Abstract
The premise of this review is that apolipoprotein (apo) E4 is much more than a contributing factor to neurodegeneration. ApoE has critical functions in redistributing lipids among CNS cells for normal lipid homeostasis, repairing injured neurons, maintaining synapto-dendritic connections, and scavenging toxins. In multiple pathways affecting neuropathology, including Alzheimer’s disease, apoE acts directly or in concert with age, head injury, oxidative stress, ischemia, inflammation, and excess amyloid β peptide production to cause neurological disorders, accelerating progression, altering prognosis, or lowering age of onset. We envision that unique structural features of apoE4 are responsible for apoE4-associated neuropathology. Although the structures of apoE2, apoE3, and apoE4 are in dynamic equilibrium, apoE4, which is detrimental in a variety of neurological disorders, is more likely to assume a pathological conformation. Importantly, apoE4 displays domain interaction (an interaction between the N- and C-terminal domains of the protein that results in a compact structure) and molten globule formation (the formation of stable, reactive intermediates with potentially pathological activities). In response to CNS stress or injury, neurons can synthesize apoE. ApoE4 uniquely undergoes neuron-specific proteolysis, resulting in bioactive toxic fragments that enter the cytosol, alter the cytoskeleton, disrupt mitochondrial energy balance, and cause cell death. Our findings suggest potential therapeutic strategies, including the use of “structure correctors” to convert apoE4 to an “apoE3-like” molecule, protease inhibitors to prevent the generation of toxic apoE4 fragments, and “mitochondrial protectors” to prevent cellular energy disruption.
Keywords: mitochondria, neurodegeneration, cytoskeleton, protein folding
Apolipoprotein (apo) E plays a fundamental role in the maintenance and repair of neurons, but its three isoforms differ in their abilities to accomplish these critical tasks (1–3). ApoE4 is associated with a wide variety of neuropathological processes. We hypothesize that different insults associated with a variety of disorders, in concert with apoE4, can lead to neuropathology. We believe that those processes are mediated by the cellular origin of apoE (astrocytes, neurons, or microglia), the nature of various injurious factors (“second hits”), and the structure of apoE4. Thus, understanding the structure and function of the apoE isoforms will yield new strategies for treating neuropathologies.
Although apoE is involved in many neuropathologies, we will focus on its role in Alzheimer’s disease (AD). We will consider the unique structural features that distinguish apoE4 from apoE3 and apoE2, the sites of synthesis and normal roles of apoE in the nervous system, and the pathological roles of apoE4 with or without amyloid β (Aβ) peptide. The evidence suggests that apoE4 is considerably more than a simple contributing factor in AD pathogenesis.
ApoE and Neuropathology
ApoE4’s involvement in neuropathology is well documented (4, 5) and is the major known genetic risk factor for AD in many populations (6–12). In some, 40–80% of patients with AD possess at least one apoE4 allele (12). Likewise, apoE4 is associated with earlier onset, progression, or severity of head trauma (13–19), stroke (20, 21), complications after coronary artery bypass surgery (22, 23), Parkinson’s disease (24–27), amyotrophic lateral sclerosis (28–32), multiple sclerosis (33, 34), diabetic neuropathy (35), sleep apnea (36), Lewy body disorders (37), and CNS ischemia (38).
Throughout life and increasing with age, neurons must be remodeled and repaired to maintain synapto-dendritic connections. Through its lipid transport function, apoE is an important factor in these processes. ApoE3 and apoE2 are effective in maintaining and repairing neuronal cells (1, 3, 39), but apoE4 is much less so. The injurious insults or stressors could include oxidative stress, ischemia, excess Aβ production, SOD1 mutations, inflammation, and the aging process itself.
Impaired cognition in “normal” individuals carrying the apoE4 allele worsens with age, suggesting a global detrimental effect on the CNS (40). ApoE4 is also associated with impaired CNS glucose utilization in normal and AD patients (41–44). In both 65- to 75-year-old and 29- to 39-year-old subjects, the apoE4 allele was associated with lower glucose utilization than the apoE3 allele and affected the hippocampus and cortex, the same areas affected by AD (43, 44). Consistent with these observations, the effect of apoE on mitochondrial metabolism is considered a critical target for cognitive decline in AD (45–47).
ApoE and AD
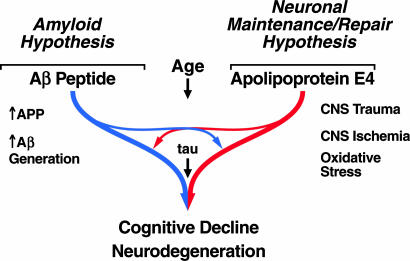
Although apoE4 is strongly linked to AD pathology, its mode of action is unknown. Several mechanisms have been proposed (48–76) (Table 1). Through interactions with the Aβ peptide, apoE4 may increase Aβ deposition in plaques and impair its clearance. However, apoE may act through other pathways that may or may not involve Aβ (Fig. 1).
Table 1.
Potential roles for apoE in neuropathology
| Effects | References |
|---|---|
| Protective effects of apoE3 | |
| Stimulates neurite outgrowth | 48–52 |
| Protects from neurodegeneration | 53–54 |
| Protects from cognitive decline | 55 |
| Protects tau from phosphorylation | 56–57 |
| Antioxidative effect | 58–60 |
| Stimulates cholesterol efflux | 61–62 |
| Stimulates Aβ clearance | 63–64 |
| Stimulates synaptic plasticity | 77 |
| Detrimental effects of apoE4 | |
| Inhibits neurite outgrowth | 48–49 |
| Disrupts neuronal cytoskeleton | 48–50 |
| Stimulates tau phosphorylation | 65–67 |
| Causes neurodegeneration | 53, 54, 68 |
| Causes cognitive decline | 55, 69, 70 |
| Causes neurodegeneration by apoE4 fragments | 67, 71–73 |
| Potentiates Aβ-induced lysosomal leakage and apoptosis | 74 |
| Decreases androgen receptor | 70 |
| Enhances Aβ deposition | 64, 75, 76 |
Fig. 1.
Multiple factors interact through various pathways to cause cognitive decline and neurodegeneration. ApoE4 may be associated with neuropathology through interactions with Aβ peptide that converge on the amyloid cascade or may act independently of Aβ.
Insights into the role of apoE in neuropathology have come from studies of transgenic mice expressing human apoE3 or apoE4 in neurons or astrocytes. Features of AD pathology in these animals include reduced numbers of presynaptic terminals in mice expressing apoE4 with (54, 64) or without (53, 66, 68) expression of human amyloid precursor protein (APP), increased plaque deposition in apoE4 mice expressing APP (64), increased phosphorylation of tau (65–67), impaired learning and memory (55, 69), and altered long-term potentiation (77). Especially impressive is the impact of apoE4 on memory in transgenic mice lacking endogenous mouse apoE and expressing human apoE in neurons under the control of the neuron-specific enolase (NSE) promoter. NSE-apoE4 mice showed significant learning impairment in a water maze and in vertical exploratory behavior (55, 69). Susceptibility to these effects was influenced by age and gender. Moreover, morphological studies demonstrated a loss of synapto-dendritic connections at 6 months of age that correlated with the onset of the learning deficits. Working memory was also impaired in transgenic mice expressing apoE4 in astrocytes (62). Thus, the neuropathological effects of human apoE4 with or without the involvement of Aβ are clearly demonstrated in mice.
Structural Differences Among the ApoE Isoforms
ApoE is a polymorphic 299-aa protein (Mr = 34,200) (1–3, 39, 78, 79). The gene, located on chromosome 19, encodes three alleles: apoE2 (frequency in populations, 5–10%), apoE3 (60–70%), and apoE4 (15–20%). The isoforms differ only at residues 112 and 158. ApoE3 has Cys-112 and Arg-158, whereas apoE4 has arginine at both sites, and apoE2 has cysteines. In apoE2, Cys-158 results in defective receptor binding and type III hyperlipoproteinemia (1–3, 80).
Arg-112 in apoE4 mediates two key properties, domain interaction and reduced protein stability or molten globule formation (81, 82), that likely contribute to apoE4-associated neuropathology. Although the dynamics of apoE2 and apoE3 structure allow them to display these same features to a lesser extent, apoE4 is predisposed to assume the pathological conformation.
ApoE4 Domain Interaction.
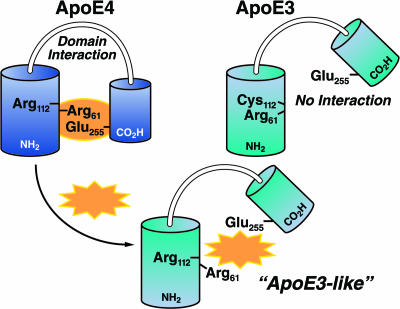
ApoE has two structural domains: a 22-kDa N-terminal domain (residues 1–191) containing the low-density lipoprotein (LDL) receptor binding site (residues 136–150) and a 10-kDa C-terminal domain (residues 216–299) containing the major lipid binding site (residues ≈240–270) (1–3, 39, 78). In apoE4, but not to the same extent in apoE2 or apoE3, the two domains interact (Fig. 2) (83, 84). X-ray crystallographic studies indicate that Arg-112 in apoE4 allows the side chain of Arg-61 to extend away from the helical bundle (83); the side chain of Arg-61 in apoE3 and apoE2 has a different orientation (tucked between helices 2 and 3) (85). We hypothesized that Arg-61 in apoE4 interacts with Glu-255 in the C-terminal region (81). Mutating Arg-61 to threonine or Glu-255 to alanine abolished domain interaction (84). Thus, apoE4 domain interaction may be mediated by ionic binding between Arg-61 and Glu-255. ApoE3 and apoE2 are much less likely to undergo domain interaction.
Fig. 2.
ApoE4 domain interaction can be disrupted by small molecules (represented by gold symbol). In apoE4, Arg-61 in the N-terminal domain interacts with Glu-255 in the C-terminal domain. Small molecules that are predicted to interact with apoE4 in the region of Arg-61 would disrupt domain interaction and convert apoE4 to an “apoE3-like” molecule.
ApoE4 domain interaction has been demonstrated in cultured cells by fluorescence resonance energy transfer (86). Mutation of Arg-61 to Thr or Glu-255 to Ala in apoE4 abolished domain interaction. Structural studies demonstrated that, in lipid-free and phospholipid-bound apoE4, the two domains were closer together than in apoE3 and that Arg-61 and Glu-255 were in close proximity (87).
“Humanizing” the mouse apoE gene by gene targeting introduces domain interaction and converts mouse apoE from an “apoE3-like” to an “apoE4-like” functional molecule in a mouse. Mouse apoE contains arginine at a position equivalent to 112 in human apoE4 but lacks Arg-61, which mediates domain interaction. Replacing Thr-61 with arginine introduces domain interaction and allows mouse “Arg-61” apoE to behave like human apoE4 (88).
Domain interaction mediates several neuropathological effects of apoE4, including increases in Aβ production, potentiation of Aβ-induced lysosomal leakage and apoptosis, and enhanced proteolytic cleavage in neurons. Inhibiting apoE4 domain interaction with small-molecule “structure correctors” represents a new therapeutic strategy.
ApoE4 Molten Globule Formation.
Protein instability is an important component of several neurodegenerative disorders (89). ApoE4 is the least stable isoform. ApoE4 denatures at lower concentrations of guanidine·HCl and urea and at lower temperatures (instability: apoE4 > apoE3 > apoE2) (82, 90). Furthermore, the denaturation pattern of apoE4 does not fit a two-state equilibrium (native versus fully unfolded), suggesting that apoE4 exists as a partially folded intermediate or a molten globule. Reactive intermediates have several pathophysiological activities (91, 92), including altered intradomain interactions, increased lipid and membrane binding, membrane disruption, translocation across membranes, and increased susceptibility to proteolysis. ApoE4 is most likely to form molten globules, but apoE3 tends to do so to a lesser degree. Domain interaction appears to contribute to the instability of apoE4.
Sites of Synthesis in the Nervous System
The brain is second only to the liver in synthesizing apoE (93), and apoE is synthesized in a variety of cell types in the nervous system (1). The site of origin of apoE undoubtedly affects its function.
Peripheral Nervous System.
ApoE is produced in glia surrounding sensory and motor neurons and in nonmyelinating Schwann cells. Resident macrophages and those recruited to injured peripheral nerves secrete large quantities of apoE, which accumulates in the extracellular matrix of the degenerating stump and regenerating nerve (1).
One of the first important links between apoE and neurobiology was the observation that apoE concentration increases 200-fold in an injured rat sciatic nerve (94–96) and then returns to baseline by 8 weeks when sciatic nerve regeneration is largely complete. Studies of peripheral nerve injury in Apoe−/− mice suggested that normal regeneration can occur without apoE (97). However, careful ultrastructural examination demonstrated a reduced number of axons and defects in their morphology (98). Other apolipoproteins (apoA-I, apoA-II) abundant in the periphery might act as less effective substitutes. Furthermore, apoE has isoform-specific effects on neurite outgrowth in dorsal root ganglion cells and Neuro-2a cells in culture (48, 49).
CNS.
In the CNS, astrocytes are the major cell type that produces apoE (99, 100) (Fig. 3). However, CNS neurons express apoE under physiological and pathological conditions (101–110). ApoE mRNA is found in cortical and hippocampal neurons in humans (106) and in transgenic mice expressing human apoE under the control of the human apoE promoter (109). Treatment with kainic acid induces apoE synthesis in hippocampal neurons in rats (111), and apoE is expressed in neurons in cerebral infarct patients (112). ApoE synthesis and secretion can be modulated in cultured neurons (110, 113–116). ApoE expression in neuronal cells is regulated by the mitogen-activated protein kinase signaling pathways (110).
Fig. 3.
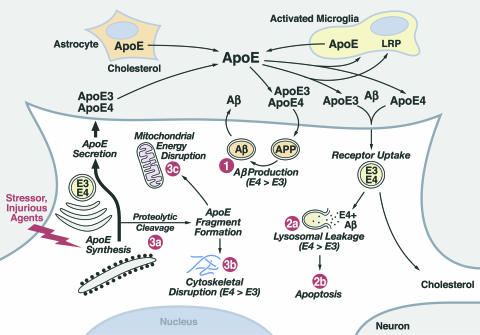
Roles of apoE in lipid redistribution among cells in the CNS and apoE isoform-specific differences in neuropathology. ApoE is synthesized by astrocytes, activated microglia, and neurons. Three potential detrimental roles for apoE4: (1) enhanced Aβ production, (2) potentiation of Aβ1–42-induced lysosomal leakage and apoptosis, and (3) enhanced neuron-specific proteolysis resulting in translocation of neurotoxic apoE4 fragments in the cytosol, where they are associated with cytoskeletal disruption and mitochondrial dysfunction.
We have hypothesized that neuronal expression of apoE is induced to promote neuron protection or to repair injured neurons (1–3, 78, 79). However, apoE4 appears to be less effective in this process; in fact, it becomes pathological because of its susceptibility to intraneuronal proteolysis.
Role of ApoE in the CNS
ApoE plays important roles in neurobiology (1–3, 78, 79). Its role in the redistribution of lipids among cells throughout the body, including the CNS, is well established (1) (Fig. 3). ApoE is the major apolipoprotein in the CNS (LDL and apoB are absent) capable of redistributing lipids through the LDL receptor-related family of receptors. ApoE occurs in the cerebrospinal fluid as small high-density lipoprotein-like particles or phospholipid disks (117). ApoE-containing lipoproteins can deliver lipids, including cholesterol, to sites of injury for repair of cells. Again, apoE3 (and apoE2) seems more effective in the normal maintenance and repair of cells than apoE4, and apoE4 may be detrimental in the process.
Isoform-specific effects of apoE have been demonstrated on neurite extension in culture systems (48–52). In the presence of a source of lipid, apoE3 stimulates neurite outgrowth, whereas apoE4 does not. Astrocyte-derived apoE3, but not apoE4, also caused neurite extension in rat hippocampal neurons (52). The inhibition of neurite extension by apoE4 appears to be related to alterations in the cytoskeleton, especially an effect on microtubule stability (118). These effects may be mediated through tau (a microtubule-stabilizing protein). ApoE3, but not apoE4, binds to tau in vitro, and may protect tau from hyperphosphorylation, which inhibits tau’s ability to stabilize microtubules (56, 57, 65). The isoform-specific effect on neurite extension is mediated through the apoE receptor binding region and can be blocked by inhibiting LDL receptor-related protein binding or by removing cell-surface heparan sulfate proteoglycans to disrupt the heparan sulfate proteoglycan/LDL receptor-related protein pathway (50).
ApoE4 Neuropathology in the Context of Aβ
Effects of ApoE4 on Aβ Clearance and Deposition.
Decreased Aβ clearance or increased Aβ deposition has been suggested to play an important role in AD pathogenesis (119, 120). Both in vitro and in vivo studies demonstrate that apoE4 inhibits Aβ clearance and/or stimulates Aβ deposition (63, 121–124), leading to plaque formation (64, 75). A recent review summarizes the effects of apoE4 on Aβ clearance and deposition (79).
ApoE4 Increases Aβ Production.
Many studies have focused on the role of apoE in stimulating Aβ deposition or clearance (63, 64, 75, 76). However, apoE4 also enhances Aβ production; apoE3 does so to a lesser extent (125). In rat neuroblastoma B103 cells stably transfected with human wild-type APP695, lipid-poor apoE4 stimulated Aβ production 60%, compared with only 30% by apoE3, and robustly stimulated APP recycling as well. The latter effect was inhibited by blocking the LDL receptor-related protein pathway using the receptor-associated protein or LDL receptor-related protein siRNA (125), suggesting that this pathway is involved in APP recycling and Aβ production.
The unique structure of apoE4 contributed to its ability to enhance Aβ production. Replacing Arg-61 with Thr in apoE4, which abolishes domain interaction, completely inhibited apoE4-mediated stimulation of Aβ production (125). Excitingly, the apoE4 effect was abolished by treating the apoE4 with small molecules predicted to interact with the N-terminal region of apoE4 (but not apoE3) in the vicinity of Arg-61 and Arg-112 and disrupt domain interaction (125).
ApoE4 Potentiation of Aβ-Induced Lysosomal Leakage and Apoptosis.
Aβ1–42 causes lysosomal leakage (126). In cultured Neuro-2a cells, we showed that apoE4 enhances Aβ-induced lysosomal leakage and apoptotic cell death to a much greater extent (2- to 4-fold) than apoE3 (74, 127).
We hypothesized that apoE4, which is more unstable, was more likely to form a reactive intermediate (molten globule) when it reached the acidic pH of late endosomes or lysosomes and assume detrimental activities, including membrane destabilization. To test this possibility, we blocked lysosomal acidification with bafilomycin or NH4Cl (127). The enhancement of lysosomal leakage and apoptosis was abolished, suggesting slower formation of the unstable apoE4 intermediates. The activity of apoE3 in the context of Aβ1–42 was not affected by neutralization of the lysosomal pH. ApoE4 also altered membrane stability in a model membrane system to a greater extent at pH 4.0 than at pH 7.4 (127).
ApoE4 Neuropathology Independent of Aβ
ApoE Cleavage in Neurons.
The induction of apoE synthesis by neurons has been suggested to protect neurons from injury or to promote intraneuronal repair and maintenance of synapto-dendritic connections (1–3, 78, 79). However, when synthesized by neurons, apoE can be cleaved by a protease, and the fragments are detrimental to the repair/maintenance process (67, 128, 129). Because of its unique conformation and reactivity, apoE4 is much more susceptible to the proteolysis than apoE3.
Brains of AD patients and transgenic mice expressing apoE in CNS neurons possess C-terminal-truncated fragments (29 and 15–20 kDa) of apoE (Fig. 5 A–C, which is published as supporting information on the PNAS web site). In mice, the accumulation of apoE4 fragments peaks at 6–7 months of age, coinciding with the appearance of neurodegenerative changes in the brain and significant deficits in learning and memory (129). A first cleavage site yielded fragments of 29–30 kDa lacking the C-terminal 27 aa. In brains of AD patients and neuron-specific enolase-apoE4 mice, fragments of 15–20 kDa, all of which lacked the C-terminal 27 aa, were also observed.
We have shown that the apoE cleaving enzyme is a neuron-specific, chymotrypsin-like serine protease that cuts apoE at Met-272 and/or Leu-268. ApoE4 is highly susceptible to proteolysis; apoE3 is less so (128, 129). Interestingly, interfering with domain interaction by mutating Arg-61 to threonine or Glu-255 to alanine markedly reduces the susceptibility of apoE4 to proteolysis (Fig. 5D).
From immunocytochemical studies of neurons in culture, apoE appears in a pattern indicative of the endoplasmic reticulum (ER) and Golgi apparatus (as would be expected for a secretory protein) (130). However, undetectable amounts of full-length apoE3 or apoE4 may enter the cytosol. In contrast, expression of apoE4(1–272) in neurons has several effects: neurotoxicity, translocation of the fragments into the cytosol, and accumulation of the fragments in filamentous cytoplasmic structures (phosphorylated tau and neurofibrillary tangle-like structures) and in mitochondria.
Neurotoxicity.
We identified the structural features and domains of apoE responsible for its neurotoxicity. The conformation of the C terminus is a key element. ApoE4(1–272) is toxic to cultured neurons (67, 130) (Fig. 4) and causes neurodegenerative changes and neurofibrillary tangle-like structures in CNS neurons in the hippocampus and cortical regions in transgenic mice (128). However, expression of an apoE4 fragment lacking both the C-terminal 27 aa and the lipid binding region [apoE4(Δ241–299)] in mice (128) did not result in neurodegeneration. Thus, only fragments generated by the apoE cleaving enzyme cause neuropathology.
Fig. 4.
Regions of apoE4 involved in causing neurotoxicity, translocation into the cytosol, and mitochondrial targeting. The receptor binding region (amino acids 136–150) plays a key role in translocation, as well as in binding to the LDL receptor and tau. The lipid binding region (amino acids 240–270) plays a role in translocation and mitochondrial localization, in addition to possessing lipid and Aβ binding.
In Neuro-2a cells, apoE4(127–272) is also neurotoxic (Fig. 4). It contains both the positively charged receptor binding region (amino acids 136–150) and the hydrophobic lipid binding region (amino acids 240–270). Mutagenesis of four conserved residues in the lipid binding region or three conserved positively charged residues in the receptor binding region abolished the neurotoxicity of apoE4(127–272). Furthermore, apoE4(171–272), which lacks the receptor binding region, was not neurotoxic (130). Interestingly, although full-length apoE4 is not neurotoxic, mutation of one of three highly conserved C-terminal residues converts it to a neurotoxic form (Fig. 4).
Translocation into the Cytosol.
Neurotoxicity correlated with the ability of the apoE fragments to enter the cytosol, where they interacted with the cytoskeleton or mitochondria (Fig. 4). ApoE4 could escape the secretory pathway by translocating through the ribosome–membrane junction during protein synthesis, being proteolytically clipped, and then entering the cytosol. Alternatively, apoE4 or the apoE4 fragment within the ER/Golgi compartment may assume a conformation conducive to membrane translocation. Two structural domains are important in membrane-penetrating proteins: a positively charged region enriched in arginine, lysine, and histidine and a protein transduction domain enriched in hydrophobic residues. ApoE has both a positively charged receptor binding region (residues 136–150) and a hydrophobic lipid binding region (residues 240–270) (Fig. 4).
Several membrane-penetrating proteins have been identified (131–134). For example, Tat released from HIV-1-infected cells can be taken up by neighboring cells and ultimately enter the cytosol (135, 136). The VP22 structural protein of herpes simplex virus closely resembles apoE in several ways. It is of similar size (38 kDa), enters the cytosol, and binds to cytoskeletal elements (microtubules and microfilaments) (137, 138). Interestingly, the protein-transduction domain of Tat has been used to deliver peptides, proteins, and oligodeoxynucleotides to cells (134, 139, 140), and the receptor binding region of apoE has also been used to deliver oligodeoxynucleotides to cells (141, 142).
Cell-penetrating peptides have common structural features in their protein-transduction domains (131–134) (Table 2, which is published as supporting information on the PNAS web site). The positively charged receptor binding region and the hydrophobic lipid binding region of apoE are required for apoE4(1–272) and apoE4(127–272) to enter the cytosol (130). Mutations of three positive residues in the receptor binding region or four residues in the hydrophobic lipid binding region prevent apoE4(1–272) from entering the cytosol (Fig. 4). Although apoE4(1–191), which lacks the lipid binding region, enters the cytosol, efficient translocation likely requires both the receptor and lipid binding regions.
Mitochondrial Binding.
Mitochondrial dysfunction was described in patients with AD, especially in those with the apoE4 genotype (143). Previously, we showed that apoE avidly binds to the α- and β-subunits of mitochondrial F1-ATPase (144), suggesting a possible role for apoE in intracellular transport within or between cell organelles. More recently, when we expressed apoE4 (127–272) in Neuro-2a cells, it localized to the mitochondria (130) (Fig. 4). Because translocation across the membrane required the receptor and lipid binding regions, this is most likely the smallest fragment to translocate and associate with mitochondria. Fragments lacking the signal peptide and causing them to enter the cytosol were examined. The mitochondrial binding region was localized to the lipid binding region, as demonstrated by the association of apoE4(171–272) with the mitochondria (Fig. 4) (130).
The disruption of the electropotential of the mitochondria by the apoE fragments in cultured neurons may affect neuronal function in several ways. Mitochondria play a critical role in synaptogenesis (145), and apoE4 expression in mice results in a significant loss of synapto-dendritic connections within the brain (53).
In addition, the apoE fragment associated with the mitochondria may induce the mitochondrial-apoptotic pathway. ApoE4 potentiated staurosporine- and H2O2-induced apoptosis in cultured neurons, and apoE3 protected the cells from DNA fragmentation (74). On the other hand, apoE4-associated neuropathology may occur through a disruption of mitochondrial regulation of energy and glucose metabolism in neurons. Association of apoE4 genotype with altered CNS glucose metabolism has been demonstrated in both normal and AD patients (41–44). Even in young subjects with no signs of dementia and unlikely to possess Aβ deposits, apoE4 is associated with reduced neuronal glucose utilization, reflecting altered mitochondrial activity.
The apoE fragment could also disrupt mitochondrial trafficking, resulting in failure to deliver these organelles to appropriate sites in neurons and causing energy depletion and disruption of calcium homeostasis (reviewed in ref. 146). The disruption of microtubules in cultured neurons by apoE4 may accentuate the mitochondrial dysfunction, or the mitochondrial dysfunction may in part cause the cytoskeletal abnormality observed with apoE4. The association of apoE4 with hyperphosphorylation of tau (129) and the occurrence of neurofibrillary tangle-like structures in neurons (67) could result in an abnormal distribution of mitochondria in neurons.
We do not know how the apoE fragments are transferred to the mitochondria or how they associate with the mitochondria. The unique lipid composition of mitochondria could enable the apoE fragments to bind directly through a hydrophobic lipid interaction. Alternatively, the fragments may interact with cytoplasmic factors (chaperones) that target the apoE to the import channels (147, 148). Alternatively, apoE may undergo proteolytic cleavage in the ER and translocate by lateral diffusion through mitochondrion-associated ER membranes that exist transiently to allow proteins to move between the ER and the mitochondria (149, 150). The hepatitis C virus core protein synthesized in the ER of HeLa cells is targeted to the mitochondria (after it is processed by a peptidase) along such membranes (151).
We hypothesize that mitochondrial and cytoskeletal alterations caused by apoE4 fragments are a key mechanism for the Aβ-independent neuropathology involving apoE4. Our observations suggest several therapeutic approaches. An inhibitor of the protease could prevent the generation of the detrimental apoE fragments. Alternatively, a small molecule that could enter the ER and alter apoE4 conformation to prevent domain interaction and molten globule formation may reduce the affinity of apoE for the protease. Blocking the interaction of the apoE4 fragments with the mitochondria could prevent the detrimental effects on mitochondrial function, and increasing the number and activity of neuronal mitochondria might negate some of the neuropathological effects of apoE4. Consistent with our data related to apoE and mitochondrial damage (46), treatment of AD patients with rosiglitazone maleate, an insulin sensitizer and mitochondrial stimulator, appears to attenuate the rapid loss of cognitive function in apoE4 patients and to improve cognition in those without apoE4 (45). Based on these data and the studies of apoE fragments, the mitochondrial metabolism hypothesis, proposed by Roses and Saunders, opens the door for new therapeutic approaches for AD (see ref. 47 for a review of mitochondrial energetics and the mitochondrial hypothesis) in combination with other ways to negate the detrimental effects of apoE4.
ApoE4 Effects on Tau Phosphorylation.
ApoE3 and apoE4 appear to differ in their effects on the phosphorylation and aggregation of tau, which is independent of Aβ. In vitro, apoE3 forms an SDS-stable complex with tau in a 1:1 ratio, whereas apoE4 does not interact significantly (56). Phosphorylation of tau by a crude brain extract inhibited the interaction of apoE3 with tau (56), suggesting that apoE3 binds to nonphosphorylated tau. Furthermore, the N-terminal domain of apoE3 is responsible for irreversibly binding to tau through the microtubule-binding repeat regions (56, 152). Increased phosphorylation of tau has been observed in transgenic mice expressing human apoE4 in neurons but not in mice expressing apoE4 in astrocytes (65, 66, 129). Thus, apoE4 may have a neuron-specific effect on tau phosphorylation. Our studies suggest that C-terminal-truncated apoE stimulates tau phosphorylation and intracellular neurofibrillary tangle-like inclusion formation in transgenic mice (128).
New Therapeutic Approaches: Small Molecules to Block Domain Interaction
Using the dock screening program (153–157), we identified 65 small molecules that were predicted to bind preferentially to apoE4. Nine inhibited the preferential binding of apoE4 to emulsion particles, an assay for domain interaction, and converted apoE4 to an apoE3-like molecule (125). Two compounds, GIND-25, a disulfonate, and GIND-105, a monosulfoalkyl, decreased Aβ production induced by apoE4 to levels very similar to those induced by apoE3 (Fig. 6, which is published as supporting information on the PNAS web site).
We have demonstrated that inhibiting apoE4 domain interaction, and presumably altering its conformation, can modulate its neuropathologic activity. Disruption of the intramolecular interaction within apoE4 and its conformation represents a reasonable therapeutic strategy. Recently, small molecules were shown to bind to the hepatitis C virus polymerase and inhibit RNA synthesis by altering intramolecular structure (158). Furthermore, small molecules correct the phenylalanine deletion mutation (ΔF508) in the cystic fibrosis transmembrane conductance regulator chloride channel, which is the common cause of cystic fibrosis (159). Small molecules increase halide flux across membranes in the ΔF508-transfected cells and increase plasma membrane expression of the regulator, suggesting improved folding in the ER and stability on the cell surface.
Small molecules can also stabilize transthyretin homotetramers, reducing their susceptibility to misfold and form monomeric transthyretin that undergoes amyloidogenesis (160). Further proof-of-concept, suggesting that alteration in protein conformation within cells in vitro and in vivo may be a valid approach to alter protein function, comes from studies of Fabry disease (161, 162). A recent review summarizes the use of small molecules to modulate the structure and function of various proteins of biological importance (163).
We asked whether apoE4 with Thr-61, which prevents domain interaction, or the small molecules that block domain interaction could prevent the apoE4 potentiation of Aβ-induced lysosomal leakage and apoptosis. The answer was yes. Both the mutant apoE4 and several small molecules identified by dock screening did just that (Z.-S. Ji, Y.H., and R.W.M., unpublished data).
Conclusions
Increasing evidence points to an important role for apoE4 in neurodegeneration in general and AD in particular. ApoE4 is more than a susceptibility factor for AD. Like Aβ and other molecules, it is a causative agent. The deceptively small differences between the isoforms cause significant differences in structure and ultimately function. In fact, the continuum represented by the structural equilibrium of the three isoforms is consistent with the continuum of the incidence of AD. ApoE3 assumes the pathological conformation less readily, and fewer individuals with two apoE3 alleles develop AD. ApoE4 more readily assumes the pathological conformations (domain interaction and molten globule formation), and individuals with one or two apoE4 alleles are at much greater risk for AD.
The structural features of apoE4 also suggest excellent therapeutic strategies that are amenable to testing. Agents capable of converting apoE4 to an apoE3-like molecule (structure “correctors”), blocking apoE4 proteolytic processing (protease “inhibitors”), or preventing mitochondrial dysfunction (mitochondrial “protectors”) offer promise of reducing or eliminating the detrimental effects of apoE.
Supplementary Material
Acknowledgments
We thank Dr. Lennart Mucke for critical reading of the manuscript, Sylvia Richmond for manuscript preparation, Stephen Ordway and Gary Howard for editorial assistance, John C. W. Carroll and John Hull for graphics, and Stephen Gonzales and Chris Goodfellow for photography. This work was supported in part by National Institutes of Health Program Project Grant P01 AG022074.
Abbreviations
- apo
apolipoprotein
- AD
Alzheimer’s disease
- Aβ
amyloid β
- APP
amyloid precursor protein
- LDL
low-density lipoprotein
- ER
endoplasmic reticulum.
Conflict of interest statement: No conflicts declared.
See accompanying Profile on page 5641.
References
- 1.Mahley R. W. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Weisgraber K. H. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 3.Mahley R. W., Rall S. C., Jr. Annu. Rev. Genomics Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 4.Corder E. H., Saunders A. M., Risch N. J., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Jr., Rimmler J. B., Locke P. A., Conneally P. M., Schmader K. E., et al. Nat. Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 5.Lippa C. F., Smith T. W., Saunders A. M., Hulette C., Pulaski-Salo D., Roses A. D. Neurology. 1997;48:515–519. doi: 10.1212/wnl.48.2.515. [DOI] [PubMed] [Google Scholar]
- 6.Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., Pericak-Vance M. A. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 7.Roses A. D. Annu. Rev. Med. 1996;47:387–400. doi: 10.1146/annurev.med.47.1.387. [DOI] [PubMed] [Google Scholar]
- 8.Strittmatter W. J., Saunders A. M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G. S., Roses A. D. Proc. Natl. Acad. Sci. USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang M.-X., Stern Y., Marder K., Bell K., Gurland B., Lantigua R., Andrews H., Feng L., Tycko B., Mayeux R. J. Am. Med. Assoc. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 10.Romas S. N., Santana V., Williamson J., Ciappa A., Lee J. H., Rondon H. Z., Estevez P., Lantigua R., Medrano M., Torres M., et al. Arch. Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 11.Saunders A. M., Strittmatter W. J., Schmechel D., St. George-Hyslop P. H., Pericak-Vance M. A., Joo S. H., Rosi B. L., Gusella J. F., Crapper-MacLachlan D. R., Alberts M. J., et al. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 12.Farrer L. A., Cupples L. A., Haines J. L., Hyman B., Kukull W. A., Mayeux R., Myers R. H., Pericak-Vance M. A., Risch N., Van Duijn C. M. J. Am. Med. Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 13.Nicoll J. A. R., Roberts G. W., Graham D. I. Ann. N.Y. Acad. Sci. 1996;777:271–275. doi: 10.1111/j.1749-6632.1996.tb34431.x. [DOI] [PubMed] [Google Scholar]
- 14.Teasdale G. M., Nicoll J. A. R., Murray G., Fiddes M. Lancet. 1997;350:1069–1071. doi: 10.1016/S0140-6736(97)04318-3. [DOI] [PubMed] [Google Scholar]
- 15.Jordan B. D., Relkin N. R., Ravdin L. D., Jacobs A. R., Bennett A., Gandy S. J. Am. Med. Assoc. 1997;278:136–140. [PubMed] [Google Scholar]
- 16.McCarron M. O., Delong D., Alberts M. J. Neurology. 1999;53:1308–1311. doi: 10.1212/wnl.53.6.1308. [DOI] [PubMed] [Google Scholar]
- 17.Friedman G., Froom P., Sazbon L., Grinblatt I., Shochina M., Tsenter J., Babaey S., Yehuda A. B., Groswasser Z. Neurology. 1999;52:244–248. doi: 10.1212/wnl.52.2.244. [DOI] [PubMed] [Google Scholar]
- 18.Crawford F. C., Vanderploeg R. D., Freeman M. J., Singh S., Waisman M., Michaels L., Abdullah L., Warden D., Lipsky R., Salazar A., et al. Neurology. 2002;58:1115–1118. doi: 10.1212/wnl.58.7.1115. [DOI] [PubMed] [Google Scholar]
- 19.Chamelian L., Reis M., Feinstein A. Brain. 2004;127:2621–2628. doi: 10.1093/brain/awh296. [DOI] [PubMed] [Google Scholar]
- 20.Alberts M. J., Graffagnino C., McClenny C., DeLong D., Strittmatter W., Saunders A. M., Roses A. D. Lancet. 1995;346:575. doi: 10.1016/s0140-6736(95)91411-0. [DOI] [PubMed] [Google Scholar]
- 21.Slooter A. J. C., Tang M.-X., van Duijn C. M., Stern Y., Ott A., Bell K., Breteler M. M. B., Van Broeckhoven C., Tatemichi T. K., Tycko B., et al. J. Am. Med. Assoc. 1997;277:818–821. doi: 10.1001/jama.277.10.818. [DOI] [PubMed] [Google Scholar]
- 22.Newman M. F., Croughwell N. D., Blumenthal J. A., Lowry E., White W. D., Spillane W., Davis R. D., Jr., Glower D. D., Smith L. R., Mahanna E. P., et al. Ann. Thorac. Surg. 1995;59:1326–1330. doi: 10.1016/0003-4975(95)00076-w. [DOI] [PubMed] [Google Scholar]
- 23.Tardiff B. E., Newman M. F., Saunders A. M., Strittmatter W. J., Blumenthal J. A., White W. D., Croughwell N. D., Davis R. D., Jr., Roses A. D., Reves J. G., et al. Ann. Thorac. Surg. 1997;64:715–720. doi: 10.1016/s0003-4975(97)00757-1. [DOI] [PubMed] [Google Scholar]
- 24.Harhangi B. S., de Rijk M. C., van Duijn C. M., Van Broeckhoven C., Hofman A., Breteler M. M. B. Neurology. 2000;54:1272–1276. doi: 10.1212/wnl.54.6.1272. [DOI] [PubMed] [Google Scholar]
- 25.Parsian A., Racette B., Goldsmith L. J., Perlmutter J. S. Genomics. 2002;79:458–461. doi: 10.1006/geno.2002.6707. [DOI] [PubMed] [Google Scholar]
- 26.Li Y. J., Hauser M. A., Scott W. K., Martin E. R., Booze M. W., Qin X. J., Walter J. W., Nance M. A., Hubble J. P., Koller W. C., et al. Neurology. 2004;62:2005–2009. doi: 10.1212/01.wnl.0000128089.53030.ac. [DOI] [PubMed] [Google Scholar]
- 27.Martinez M., Brice A., Vaughan J. R., Zimprich A., Breteler M. M. B., Meco G., Filla A., Farrer M. J., Bétard C., Singleton A., et al. Am. J. Med. Genet. 2005;136B:72–74. doi: 10.1002/ajmg.b.30196. [DOI] [PubMed] [Google Scholar]
- 28.Al-Chalabi A., Enayat Z. E., Bakker M. C., Sham P. C., Ball D. M., Shaw C. E., Lloyd C. M., Powell J. F., Leigh P. N. Lancet. 1996;347:159–160. doi: 10.1016/s0140-6736(96)90343-8. [DOI] [PubMed] [Google Scholar]
- 29.Moulard B., Sefiani A., Laamri A., Malafosse A., Camu W. J. Neurol. Sci. 1996;139(Suppl.):34–37. doi: 10.1016/0022-510x(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 30.Smith R. G., Haverkamp L. J., Case S., Appel V., Appel S. H. Lancet. 1996;348:334–335. doi: 10.1016/s0140-6736(05)64502-3. [DOI] [PubMed] [Google Scholar]
- 31.Drory V. E., Birnbaum M., Korczyn A. D., Chapman J. J. Neurol. Sci. 2001;190:17–20. doi: 10.1016/s0022-510x(01)00569-x. [DOI] [PubMed] [Google Scholar]
- 32.Lacomblez L., Doppler V., Beucler I., Costes G., Salachas F., Raisonnier A., Le Forestier N., Pradat P.-F., Bruckert E., Meininger V. Neurology. 2002;58:1112–1114. doi: 10.1212/wnl.58.7.1112. [DOI] [PubMed] [Google Scholar]
- 33.Fazekas F., Strasser-Fuchs S., Kollegger H., Berger T., Kristoferitsch W., Schmidt H., Enzinger C., Schiefermeier M., Schwarz C., Kornek B., et al. Neurology. 2001;57:853–857. doi: 10.1212/wnl.57.5.853. [DOI] [PubMed] [Google Scholar]
- 34.Chapman J., Vinokurov S., Achiron A., Karussis D. M., Mitosek-Szewczyk K., Birnbaum M., Michaelson D. M., Korczyn A. D. Neurology. 2001;56:312–316. doi: 10.1212/wnl.56.3.312. [DOI] [PubMed] [Google Scholar]
- 35.Bedlack R. S., Edelman D., Gibbs J. W., III, Kelling D., Strittmatter W., Saunders A. M., Morgenlander J. Neurology. 2003;60:1022–1024. doi: 10.1212/01.wnl.0000056689.50682.94. [DOI] [PubMed] [Google Scholar]
- 36.Gottlieb D. J., DeStefano A. L., Foley D. J., Mignot E., Redline S., Givelber R. J., Young T. Neurology. 2004;63:664–668. doi: 10.1212/01.wnl.0000134671.99649.32. [DOI] [PubMed] [Google Scholar]
- 37.Tsuang D. W., Dalan A. M., Eugenio C. J., Poorkaj P., Limprasert P., La Spada A. R., Steinbart E. J., Bird T. D., Leverenz J. B. Arch. Neurol. 2002;59:1622–1630. doi: 10.1001/archneur.59.10.1622. [DOI] [PubMed] [Google Scholar]
- 38.Horsburgh K., Graham D. I., Stewart J., Nicoll J. A. R. J. Neuropathol. Exp. Neurol. 1999;58:227–234. doi: 10.1097/00005072-199903000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Weisgraber K. H., Mahley R. W. FASEB J. 1996;10:1485–1494. doi: 10.1096/fasebj.10.13.8940294. [DOI] [PubMed] [Google Scholar]
- 40.Deary I. J., Whiteman M. C., Pattie A., Starr J. M., Hayward C., Wright A. F., Carothers A., Whalley L. J. Nature. 2002;418:932. doi: 10.1038/418932a. [DOI] [PubMed] [Google Scholar]
- 41.Small G. W., Mazziotta J. C., Collins M. T., Baxter L. R., Phelps M. E., Mandelkern M. A., Kaplan A., La Rue A., Adamson C. F., Chang L., et al. J. Am. Med. Assoc. 1995;273:942–947. [PubMed] [Google Scholar]
- 42.Small G. W., Ercoli L. M., Silverman D. H. S., Huang S.-C., Komo S., Bookheimer S. Y., Lavretsky H., Miller K., Siddarth P., Rasgon N. L., et al. Proc. Natl. Acad. Sci. USA. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiman E. M., Caselli R. J., Chen K., Alexander G. E., Bandy D., Frost J. Proc. Natl. Acad. Sci. USA. 2001;98:3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiman E. M., Chen K., Alexander G. E., Caselli R. J., Bandy D., Osborne D., Saunders A. M., Hardy J. Proc. Natl. Acad. Sci. USA. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risner M. E., Saunders A. M., Altman J. F. B., Ormandy G. C., Craft S., Foley I. M., Zvartau-Hind M. E., Hosford D. A., Roses A. D. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500369. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y., Mahley R. W. Alzheimer’s Dementia. 2006. in press. [DOI] [PubMed] [Google Scholar]
- 47.Roses A. D., Saunders A. M. Alzheimer’s Dementia. 2006. in press. [Google Scholar]
- 48.Nathan B. P., Bellosta S., Sanan D. A., Weisgraber K. H., Mahley R. W., Pitas R. E. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 49.Bellosta S., Nathan B. P., Orth M., Dong L.-M., Mahley R. W., Pitas R. E. J. Biol. Chem. 1995;270:27063–27071. doi: 10.1074/jbc.270.45.27063. [DOI] [PubMed] [Google Scholar]
- 50.Holtzman D. M., Pitas R. E., Kilbridge J., Nathan B., Mahley R. W., Bu G., Schwartz A. L. Proc. Natl. Acad. Sci. USA. 1995;92:9480–9484. doi: 10.1073/pnas.92.21.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeMattos R. B., Curtiss L. K., Williams D. L. J. Biol. Chem. 1998;273:4206–4212. doi: 10.1074/jbc.273.7.4206. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y., Wu S., Bu G., Onifade M. K., Patel S. N., LaDu M. J., Fagan A. M., Holtzman D. M. J. Neurosci. 1998;18:3261–3272. doi: 10.1523/JNEUROSCI.18-09-03261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buttini M., Orth M., Bellosta S., Akeefe H., Pitas R. E., Wyss-Coray T., Mucke L., Mahley R. W. J. Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buttini M., Yu G.-Q., Shockley K., Huang Y., Jones B., Masliah E., Mallory M., Yeo T., Longo F. M., Mucke L. J. Neurosci. 2002;22:10539–10548. doi: 10.1523/JNEUROSCI.22-24-10539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raber J., Wong D., Yu G.-Q., Buttini M., Mahley R. W., Pitas R. E., Mucke L. Nature. 2000;404:352–354. doi: 10.1038/35006165. [DOI] [PubMed] [Google Scholar]
- 56.Strittmatter W. J., Weisgraber K. H., Goedert M., Saunders A. M., Huang D., Corder E. H., Dong L.-M., Jakes R., Alberts M. J., Gilbert J. R., et al. Exp. Neurol. 1994;125:163–171. doi: 10.1006/exnr.1994.1019. [DOI] [PubMed] [Google Scholar]
- 57.Lovestone S., Anderton B. H., Hartley K., Jensen T. G., Jorgensen A. L. NeuroReport. 1996;7:1005–1008. doi: 10.1097/00001756-199604100-00010. [DOI] [PubMed] [Google Scholar]
- 58.Miyata M., Smith J. D. Nat. Genet. 1996;14:55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- 59.Lauderback C. M., Kanski J., Hackett J. M., Maeda N., Kindy M. S., Butterfield D. A. Brain Res. 2002;924:90–97. doi: 10.1016/s0006-8993(01)03228-0. [DOI] [PubMed] [Google Scholar]
- 60.Mazur-Kolecka B., Frackowiak J., Kowal D., Krzeslowska J., Dickson D. NeuroReport. 2002;13:465–468. doi: 10.1097/00001756-200203250-00021. [DOI] [PubMed] [Google Scholar]
- 61.Gong J.-S., Kobayashi M., Hayashi H., Zou K., Sawamura N., Fujita S. C., Yanagisawa K., Michikawa M. J. Biol. Chem. 2002;277:29919–29926. doi: 10.1074/jbc.M203934200. [DOI] [PubMed] [Google Scholar]
- 62.Fagan A. M., Holtzman D. M., Munson G., Mathur T., Schneider D., Chang L. K., Getz G. S., Reardon C. A., Lukens J., Shah J. A., et al. J. Biol. Chem. 1999;274:30001–30007. doi: 10.1074/jbc.274.42.30001. [DOI] [PubMed] [Google Scholar]
- 63.LaDu M. J., Falduto M. T., Manelli A. M., Reardon C. A., Getz G. S., Frail D. E. J. Biol. Chem. 1994;269:23403–23406. [PubMed] [Google Scholar]
- 64.Holtzman D. M., Bales K. R., Tenkova T., Fagan A. M., Parsadanian M., Sartorius L. J., Mackey B., Olney J., McKeel D., Wozniak D., et al. Proc. Natl. Acad. Sci. USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tesseur I., Van Dorpe J., Spittaels K., Van den Haute C., Moechars D., Van Leuven F. Am. J. Pathol. 2000;156:951–964. doi: 10.1016/S0002-9440(10)64963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tesseur I., Van Dorpe J., Bruynseels K., Bronfman F., Sciot R., Van Lommel A., Van Leuven F. Am. J. Pathol. 2000;157:1495–1510. doi: 10.1016/S0002-9440(10)64788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y., Liu X. Q., Wyss-Coray T., Brecht W. J., Sanan D. A., Mahley R. W. Proc. Natl. Acad. Sci. USA. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buttini M., Akeefe H., Lin C., Mahley R. W., Pitas R. E., Wyss-Coray T., Mucke L. Neuroscience. 2000;97:207–210. doi: 10.1016/s0306-4522(00)00069-5. [DOI] [PubMed] [Google Scholar]
- 69.Raber J., Wong D., Buttini M., Orth M., Bellosta S., Pitas R. E., Mahley R. W., Mucke L. Proc. Natl. Acad. Sci. USA. 1998;95:10914–10919. doi: 10.1073/pnas.95.18.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raber J., Bongers G., LeFevour A., Buttini M., Mucke L. J. Neurosci. 2002;22:5204–5209. doi: 10.1523/JNEUROSCI.22-12-05204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marques M. A., Tolar M., Harmony J. A. K., Crutcher K. A. NeuroReport. 1996;7:2529–2532. doi: 10.1097/00001756-199611040-00025. [DOI] [PubMed] [Google Scholar]
- 72.Tolar M., Marques M. A., Harmony J. A. K., Crutcher K. A. J. Neurosci. 1997;17:5678–5686. doi: 10.1523/JNEUROSCI.17-15-05678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolar M., Keller J. N., Chan S., Mattson M. P., Marques M. A., Crutcher K. A. J. Neurosci. 1999;19:7100–7110. doi: 10.1523/JNEUROSCI.19-16-07100.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ji Z.-S., Miranda R. D., Newhouse Y. M., Weisgraber K. H., Huang Y., Mahley R. W. J. Biol. Chem. 2002;277:21821–21828. doi: 10.1074/jbc.M112109200. [DOI] [PubMed] [Google Scholar]
- 75.Bales K. R., Verina T., Cummins D. J., Du Y., Dodel R. C., Saura J., Fishman C. E., DeLong C. A., Piccardo P., Petegnief V., et al. Proc. Natl. Acad. Sci. USA. 1999;96:15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irizarry M. C., Cheung B. S., Rebeck G. W., Paul S. M., Bales K. R., Hyman B. T. Acta Neuropathol. 2000;100:451–458. doi: 10.1007/s004010000263. [DOI] [PubMed] [Google Scholar]
- 77.Trommer B. L., Shah C., Yun S. H., Gamkrelidze G., Pasternak E. S., Ye G. L., Sotak M., Sullivan P. M., Pasternak J. F., LaDu M. J. NeuroReport. 2004;15:2655–2658. doi: 10.1097/00001756-200412030-00020. [DOI] [PubMed] [Google Scholar]
- 78.Mahley R. W., Huang Y. Curr. Opin. Lipidol. 1999;10:207–217. doi: 10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 79.Huang Y., Weisgraber K. H., Mucke L., Mahley R. W. J. Mol. Neurosci. 2004;23:189–204. doi: 10.1385/JMN:23:3:189. [DOI] [PubMed] [Google Scholar]
- 80.Mahley R. W., Huang Y., Rall S. C., Jr. J. Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- 81.Weisgraber K. H. J. Lipid Res. 1990;31:1503–1511. [PubMed] [Google Scholar]
- 82.Morrow J. A., Hatters D. M., Lu B., Höchtl P., Oberg K. A., Rupp B., Weisgraber K. H. J. Biol. Chem. 2002;277:50380–50385. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 83.Dong L.-M., Wilson C., Wardell M. R., Simmons T., Mahley R. W., Weisgraber K. H., Agard D. A. J. Biol. Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 84.Dong L.-M., Weisgraber K. H. J. Biol. Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 85.Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 86.Xu Q., Brecht W. J., Weisgraber K. H., Mahley R. W., Huang Y. J. Biol. Chem. 2004;279:25511–25516. doi: 10.1074/jbc.M311256200. [DOI] [PubMed] [Google Scholar]
- 87.Hatters D. M., Budamagunta M. S., Voss J. C., Weisgraber K. H. J. Biol. Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- 88.Raffaï R. L., Dong L.-M., Farese R. V., Jr., Weisgraber K. H. Proc. Natl. Acad. Sci. USA. 2001;98:11587–11591. doi: 10.1073/pnas.201279298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taylor J. P., Hardy J., Fischbeck K. H. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 90.Morrow J. A., Segall M. L., Lund-Katz S., Phillips M. C., Knapp M., Rupp B., Weisgraber K. H. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 91.Dobson C. M. Philos. Trans. R. Soc. London B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ptitsyn O. B. Adv. Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 93.Elshourbagy N. A., Liao W. S., Mahley R. W., Taylor J. M. Proc. Natl. Acad. Sci. USA. 1985;82:203–207. doi: 10.1073/pnas.82.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ignatius M. J., Gebicke-Härter P. J., Skene J. H. P., Schilling J. W., Weisgraber K. H., Mahley R. W., Shooter E. M. Proc. Natl. Acad. Sci. USA. 1986;83:1125–1129. doi: 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boyles J. K., Zoellner C. D., Anderson L. J., Kosik L. M., Pitas R. E., Weisgraber K. ., Hui D. Y., Mahley R. W., Gebicke-Haerter P. J., Ignatius M. J., et al. J. Clin. Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ignatius M. J., Shooter E. M., Pitas R. E., Mahley R. W. Science. 1987;236:959–962. doi: 10.1126/science.3576212. [DOI] [PubMed] [Google Scholar]
- 97.Popko B., Goodrum J. F., Bouldin T. W., Zhang S. H., Maeda N. J. Neurochem. 1993;60:1155–1158. doi: 10.1111/j.1471-4159.1993.tb03268.x. [DOI] [PubMed] [Google Scholar]
- 98.Fullerton S. M., Strittmatter W. J., Matthew W. D. Exp. Neurol. 1998;153:156–163. doi: 10.1006/exnr.1998.6872. [DOI] [PubMed] [Google Scholar]
- 99.Boyles J. K., Pitas R. E., Wilson E., Mahley R. W., Taylor J. M. J. Clin. Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pitas R. E., Boyles J. K., Lee S. H., Foss D., Mahley R. W. Biochim. Biophys. Acta. 1987;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- 101.Diedrich J. F., Minnigan H., Carp R. I., Whitaker J. N., Race R., Frey W., Jr., Haase A. T. J. Virol. 1991;65:4759–4768. doi: 10.1128/jvi.65.9.4759-4768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han S.-H., Einstein G., Weisgraber K. H., Strittmatter W. J., Saunders A. M., Pericak-Vance M., Roses A. D., Schmechel D. E. J. Neuropathol. Exp. Neurol. 1994;53:535–544. doi: 10.1097/00005072-199409000-00013. [DOI] [PubMed] [Google Scholar]
- 103.Bao F., Arai H., Matsushita S., Higuchi S., Sasaki H. NeuroReport. 1996;7:1733–1739. doi: 10.1097/00001756-199607290-00008. [DOI] [PubMed] [Google Scholar]
- 104.Beffert U., Poirier J. Ann. N.Y. Acad. Sci. 1996;777:166–174. doi: 10.1111/j.1749-6632.1996.tb34415.x. [DOI] [PubMed] [Google Scholar]
- 105.Metzger R. E., LaDu M. J., Pan J. B., Getz G. S., Frail D. E., Falduto M. T. J. Neuropathol. Exp. Neurol. 1996;55:372–380. doi: 10.1097/00005072-199603000-00013. [DOI] [PubMed] [Google Scholar]
- 106.Xu P.-T., Gilbert J. R., Qiu H.-L., Ervin J., Rothrock-Christian T. R., Hulette C., Schmechel D. E. Am. J. Pathol. 1999;154:601–611. doi: 10.1016/S0002-9440(10)65305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu P.-T., Gilbert J. R., Qiu H.-L., Rothrock-Christian T., Settles D. L., Roses A. D., Schmechel D. E. Neurosci. Lett. 1998;246:65–68. doi: 10.1016/s0304-3940(98)00247-x. [DOI] [PubMed] [Google Scholar]
- 108.Xu P.-T., Schmechel D., Qiu H.-L., Herbstreith M., Rothrock-Christian T., Eyster M., Roses A. D., Gilbert J. R. Neurobiol. Dis. 1999;6:63–75. doi: 10.1006/nbdi.1998.0213. [DOI] [PubMed] [Google Scholar]
- 109.Xu P.-T., Schmechel D., Rothrock-Christian T., Burkhart D. S., Qiu H.-L., Popko B., Sullivan P., Maeda N., Saunders A. M., Roses A. D., et al. Neurobiol. Dis. 1996;3:229–245. doi: 10.1006/nbdi.1996.0023. [DOI] [PubMed] [Google Scholar]
- 110.Harris F. M., Tesseur I., Brecht W. J., Xu Q., Mullendorff K., Chang S., Wyss-Coray T., Mahley R. W., Huang Y. J. Biol. Chem. 2004;279:3862–3868. doi: 10.1074/jbc.M309475200. [DOI] [PubMed] [Google Scholar]
- 111.Boschert U., Merlo-Pich E., Higgins G., Roses A. D., Catsicas S. Neurobiol. Dis. 1999;6:508–514. doi: 10.1006/nbdi.1999.0251. [DOI] [PubMed] [Google Scholar]
- 112.Aoki K., Uchihara T., Sanjo N., Nakamura A., Ikeda K., Tsuchiya K., Wakayama Y. Stroke. 2003;34:875–880. doi: 10.1161/01.STR.0000064320.73388.C6. [DOI] [PubMed] [Google Scholar]
- 113.Dekroon R. M., Armati P. J. Glia. 2001;33:298–305. doi: 10.1002/1098-1136(20010315)33:4<298::aid-glia1028>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 114.Dupont-Wallois L., Soulié C., Sergeant N., Wavrant-de Wrieze N., Chartier-Harlin M.-C., Delacourte A., Caillet-Boudin M.-L. Neurobiol. Dis. 1997;4:356–364. doi: 10.1006/nbdi.1997.0155. [DOI] [PubMed] [Google Scholar]
- 115.Ferreira S., Dupire M.-J., Delacourte A., Najib J., Caillet-Boudin M.-L. Exp. Neurol. 2000;166:415–421. doi: 10.1006/exnr.2000.7510. [DOI] [PubMed] [Google Scholar]
- 116.Soulié C., Mitchell V., Dupont-Wallois L., Chartier-Harlin M.-C., Beauvillain J.-C., Delacourte A., Caillet-Boudin M.-L. Neurosci. Lett. 1999;265:147–150. doi: 10.1016/s0304-3940(99)00167-6. [DOI] [PubMed] [Google Scholar]
- 117.Pitas R. E., Boyles J. K., Lee S. H., Hui D., Weisgraber K. H. J. Biol. Chem. 1987;262:14352–14360. [PubMed] [Google Scholar]
- 118.Nathan B. P., Chang K.-C., Bellosta S., Brisch E., Ge N., Mahley R. W., Pitas R. E. J. Biol. Chem. 1995;270:19791–19799. doi: 10.1074/jbc.270.34.19791. [DOI] [PubMed] [Google Scholar]
- 119.Selkoe D. J. Neuron. 2001;32:177–180. doi: 10.1016/s0896-6273(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 120.Hardy J., Selkoe D. J. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 121.Strittmatter W. J., Weisgraber K. H., Huang D. Y., Dong L.-M., Salvesen G. S., Pericak-Vance M., Schmechel D., Saunders A. M., Goldgaber D., Roses A. D. Proc. Natl. Acad. Sci. USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sanan D. A., Weisgraber K. H., Russell S. J., Mahley R. W., Huang D., Saunders A., Schmechel D., Wisniewski T., Frangione B., Roses A. D., et al. J. Clin. Invest. 1994;94:860–869. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma J., Yee A., Brewer H. B., Jr., Das S., Potter H. Nature. 1994;372:92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- 124.Wisniewski T., Castaño E. M., Golabek A., Vogel T., Frangione B. Am. J. Pathol. 1994;145:1030–1035. [PMC free article] [PubMed] [Google Scholar]
- 125.Ye S., Huang Y., Müllendorff K., Dong L., Giedt G., Meng E. C., Cohen F. E., Kuntz I. D., Weisgraber K. H., Mahley R. W. Proc. Natl. Acad. Sci. USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ditaranto K., Tekirian T. L., Yang A. J. Neurobiol. Dis. 2001;8:19–31. doi: 10.1006/nbdi.2000.0364. [DOI] [PubMed] [Google Scholar]
- 127.Ji Z.-S., Müllendorff K., Cheng I. H., Miranda R. D., Huang Y., Mahley R. W. J. Biol. Chem. 2006;281:2683–2692. doi: 10.1074/jbc.M506646200. [DOI] [PubMed] [Google Scholar]
- 128.Harris F. M., Brecht W. J., Xu Q., Tesseur I., Kekonius L., Wyss-Coray T., Fish J. D., Masliah E., Hopkins P. C., Scearce-Levie K., et al. Proc. Natl. Acad. Sci. USA. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brecht W. J., Harris F. M., Chang S., Tesseur I., Yu G.-Q., Xu Q., Fish J. D., Wyss-Coray T., Buttini M., Mucke L., et al. J. Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang S., Ma T. R., Miranda R. D., Balestra M. E., Mahley R. W., Huang Y. Proc. Natl. Acad. Sci. USA. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Derossi D., Chassaing G., Prochiantz A. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 132.Hawiger J. Curr. Opin. Chem. Biol. 1999;3:89–94. doi: 10.1016/s1367-5931(99)80016-7. [DOI] [PubMed] [Google Scholar]
- 133.Lindgren M., Hällbrink M., Prochiantz A., Langel Uuml;. Trends Pharmacol. Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 134.Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 135.Mann D. A., Frankel A. D. EMBO J. 1991;10:1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vivès E., Brodin P., Lebleu B. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 137.Aints A., Güven H., Gahrton G., Smith C. I. E., Dilber M. S. Gene Ther. 2001;8:1051–1056. doi: 10.1038/sj.gt.3301493. [DOI] [PubMed] [Google Scholar]
- 138.Martin A., O’Hare P., McLauchlan J., Elliott G. J. Virol. 2002;76:4961–4970. doi: 10.1128/JVI.76.10.4961-4970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fawell S., Seery J., Daikh Y., Moore C., Chen L. L., Pepinsky B., Barsoum J. Proc. Natl. Acad. Sci. USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cao G., Pei W., Ge H., Liang Q., Luo Y., Sharp F. R., Lu A., Ran R., Graham S. H., Chen J. J. Neurosci. 2002;22:5423–5431. doi: 10.1523/JNEUROSCI.22-13-05423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miura S.-I., Okamoto T., Via D. P., Saku K. Circ. J. 2002;66:1054–1056. doi: 10.1253/circj.66.1054. [DOI] [PubMed] [Google Scholar]
- 142.Liu K., Ou J., Saku K., Jimi S., Via D. P., Sparrow J. T., Zhang B., Pownall H. J., Smith L. C., Arakawa K. Arterioscler. Thromb. Vasc. Biol. 1999;19:2207–2213. doi: 10.1161/01.atv.19.9.2207. [DOI] [PubMed] [Google Scholar]
- 143.Gibson G. E., Haroutunian V., Zhang H., Park L. C. H., Shi Q., Lesser M., Mohs R. C., Sheu R. K.-F., Blass J. P. Ann. Neurol. 2000;48:297–303. [PubMed] [Google Scholar]
- 144.Mahley R. W., Hui D. Y., Innerarity T. L., Beisiegel U. Arteriosclerosis. 1989;9:I-14–I-18. [PubMed] [Google Scholar]
- 145.Li Z., Okamoto K.-I., Hayashi Y., Sheng M. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 146.Reynolds I. J., Rintoul G. L. Sci. STKE. 2004:pe46. doi: 10.1126/stke.2512004pe46. [DOI] [PubMed] [Google Scholar]
- 147.Pfanner N., Wiedemann N. Curr. Opin. Cell Biol. 2002;14:400–411. doi: 10.1016/s0955-0674(02)00355-1. [DOI] [PubMed] [Google Scholar]
- 148.Young J. C., Hoogenraad N. J., Hartl F. U. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 149.Vance J. E. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 150.Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., Pozzan T. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 151.Schwer B., Ren S., Pietschmann T., Kartenbeck J., Kaehlcke K., Bartenschlager R., Yen T. S. B., Ott M. J. Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Strittmatter W. J., Saunders A. M., Goedert M., Weisgraber K. H., Dong L.-M., Jakes R., Huang D. Y., Pericak-Vance M., Schmechel D., Roses A. D. Proc. Natl. Acad. Sci. USA. 1994;91:11183–11186. doi: 10.1073/pnas.91.23.11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kuntz I. D. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 154.Meng E. C., Shoichet B. K., Kuntz I. D. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- 155.Ewing T. J. A., Kuntz I. D. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- 156.Knegtel R. M. A., Kuntz I. D., Oshiro C. M. J. Mol. Biol. 1997;266:424–440. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- 157.Ewing T. J. A., Makino S., Skillman A. G., Kuntz I. D. J. Comput. Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 158.Di Marco S., Volpari C., Tomei L., Altamura S., Harper S., Narjes F., Koch U., Rowley M., De Francesco R., Migliaccio G., et al. J. Biol. Chem. 2005;280:29765–29770. doi: 10.1074/jbc.M505423200. [DOI] [PubMed] [Google Scholar]
- 159.Pedemonte N., Lukacs G. L., Du K., Caci E., Zegarra-Moran O., Galietta L. J. V., Verkman A. S. J. Clin. Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wiseman R. L., Green N. S., Kelly J. W. Biochemistry. 2005;44:9265–9274. doi: 10.1021/bi050352o. [DOI] [PubMed] [Google Scholar]
- 161.Fan J.-Q., Ishii S., Asano N., Suzuki Y. Nat. Med. 1999;5:112–115. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 162.Ishii S., Yoshioka H., Mannen K., Kulkarni A. B., Fan J.-Q. Biochim. Biophys. Acta. 2004;1690:250–257. doi: 10.1016/j.bbadis.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 163.Pagliaro L., Felding J., Audouze K., Nielsen S. J., Terry R. B., Krog-Jensen C., Butcher S. Curr. Opin. Chem. Biol. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.