed] [Google Scholar]
- 35.Dufort, P. A., and C. J. Lumsden. 1993. High microfilament concentration results in barbed-end ADP caps. Biophys. J. 65:1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keiser, T., A. Schiller, and A. Wegner. 1986. Nonlinear increase of elongation rate of actin filaments with actin monomer concentration. Biochemistry. 25:4899–4906. [DOI] [PubMed] [Google Scholar]
- 37.Attri, A. K., M. S. Lewis, and E. D. Korn. 1991. The formation of actin oligomers studied by analytical ultracentrifugation. J. Biol. Chem. 266:6815–6824. [PubMed] [Google Scholar]
- 38.Hill, T. L. 1986. Theoretical study of a model for the ATP cap at the end of an actin filament. Biophys. J. 49:981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill, T. L. 1987. Linear Aggregation Theory in Cell Biology. Springer-Verlag, New York.
- 40.Derrida, B. 1983. Velocity and diffusion constant of a periodic one-dimensional hopping model. J. Stat. Phys. 31:433–450. [Google Scholar]
- 41.Erickson, H. P. 1989. Cooperativity in protein-protein association—the structure and stability of the actin filament. J. Mol. Biol. 206:465–474. [DOI] [PubMed] [Google Scholar]
- 42.Kinosian, H., J. Selden, L. C. Gershman, and J. E. Estes. 2002. Actin filament barbed end elongation with non-muscle MgATP-actin and MgADP-actin in the presence of profilin. Biochemistry. 41:6734–6743. [DOI] [PubMed] [Google Scholar]