Abstract
NOTCH1 is frequently mutated in human precursor T-cell lymphoblastic leukemia/lymphoma (pre-T LBL). In the current study, we found that 13 of 19 cell lines and 29 of 49 primary tumors from SCL/LMO1, OLIG2/LMO1, OLIG2, LMO1, NUP98/HOXD13, and p27-/-/SMAD3+/- mice had Notch1 mutations in either the heterodimerization (HD) or the glutamic acid/serine/threonine (PEST) domain but not both. Thymocytes from clinically healthy SCL/LMO1 mice aged 5 weeks did not have Notch1 mutations, whereas thymocytes from clinically healthy SCL/LMO1 mice aged 8 to 12 weeks did have Notch1 mutations and formed tumors upon transplantation into nude mice. Remarkably, all of the HD domain mutations that we identified were single-base substitutions, whereas all of the PEST domain mutations were insertions or deletions, half of which mapped to 1 of 2 mutational “hot spots.” Taken together, these findings indicate that Notch1 mutations are very frequent events that are acquired relatively early in the process of leukemic transformation and are important for leukemic cell growth. (Blood. 2006;107: 2540-2543)
Introduction
NOTCH1 has been implicated in the pathogenesis of precursor T-cell lymphoblastic leukemia/lymphoma (pre-T LBL).1-5 It has recently been shown that over 50% of human pre-T LBL samples that did not have chromosomal aberrations involving NOTCH1 had activating mutations in the heterodimerization (HD) and/or the PEST domain of NOTCH1.6 We searched for activating Notch1 mutations in pre-T LBL from SCL/LMO1,7 OLIG2, OLIG2/LMO1,8 LMO1,9 and NUP98/HOXD13 (NHD13) transgenic mice10 and p27-/-/SMAD3-/+ mice.11 We chose to focus our studies on SCL/LMO1 mice, since SCL is frequently activated by chromosomal rearrangement in human pre-T LBL.12 To investigate the timing of Notch1 mutation, we studied thymocytes harvested from clinically healthy SCL/LMO1 mice aged 4 to 12 weeks.
Study design
Cell lines
Cell lines were established as previously described.8 F4-6 is a Friend virus-induced erythroleukemia cell line.13 Cell lines were seeded at a concentration of 1 × 104 cells/mL and treated with 5 μM of Z-IL-CHO (γ-secretase inhibitor XII; GSI) or DMSO for 72 hours and evaluated by trypan blue exclusion. Rescue of GSI treatment was performed on a SCL/LMO1 cell line stably transfected with a human ICN1 (intracellular NOTCH1) vector.14 Notch1 protein expression was determined by Western blot using an anti-Notch1 monoclonal antibody (mN1A; Santa Cruz Biotechnology, Santa Cruz, CA). Statistical significance was determined using a Student t test.
Sequence analysis
The HD and PEST domains of Notch1 were amplified with exon 26 (i25F1: 5′-GGCTGAGTTTCTTTAGAGTC-3′, and i26R1: 5′-CCTCCCCTGAGGTTACACCT-3′), exon 27 (i26F1: 5′-GAGTGTCCCATTGCGGGGCT-3′, and i27R1: 5′-TGCAGAGGTCAGAAAGTGTT-3′), and PEST domain primers (PEST2: 5′-GCCTCTGGAATGTGGGTGAT-3′, and PEST1: 5′-TACCAGGGCCTGCCCAACAC-3′), respectively. The polymerase chain reaction (PCR) was performed using the same protocol as previously described.15 PCR products were gel purified and the sequence was compared with wild-type Notch1 (GenBank no. AL732541.11).
Transplantation of thymocytes
Thymocytes (5 × 106) from SCL/LMO1 transgenic mice aged 5 to 12 weeks were injected into nude mice intraperitoneally. Mice were killed when signs of illness developed.16,17 The treatment protocol was approved by the Animal Use Committee at both Roswell Park Cancer Institute and the National Cancer Institute.
Southern blot analysis for Tcrb gene rearrangements
Genomic DNA was digested with SstI, size fractionated, transferred to nitrocellulose, and hybridized to a 32P-labeled 0.4 kb Tcrb probe.17
Results and discussion
Notch1 mutations in murine pre-T LBL
We studied 19 pre-T LBL cell lines (13 SCL/LMO1, 3 OLIG2/LMO1, 1 LMO1, and 2 NHD13; Table 1). All mutations were heterozygous except for 2 cell lines (6605/4 and 6781/3) that lost the wild-type Notch1 allele. Notch1 mutations were found in 11 (85%) of 13 of the SCL/LMO1 cell lines. Of these, 10 had mutations in the PEST domain, all insertions or deletions, whereas only 1 showed a mutation in the HD domain. The mutations in the OLIG2/LMO1 and LMO1 cell lines were both insertion mutations in the PEST domain that resulted in premature stop codons.
Table 1.
Notch1 mutations in murine pre-T LBL cell lines
| Strain and mouse | HD | PEST |
|---|---|---|
| SCL-LMO1 | ||
| 2382 | None | 66666CTGAG>GAG |
| 3781A | None | 66279C>CT |
| 3917 | None | None |
| 6472/3 | None | Del 66449-61 (13 bp) |
| 6600/1 | None | None |
| 6605/4 | None | Dup of 66408-67 (60 bp) hom |
| 6645/4 | None | 66391C>CATATAGGG |
| 6775/1 | None | 66383TGTGA>GGGAGGAGGGGTG |
| 6781/3 | 59724T>C (L→P) hom | None |
| 6812/2 | None | 66280G>AAAAA |
| 6913/3 | None | 66278A>AG |
| 7114/1 | None | 66392GGCAG>CCC |
| 7298/2 | None | 66279C>GCCCC |
| OLIG2-LMO1 | ||
| 1928 | None | None |
| 1931 | None | None |
| 1939 | None | 66508AGG>AGGG |
| LMO1 | ||
| 12/1 | None | 66408C>CCCAGCCAATGGGCACC |
| NUP98-HOXD13 | ||
| 1075 | None | None |
| 1901 | None | None |
Mutations were heterozygous unless otherwise noted. For SCL-LMO1, n = 13; for OLIG2-LMO1, n = 3; for LMO1, n = 1; and for NUP98-HOXD13, n = 2.
None indicates no mutation; Del, deletion; Dup, duplication; and hom, homozygous.
Notch1 is cleaved to the oncogenic form (intracellular Notch1 or ICN1) by γ-secretase. Some human pre-T LBL cell lines that harbor NOTCH1 mutations are sensitive to GSI,6 suggesting that ongoing signals from the ICN1 are required to maintain the malignancy. Similar to findings with human pre-T LBL cell lines,6 we found that some of the murine pre-T LBL cell lines with Notch1 mutations were sensitive to GSI treatment (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). SCL/LMO1 cell line 3781 showed a decrease in ICN1 expression and growth inhibition following GSI treatment (Figure S1A). Stable transfectants (ICN2 and ICN4) of the SCL/LMO1 cell line 6812/2 that expressed human ICN1 (hICN1) were no longer sensitive to GSI, whereas the transfectants that either did not express hICN1 or carried the empty vector remained sensitive to the GSI (Figure S1B-C). Taken together, these findings suggest that Notch1 was functionally activated by Notch1 mutations.
We expanded the survey to 49 pre-T LBL thymic tumors from several lines of genetically modified mice (Table 2). Those primary tumors included 17 tumors from which we established cell lines. The mutations in the cell lines were identical to those detected in the primary tumors with 3 exceptions. Although the primary tumor from mouse 6600/1 had a PEST domain mutation, the cell line derived from this mouse did not have this mutation. The primary tumor from mouse 6605/4 showed both an insertion of AG and a direct repeat insertion of 60 bp at different regions of the PEST domain, whereas the cell line had only the direct repeat insertion and lost the wild-type allele. The mutation in 6781/3 was heterozygous in the primary tumor, whereas the cell line lost the wild-type allele. In total, Notch1 mutations were found in 22 of 28 SCL/LMO1, of 2 OLIG2, 3 of 6 OLIG2/LMO1, 1 of 2 LMO1, 3 of 9 NHD13, and 2 of 4 p27-/-/SMAD3+/- primary tumors or cell lines. Of the 32 mutations that were identified, only 6 mutations (all missense) were found in the HD domain. Three mutations (6781/3, 262, 8013) were in positions mutated in human pre-T LBL, and the remaining 3 were at positions conserved in human, mouse, chicken, frog, and fish.6 All 26 mutations within the PEST domain involved nucleotide insertions and/or deletions, as opposed to single-base substitutions. Interestingly, 13 independent mutations were found at nucleotide numbers 66278-80 or 66391-92, indicating that these positions might represent “hot spots” for insertion/deletion mutations.
Table 2.
Notch1 mutations in murine primary pre-T LBL tumors
| Strain and mouse | Mutation |
|---|---|
| SCL/LMO1 | |
| 2383 | 66494GC>G (PEST) |
| 2853 | 66391CGG>CCCG (PEST) |
| 3781 | 66279C>CT (PEST) |
| 3911 | 66280G>TC (PEST) |
| 3917 | None |
| 6180/1 | 66571CCCC>CCCCC (PEST) |
| 6385/4 | None |
| 6472/3 | Del 66449-61 (PEST) |
| 6600/1 | 66391C>CAAAA (PEST) |
| 6603/5 | None |
| 6605/4 | 66391C>CAG (PEST)/Dup of 66408-67 (60 bp) (PEST) |
| 6645/4 | 66391C>CATATAGGG (PEST) |
| 6775/1 | 66383TGTGA>GGGAGGAGGGGTG (PEST) |
| 6781/2 | None |
| 6781/3 | 59724T>C (L→P) (HD) |
| 6782/2 | 66280G>CC (PEST) |
| 6812/1 | 66279C>CAAAA (PEST) |
| 6812/2 | 66280G>AAAAA (PEST) |
| 6846/2 | 66567CTG>CTGTG (PEST) hom |
| 6858/4 | 66533C>CATCC (PEST) |
| 6860/1 | None |
| 6990/3 | Del 66451-61 (PEST) |
| 7114/1 | 66392GGCAG>CCC (PEST) |
| 7114/3 | 66392G>TGCCCCCCT (PEST) |
| 7277/3 | None |
| 7298/2 | 66279C>GCCCC (PEST) |
| OLIG2 | |
| 556 | None |
| 1115 | None |
| OLIG2/LMO1* | |
| 259 | 58374T>C (W→R) (HD) |
| 262 | 58462T>A (L→Q) (HD) |
| 1919 | None |
| 1928 | None |
| 1931 | None |
| 1939 | 66508AGG>AGGG (PEST) |
| LMO1 | |
| 255 | None |
| 12/1 | 66408C>CCCAGCCAATGGGCACC (PEST) |
| NUP98/HOXD13 | |
| L7 | None |
| 1018 | 66452A>AGTCCGGGCCCC (PEST) |
| 1075 | None |
| 1901 | None |
| 2423 | None |
| 2553 | None |
| 2738 | 66328G>GT (PEST) |
| 2918 | None |
| 8013 | 58429T>C (L→P) (HD) |
| p27−/−, SMAD3−/+ | |
| 137 | None |
| 198 | None† |
| 27-1 | None†, 59790C>G (A→G) (HD) |
| 32-6 | 59844T>G (I→S) (HD) |
Mutations were heterozygous unless otherwise noted. For SCL/LMO1, n = 26; for OLIG2, n = 2; for OLIG2/LMO1, n = 6; for LMO1, n = 2; for NUP98/HOXD13, n = 9; and for p27−/−, SMAD3−/+, n = 4.
Del indicates deletion; Dup, duplication; hom, homozygous; none, no mutation.
Previously reported (Lin et al8).
Polymorphism at 58623G>A in the HD.
Notch1 mutations in clinically healthy SCL/LMO1 mice
Since SCL/LMO1 mice do not have Notch1 mutations in the germ line, these must be acquired mutations. To investigate the timing of Notch1 mutations in SCL/LMO1 mice, we examined thymocytes from clinically healthy SCL/LMO1 mice aged 4 to 12 weeks. We previously demonstrated16-18 that thymocytes from 4- to 5-week-old SCL/LMO1 mice had decreased total thymocytes, an increased proportion of CD4-CD8- cells, an increased apoptotic index, and oligoclonal Tcrb gene rearrangements. Despite oligoclonal Tcrb gene rearrangements, suggesting the emergence of one or more clonal populations of thymocytes, these thymocytes are not fully transformed, since they do not form tumors when transplanted into immunodeficient mice.17 In contrast, thymocytes harvested from clinically healthy SCL/LMO1 mice aged 8 to 12 weeks show similar findings in terms of increased CD4-/CD8- cells, apoptosis, and clonality, but these thymocytes are tumorigenic when transplanted into immunodeficient mice. None of 3 thymocyte samples from 4- to 5-week-old SCL/LMO1 mice had Notch1 mutations. In contrast, we found PEST domain frameshift mutations in 4 of 4 thymocyte samples analyzed from clinically healthy SCL/LMO1 mice aged 8 to 12 weeks. Moreover, these thymocytes generated tumors in immunodeficient mice that showed Notch1 mutations identical to those detected in the preleukemic thymocytes (Table 3).
Table 3.
Notch1 mutations and tumorigenicity in preleukemic thymocytes
|
|
|
|
|
Nude mice
|
||
|---|---|---|---|---|---|---|
| Mouse | Age, wk | Notch1 mutation | Injection into nude mice | Tumor | Tcrb rgmt | Notch1 mutation |
| 7162/3 | 5 | None | − | NA | NA | NA |
| 7543/3 | 5 | None | + | − | − | NA |
| 7544/2 | 5 | None | + | − | − | NA |
| 7286/1 | 8 | 66601CCCC>CCCCC | + | + | + | 66601CCCC>CCCCC |
| 7193/3 | 10 | 66569GACCCC>CCATGACTCCT | − | NA | NA | NA |
| 7151/4 | 12 | 66419C>GG | + | + | + | 66419C>GG |
| 7150/3 | 12 | 66456C>CC | + | + | + | 66456C>CC |
Thymocytes from young, clinically healthy SCL/LMO1 mice were transplanted into nude mice. Thymocytes from mice 8 weeks and older had Notch1 mutations and formed tumors in nude mice. Notch1 mutations are insertion/deletions; the numbers refer to the Notch1 reference sequence (GenBank no. AL732541.11).
Tcrb rgmt indicates clonal Tcrb rearrangement by Southern blot analysis; and NA, not applicable.
Five oligoclonal Tcrb gene rearrangements, corresponding to 3 to 5 independent clones, were detected in the preleukemic thymocytes from mouse 7151/4; 2 of these Tcrb rearrangements were found in the tumor that arose following transplantation into immunodeficient mice (Figure 1A), indicating that one of clones from the preleukemic sample had become fully transformed. The electropherogram from mouse 7151/4 thymocytes revealed low amplitude peaks, suggesting that a minority of the sample had a PEST domain frameshift mutation identical to that seen in the tumor that developed following transplantation. This finding was not unexpected, since the Southern blot suggested that the clone that emerged following transplantation represented less than 20% of the total signal. Since the mutation disrupted an AluI site in the PEST domain, we digested the PCR products with AluI. The mutated allele was barely detectable in 7151/4, consistent with the Southern blot and electropherogram data, whereas the tumor in N5/2 showed primarily mutated allele (the tumor has also lost the wild-type Notch1 allele and the small amount of wild-type PCR product is derived from contaminating normal stroma; Figure 1B). Therefore, of the 3 to 5 different clones present in the preleukemic sample, the clone with a Notch1 mutation was able to expand and form a lethal, fully malignant tumor in immunodeficient mice.
Figure 1.
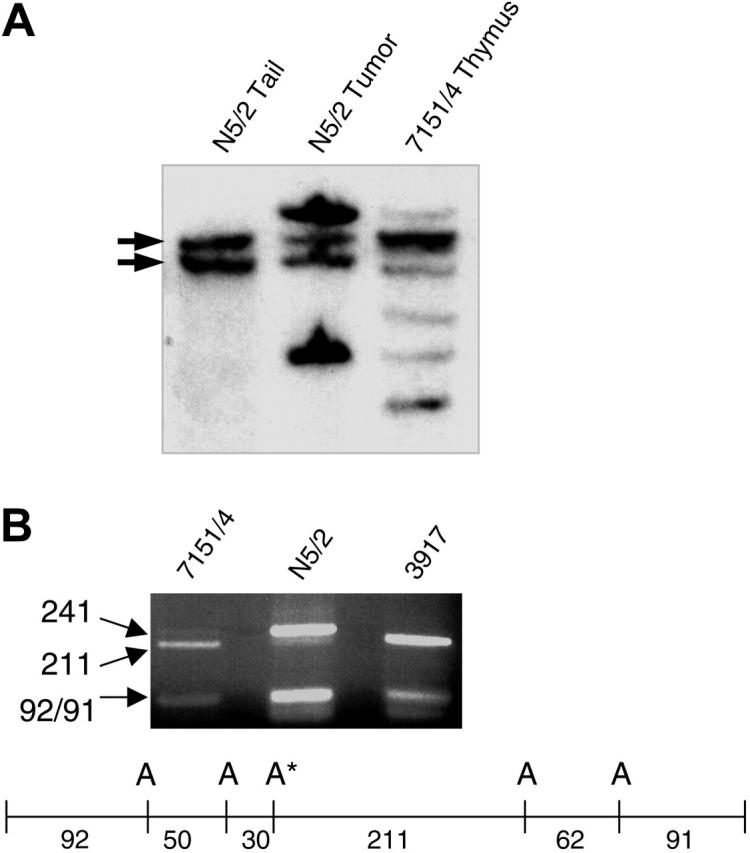
Notch1 mutations in preleukemic thymocytes. (A) Tcrb gene rearrangements in thymocytes from mouse 7151/4 and tumor resulting after transplantation into nude mouse (N5/2). Several oligoclonal rearrangements are evident in the thymocytes, whereas only 2 rearranged bands are seen in the tumor sample. Germ line bands are indicated with arrows. (B) An AluI site in the PEST domain was disrupted by the Notch1 mutation in the N5/2 sample. In 7151/4, the majority of the PCR product was wild type (211 bp), but a faint band (241 bp) indicated a small amount of the mutated allele. Sample 3917 is an SCL/LMO1 tumor that is wild type for the PEST domain used as a control. Restriction map for PEST domain PCR product. A indicates AluI sites; A*, AluI site disrupted by Notch1 mutation. Size of fragments (bp) is indicated below the map.
In contrast to human pre-T LBL, none of the 36 independent samples with Notch1 mutations had mutations in both the HD and PEST domains. In addition, all (30/30) of the PEST domain mutations were insertion or deletion mutations. In contrast, all of the 6 HD mutations were single-base substitutions. Of note, insertion/deletion mutations are characteristic of mutations seen in “stressed” cells19-21 and the SCL/LMO1 thymocytes can be regarded as stressed, given that they have increased apoptosis and proliferation compared with control thymocytes.17,22
We have shown that Notch1 mutations are very common in mouse pre-T LBL. In the studies of human pre-T LBL, many of the samples with NOTCH1 mutations had additional mutations, such as SCL or HOX11 gene rearrangements.6 It is unknown whether the NOTCH1 mutations or the gene rearrangements were the primary event in these samples. Since genetically engineered mice carry a primary mutation (such as SCL activation) in the germ line, the Notch1 mutations were not the initiating events but were acquired as the disease progressed in these mouse models and were not required for the oligoclonal expansion of SCL/LMO1 thymocytes. The mechanism(s) that cause Notch1 mutation in these mouse models remains unknown but might be related to increased metabolic stress caused by increased apoptosis and proliferation in the preleukemic thymocytes.
Supplementary Material
Acknowledgments
We thank our colleagues Drs Dave Chervinsky, Christopher Slape, and Zhenhua Zhang for technical assistance and significant discussion and Dr Masue Hayashi for significant discussion and editorial advice. We also thank Dr Frederic J. Kaye of NCI for his kind gift of the human ICN vector.
Prepublished online as Blood First Edition Paper, November 10, 2005; DOI 10.1182/blood-2005-07-3013.
Supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI).
Y.-W.L. designed and performed research, analyzed data, and wrote the first draft of the paper; R.A.N. performed research and analyzed data; J.J.L designed and performed research and analyzed data; and P.D.A. designed and performed research, analyzed data, and wrote the final draft of the paper.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66: 649-661. [DOI] [PubMed] [Google Scholar]
- 2.Hadland BK, Huppert SS, Kanungo J, et al. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 2004;104: 3097-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pui JC, Allman D, Xu L, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11: 299-308. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5: 247-253. [DOI] [PubMed] [Google Scholar]
- 5.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20: 7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306: 269-271. [DOI] [PubMed] [Google Scholar]
- 7.Aplan PD, Jones CA, Chervinsky DS, et al. An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with LMO1 to generate T-cell malignancies in transgenic mice. EMBO J. 1997;16: 2408-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YW, Deveney R, Barbara M, et al. Olig2 (NHLHB1), a HLH transcription factor, contributes to leukemogenesis in concert with LMO1. Cancer Res. 2005;65: 7151-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire EA, Rintoul CE, Sclar GM, Korsmeyer SJ. Thymic overexpression of Ttg-1 in transgenic mice results in T-cell acute lymphoblastic leukemia/lymphoma. Mol Cell Biol. 1992;12: 4186-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106: 287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfraim LA, Walz TM, James Z, Fernandez T, Letterio JJ. p21Cip1 and p27Kip1 act in synergy to alter the sensitivity of naive T cells to TGF-beta-mediated G1 arrest through modulation of IL-2 responsiveness. J Immunol. 2004;173: 3093-3102. [DOI] [PubMed] [Google Scholar]
- 12.Ferrando AA, Neuberg DS, Dodge RK, et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet. 2004;363: 535-536. [DOI] [PubMed] [Google Scholar]
- 13.Aplan PD, Nakahara K, Orkin SH, Kirsch IR. The SCL gene product: a positive regulator of erythroid differentiation. EMBO J. 1992;11: 4073-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33: 208-213. [DOI] [PubMed] [Google Scholar]
- 15.Hitoshi S, Alexson T, Tropepe V, et al. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16: 846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chervinsky DS, Lam DH, Zhao XF, Melman MP, Aplan PD. Development and characterization of T cell leukemia cell lines established from SCL/LMO1 double transgenic mice. Leukemia. 2001;15: 141-147. [DOI] [PubMed] [Google Scholar]
- 17.Chervinsky DS, Zhao XF, Lam DH, Ellsworth M, Gross KW, Aplan PD. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficient mice. Mol Cell Biol. 1999;19: 5025-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chervinsky DS, Lam DH, Melman MP, Gross KW, Aplan PD. scid thymocytes with TCRbeta gene rearrangements are targets for the oncogenic effect of SCL and LMO1 transgenes. Cancer Res. 2001;61: 6382-6387. [PubMed] [Google Scholar]
- 19.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34: 359-399. [DOI] [PubMed] [Google Scholar]
- 20.Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430: 679-682. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg SM, Hastings PJ. Genomes: worming into genetic instability. Nature. 2004;430: 625-626. [DOI] [PubMed] [Google Scholar]
- 22.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1: 138-144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


