Abstract
Variation among populations in gene expression should be related to the accumulation of random-neutral changes and evolution by natural selection. The following evolutionary analysis has general applicability to biological and medical science because it accounts for genetic relatedness and identifies patterns of expression variation that are affected by natural selection. To identify genes evolving by natural selection, we allocate the maximum among-population variation to genetic distance and then examine the remaining variation relative to a hypothesized important ecological parameter (temperature). These analyses measure the expression of metabolic genes in common-gardened populations of the fish Fundulus heteroclitus whose habitat is distributed along a steep thermal gradient. Although much of the variation in gene expression fits a null model of neutral drift, the variation in expression for 22% of the genes that regress with habitat temperature was far greater than could be accounted for by genetic distance alone. The most parsimonious explanation for among-population variation for these genes is evolution by natural selection. In addition, many metabolic genes have patterns of variation incongruent with neutral evolution: They have too much or too little variation. These patterns of biological variation in expression may reflect important physiological or ecological functions.
Keywords: evolutionary analysis, Fundulus, microarray, phylogenetic comparative approach, genomics
Gene expression has been hypothesized to be of adaptive importance (1), and heritable variation that affects fitness is necessary for evolution by natural selection. Although adaptive differences in expression have been identified in single-gene studies (2–6; see ref. 7 for review), microarray approaches offer great promise to rigorously address this hypothesis because they assay many loci at once. Furthermore, it is generally agreed that much of variation in gene expression for a particular environmental condition has a genetic basis (8, 9) according to studies in yeast (10–13), Drosophila (14–18), mice (19), and humans (20–22).
Widespread heritable variation combined with extensive natural variation in gene expression revealed by microarray studies (11, 13, 14, 23–26) provides the substrates for evolution. However, two evolutionary forces govern the variance of traits among taxa: neutral drift and natural selection. Under a neutral drift model, the variation in a trait has little biological effect§ and is a function of genetic distance: Traits will be more similar among closely related taxa than among more distantly related taxa (27–29). If natural selection has occurred, the variation in a trait affects an organism’s fitness and is a function of the ecological setting: Traits are conserved or diverge depending on the specific ecological pressures (30). Recent studies suggest that much of the extensive variation in gene expression among individuals and taxa is simply random neutral divergence (31, 32), whereas others have found extensive hallmarks of selection (16, 18, 33). However, no specific adaptive hypotheses have been applied to microarray data to refute or substantiate these claims. Here we apply a correction for genetic relatedness (i.e., the phylogenetic comparative method; refs. 34–36) to reveal variation most parsimoniously accounted for by neutral evolution. After removing the neutral phylogenetic effects, remaining variation significantly associated with an independent ecological factor, such as habitat temperature, would be most parsimoniously accounted for by natural selection. This approach is conservative because it attributes maximum among-population variation to genetic distance before considering natural selection.
This approach was applied to Fundulus heteroclitus, a teleost fish widely distributed along the Atlantic coast of North America, where there is a change of 1°C per degree latitude or ≈12.5°C between Maine and Georgia (37). In this species, there is evidence for local adaptation to this clinal variation in temperature (3, 37–39). Also, adaptation may be more prevalent in this species because of large local populations (census population size >10,000 for a single estuarine creek, Ne > 105; ref. 65) in which small selective pressures should dominate the effects of genetic drift. However, because many ecological factors may covary with temperature along the latitudinal gradient from Maine to Georgia, caution should be exercised in identifying only temperature as the specific agent of natural selection (30). Thus, temperature is considered a proxy for environmental variation that we predicted affects the evolution of gene expression.
Evolution by natural selection requires heritable variation in traits that affect fitness. Although the heritable variation among individuals in gene expression was not directly ascertained, all individuals were subject to common conditions and acclimated (see Methods) for >2 months. Acclimation is well studied (40, 41), and acclimating fish to a common environment minimizes physiological differences caused by differences in an animal’s native habitat. Thus, much of the variation in gene expression is unlikely to be due to the native habitat temperature and more likely represents both genetic and other random biological sources of variation. Although raising animals at one common temperature reduces environmental influences, it ignores gene-by-environment interactions. Thus, heritable differences due to complex interactions are not ascertained.
Results
Population Genetics.
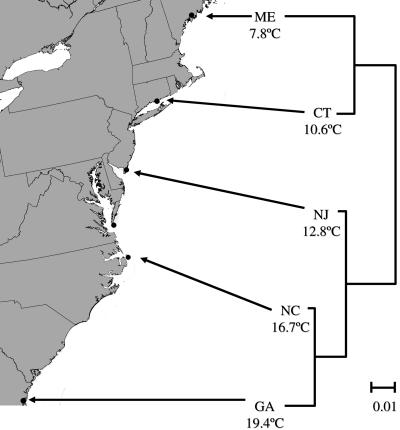
Individuals for this study were collected from Maine, Connecticut, New Jersey, North Carolina, and Georgia, and extensive structure existed among populations (Fig. 1). Population genotyping by using microsatellite markers yielded pairwise FST estimates ranging from 0.01 to 0.24 (all statistically significant, P < 0.05). The neighbor-joining tree (Fig. 1) supports previous analyses of these populations, which show a break between northern and southern groups at the Hudson River (43–45). These data also indicate significant isolation-by-distance (Mantel test, 1,000 permutations: P = 0.024, r = 0.65). Thus, the spatially separated groups appear to follow independent demographic trajectories, allowing application of phylogenetic analyses (35).
Fig. 1.
Fundulus habitat sites and microsatellite-derived neighbor-joining tree. Collection sites along the United States Atlantic seaboard: ME, Wiscasset, ME; CT, Clinton, CT; NJ, Stone Harbor, NJ; NC, Roanoke Island, NC; GA, Sapelo Island, GA; corresponding median annual habitat temperatures (°C) are averaged over 30 years. Dendrogram is a neighbor-joining tree constructed from pairwise Cavalli-Sforza and Edwards’ chord distances (42) calculated from microsatellite allele frequencies.
Gene Expression Variation Among Individuals and Populations.
Expression for 329 genes involved in central metabolic pathways was measured by using eight technical replicates for each of five males per population sampled from five populations (n = 25). The mean coefficient of variation (CV; equals SD/mean) for technical replication was 2.54%, with >95% of genes with CV < 5%. A list of all genes and results from all statistical tests can be found as Table 2, which is published as supporting information on the PNAS web site. All fish used appeared healthy and unstressed. Because fish were raised in a common laboratory setting for 2 months, differences are likely not due to acclimatization to native habitat (40, 41). Differences in body weight or sex are unlikely sources of significant variation because body weight does not affect gene expression in this species (25) and only postreproductive males were used. Additionally, when blood was sampled four times over a 6-week period from five different individuals, there was little additional variation (D.L.C., unpublished data). These results demonstrate that RNA isolation, array hybridization, and random biological differences (e.g., temporal variation in the physiological state of individuals) contribute little additional variation. Instead we suggest that much of the variation is more likely due to genetic sources of variation. Alternatively, epigenetic or early developmental effects could establish an irreversible phenotype and cannot be ruled out. However, given that much of gene expression variation is heritable (8), it seems more parsimonious to assume a genetic basis (although this basis remains to be verified). Other sources of variation (e.g., random biological differences, differences in social status, and general well being) seem unlikely to covary with temperature and, thus, would make any significant relationship more difficult to discern.
Gene expression varied extensively among individuals within populations (69%, 227 of 329 genes at P < 0.05; false discovery rate <1%). False discovery rate (FDR) estimates the proportion of tests that reject the null hypothesis that are false; thus with an FDR of <1%, ≈2 of the 227 significant differences are false positives. False discovery rates were estimated as described by Storey and Tibshirani (46) with the application of qvalue software provided at http://faculty.washington.edu/∼jstorey/qvalue/index.html.
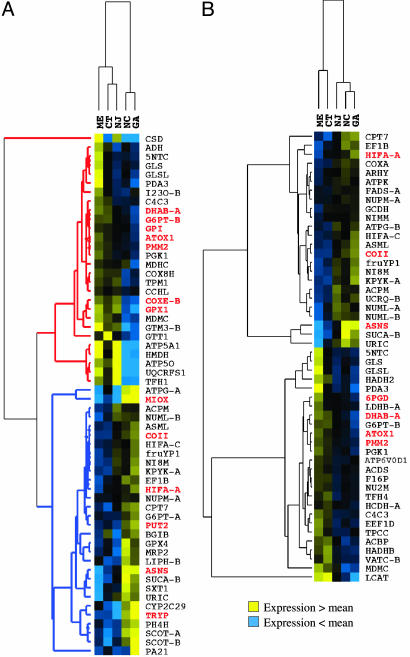
Among populations, 12%, or 41 of 329 genes, were significantly different (P < 0.05; FDR = 22%). This relatively large FDR suggests that many of the 41 differences may be false positives, and, thus, the difference among populations may be as low as 10% of all measured genes. Regression analyses suggest that habitat temperature (see Methods) accounted for significant variation for 18% of genes (58 of 329, FDR = 21%; average r2 = 0.68). Of these 58 genes, approximately equal numbers increase (28) or decrease (30) in expression with colder latitudes (Fig. 2A). Among these genes that regress significantly with temperature, the clustering of populations (top of Fig. 2A) is similar to the topology of the genetic distance neighbor-joining tree (Fig. 1). The apparent covariance between habitat temperature differences and genetic distances illustrates the problem of relatedness among taxa: Either of these factors could be responsible for the divergence in gene expression among populations.
Fig. 2.
Hierarchical clustering indicating genes with ecological (habitat temperature) and phylogenetic components to among-population variation in expression. Patterns of expression [yellow and blue represent expression greater than (yellow) and less than (blue) grand mean, respectively] for genes that correlate with habitat temperature (A) and that correlate with genetic distance (B). In A, gene clusters reflect decreases (red dendrogram) or increases (blue dendrogram) in expression with habitat temperature. Red bolded gene names are those that regress with habitat temperature after correction of observations for expected nonindependence due to phylogeny (phylogenetic generalized least squares method) and, thus, appear to be evolving by natural selection.
In contrast to the patterns of the temperature genes indicating ecological effect, genetic distance relationships (phylogeny) accounted for a significant proportion of among-population expression variation for 15% of genes (50 of 329 genes, FDR = 18%; Mantel test). Identification of genes with a phylogenetic component by the physig program (47, 48) (a permutation test that examines correlation between trait values and phylogeny branch lengths) corroborated Mantel results (Pearson’s correlation coefficient = 0.84; P < 0.001). The most parsimonious explanation for the among-population variation in expression for this subset of genes is random genetic drift because patterns of gene expression correlate with genetic distance. Not surprisingly, hierarchical clustering of the genes with a significant phylogenetic signal (Fig. 2B) produces a dendrogram identical to the genetic distance neighbor-joining tree topology (Fig. 1). Natural selection also may contribute to this phylogenetic pattern because habitat temperature covaries with genetic distance, but as in other analyses (31, 32), the most parsimonious explanation for this pattern is neutral drift.
Evolutionary Analysis.
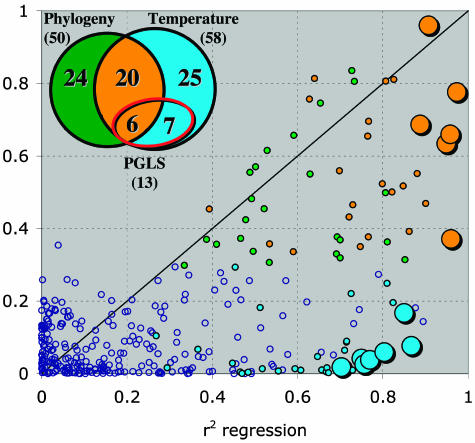
To determine whether natural selection is affecting patterns of gene expression requires evidence of departure from neutrality. This evidence would include a significant covariation between gene expression and native habitat temperature after removing gene-specific phylogenetic effects. Temperature was chosen because of the steep thermal cline in habitat temperature and previous research indicating adaptation to temperature (3, 37–39). Approximately half of the 58 genes that have a significant habitat temperate component also have a significant phylogenetic component (Fig. 3). To correct among-population expression data for nonindependence due to phylogeny for each gene separately (48, 49), a matrix of expected covariances among populations was constructed by using branch lengths of the microsatellite-derived dendrogram. After removing phylogenetic effects, the expression levels of 13 genes (22% of the 58 temperature genes) significantly regressed with habitat temperature (Fig. 3) (1,000 random permutations; FDR = 3%). Many of these genes appear related to temperature variation in other data sets. For example, 6-phosphogluconate dehydrogenase (6PGD), cold-inducible RNA-binding protein (CIRP), cytochrome C oxidase (COXE), δ-1-pyrroline-5-carboxylate dehydrogenase (PUT2), glucose-6-phosphate isomerase (GPI), glutathione peroxidase (GPX1), hypoxia-inducible factor (HIFA), NADH-ubiquinone oxidoreductase (NUAM), nucleoside diphosphate kinase (NDKB), and phosphomannomutase (PMM2) appear to be involved in temperature acclimation responses in carp (50), and glucose-6-phosphate 1-dehydrogenase (G6PT) and NUAM are associated with adaptive patterns of variation within and among Fundulus species (24). Whether these 13 loci have evolved independently or, in the extreme, are governed by a single variant at a transacting locus that only affects this subset of genes, is not addressed in this study.
Fig. 3.
Relationships between ecological and phylogenetic effects on among-population variation in gene expression. For each gene, the explained variation (r2) for phylogeny (genetic distance based on Cavalli-Sforza and Edwards’ chord distances (42) calculated from microsatellite allele frequencies) is plotted against the explained variation (r2) for habitat temperatures. Venn diagram is for the numbers of genes that have significant regression with habitat temperature (blue), phylogeny (green), or both temperature and phylogeny (orange). Colors of spots in graph correspond to Venn diagram. Enlarged spots are the 13 genes that regress significantly with habitat temperature after correcting for phylogeny (red circle; Venn diagram) by using the phylogenetic generalized least squares (PGLS) approach and, thus, appear to be evolving by natural selection.
Discussion
Using Fundulus microarray data, we describe an approach to distinguish between neutral and adaptive evolutionary processes affecting gene expression. Our approach is different from other comparative analyses of microarray data. It has recently been argued that higher variation among versus within taxa (equivalent to the F statistic) is indicative of natural selection (18). We extend this proposal by suggesting that variation among taxa accounted for by genetic distance is most parsimoniously explained by neutral drift, and only variation that exceeds this phylogenetic variance may be considered indicative of natural selection. Others have corrected the ratio of variance among versus within taxa for divergence time (33) or population size (16) with an arbitrary cutoff to identify the influence of natural selection. The analyses reported here differ from other attempts to define directional selection because the experimental design statistically tests for selection in an ecological context after accounting for the nonindependence of samples due to relatedness (34). Notice here that the detection of natural selection does not depend on a constant, an arbitrary cutoff value, or a rate. Instead, after accounting for the contribution of genetic distance for each gene separately, the residual variation must significantly regress with an ecological factor to reject of the neutral drift null hypothesis.
The identification of 22% of temperature-related genes as adaptive is conservative. Phylogenetic correction is not a technical improvement of comparative analyses per se, but rather a conceptual decision to prioritize random genetic drift over ecology as the correlate for trait variation (51). The comparative approach used here to identify these temperature-related genes assumes that ecological forces act only on the remaining residual variation after accounting for phylogenetic divergence and, thus, is highly conservative for identifying traits evolving by directional selection (51). Although half of the genes that correlate with habitat temperature also correlate with genetic distance, for the 13 genes where the variation accounted for by habitat temperature is far greater than can be accounted for by genetic distance alone, evolution by natural selection is the most parsimonious explanation.
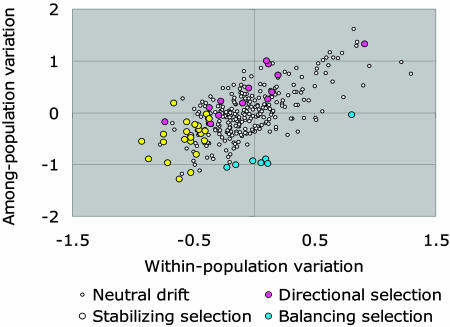
Eighty-two genes (25%) have patterns of variation accounted for by phylogeny, habitat temperature, or both. In contrast, what (if anything) accounts for patterns of expression variation in the remaining 75% of genes? The adaptive pattern referred to above for 13 temperature genes suggests directional or divergent selection, yet as others have pointed out (16, 18, 33), stabilizing and balancing selection also can influence patterns of gene expression. The neutral theory suggests that with greater constraints (stabilizing selection; ref. 52), one should expect low variation among individuals and among populations (18). A formal method to identify genes most influenced by stabilizing selection would calculate variation among all individuals across all populations, then test which genes have significantly less variation compared with all other genes. We do this calculation by comparing the variation for a gene versus the variation for all other genes: an F ratio with variance in expression among all individuals for all genes as the numerator, and variance in expression among individuals for a single gene as the denominator (expression levels are standardized such that the mean expression for each gene is equal). An additional criterion would be no correlation between among-population differences in expression and genetic distance. We identify 24 genes that have disproportionately low variance in expression among individuals and populations (Bonferroni-corrected P < 0.01; Fig. 4). These genes with the least variation in expression are disproportionately represented by genes involved in oxidative phosphorylation (10 of the 24 significant genes with low variance, P < 0.05; Fisher’s exact test), suggesting the biological importance of maintaining tight regulation for expression in this pathway. These central metabolism genes are medically important. For example, variation in oxidative phosphorylation genes contributes to inherited human diseases (53, 54).
Fig. 4.
Gene expression variation within and among populations indicating different patterns of evolutionary divergence. Plotted are the log of within- and among-population variation for gene expression. The ratios of these values are often used in statistical tests (e.g., ANOVA). Under neutral drift, within-population variation is correlated with among population variation (open circles). For other genes, different forms of selection overwhelm the general patterns indicated by drift and reject this null model. Genes under directional selection (pink circles) were identified as divergent along a habitat temperature gradient after correcting for variance due to phylogeny (phylogenetic generalized least squares method), and have higher variation among populations than within. Genes most influenced by stabilizing selection (yellow circles) have lower variation both within and among populations than most genes (F test), and genes under balancing selection (blue circles) have higher variation within than among populations (inverted F test).
In addition to directional selection, another pattern in conflict with the predictions of neutral evolution are genes for which within-population variation is higher than among-population variation (18). We propose testing for this pattern by calculating the inverse F statistic (the variation within populations versus variation among populations). Seven genes have significantly greater variation within a population than among the population (Fig. 4), indicative of balancing selection.
In microarray studies, genes with little biological variance in expression (suggestive of stabilizing selection) or genes with high variance within populations but little variance among populations (suggestive of balancing selection) are rarely considered. Two other attempts to recognize stabilizing selection examined the variation among taxa versus within (16, 33) while taking into account either divergence time or population size, and both indicated extensive stabilizing selection (up to 100% of all genes). However, stabilizing selection interacts with drift to influence the variation of traits along a continuum from high constraints minimizing the variation caused by drift (dominance of stabilizing selection) to a high variation allowable by fewer constraints (dominance of neutral drift). As such, binning traits as those influenced by drift or stabilizing selection is unnecessarily arbitrary. Rather, we propose the application of statistics that identify traits more or less affected by these evolutionary forces along the continuum. The application of evolutionary theory to identify genes with expression variation under strong constraints could be useful in medical genetics, and identifying genes under balancing selection could contribute to the debate over how variation is maintained in populations (55).
Evolutionary analyses provide a powerful approach for identifying genes with expression patterns that are of general interest to biologists or medical science. For the data presented here, among-population variation was positively correlated with within-population variation (P < 0.001; product-moment correlation), supporting the neutral prediction of gene expression evolution (32). Certainly, because of the costs of selection (summarized in the concept of genetic load; ref. 56), it seems unreasonable to expect that many, or even a significant minority of, genes can be subject to the effects of natural selection. Studies have indicated that most expression variation between humans and apes and mice is selectively neutral (31, 32), but these comparisons lacked an ecological context and, thus, could not test alternative hypotheses for the small subset of genes that may be affected by natural selection. In contrast, within the ecological context of ocean-depth gradients, natural selection has been important for shaping metabolic variation among species, genera, families, and phyla of marine organisms (57, 58), and phylogeny appears to account for little metabolic variation among these same groups. Notice also that detecting phylogenetic signals in comparative gene expression data may be best accomplished by comparing closely related taxa because the character space in which traits may vary is finite, and the influence of drift may become less obvious as divergence increases. For example, we detect expression levels that vary >2-fold among populations due to drift [e.g., asparagine synthetase (ASNS), phosphatidylcholine-sterol acyltransferase (LCAT), and succinyl-CoA ligase (SUCA)], but we would not expect variance to continuously increase with phylogenetic divergence because of eventual functional constraints. That is, ever-increasing differences in gene expression will not occur if for no other reason than because there is a limit to the amount of mRNA that can be produced. With increasing genetic distance, phenotypic divergence among taxa may become nonlinear (59).
Although phylogeny accounted for much of the variation among populations of F. heteroclitus distributed along a strong habitat gradient, variation for 13 of 58 genes that regress with habitat temperature exceeded that which could be accounted for by phylogeny alone and is most parsimoniously explained by directional selection. In total, our data suggest that natural selection is acting on the expression of 44 of the 329 genes (directional selection acting on 13 genes, stabilizing selection acting on 24 genes, and balancing selection acting on 7 genes). This conclusion required analysis within the appropriate ecological context among closely related taxa. The effect of natural selection along an environmental gradient in this organism is exemplary of what one would expect to find in any organism, including humans. That is, similar to the influence of malaria on human hemoglobin (60) or G6PDH (61), defining the functional importance of variation is difficult to achieve without evolutionary analyses.
Methods
Animals and Maintenance.
The teleost fish F. heteroclitus were collected from the field in June 2003 and acclimated in the laboratory to common controlled conditions (20°C, 15 parts per thousand salinity) in recirculating 100-gallon tanks for at least 2 months before experiments. The acclimation temperature is experienced by all populations from spring to fall and, thus, is ecologically relevant. Fish were killed by cervical dislocation, and livers were excised and stored in RNAlater (Ambion, Austin, TX) at −20°C. Fish were collected from the following five populations: Wiscasset, ME; Clinton, CT; Stone Harbor, NJ; Roanoke Island, NC; and Sapelo Island, GA. Only healthy adult male fish were used for the following experiments.
Habitat temperatures were derived from a minimum of 30 years mean annual surface temperatures. Coastal surface temperatures were considered more appropriate estimators of shallow estuarine habitat temperatures than temperatures logged by nearby open-ocean buoys.
Population Genetics.
Individuals from Georgia (n = 209), North Carolina (n = 54), New Jersey (n = 98), Connecticut (n = 50), and Maine (n = 50) were genotyped by using five microsatellite markers. Markers B101 (trimer), B128 (trimer), B4 (trimer), B7 (trimer), and C1 (tetramer) (62) had, respectively, 19, 13, 24, 21, and 21 alleles per locus among the 461 individuals. Weir and Cockerham’s theta (θ) estimator of FST was calculated for each population pair from genotype frequencies in genetix (www.univ-montp2.fr/∼genetix/genetix/genetix.htm), and significance of θ was tested by using 10,000 random permutations. Neighbor-joining trees were constructed in ntsys-pc (Exeter Software, Setauket, NY) by using Cavalli-Sforza and Edwards’ chord distance (CSE) as the genetic distance metric (42). Isolation by distance was tested by using a Mantel test (1,000 permutations) for correlation between CSE and shoreline distance (gapped across major bays) matrices.
Microarrays.
Microarrays were printed by using 329 cDNAs that encode essential proteins for cellular metabolism. All ESTs with enzyme commission numbers or associated with central metabolic pathways from a F. heteroclitus EST collection of >42,000 expressed sequences (http://genomics.rsmas.miami.edu/funnybase/super_craw4) were included on the array. These cDNAs were amplified with amine-linked primers and printed on 3D-Link activated slides (Surmodics, Eden Prairie, MN) at the University of Miami core microarray facility. The suite of 329 amplified cDNAs was printed as a group in four spatially separated replicates.
Microarray analyses were applied to livers from five individuals collected from each of the five populations of F. heteroclitus. RNA was extracted from tissue homogenate in a chaotropic buffer by using phenol/chloroform/isoamyl alcohol, and purified RNA was amplified to make amplified RNA (aRNA) by using a modified Eberwine protocol as described in ref. 26. Each labeled aRNA sample was suspended in 1.5 pmol/μl hybridization buffer, applied to the slide, and incubated 12–18 h at 42°C. Each of the 25 samples was hybridized twice, once with Cy3 and once with Cy5. Because a hybridization zone covered four replicate printed arrays, total experimental replication per sample per gene was 8-fold. A total of 50 hybridizations (25 × 2) were balanced among replicate individuals and populations in a loop design. Slides were scanned by using the Packard Bioscience ScanArray Express microarray scanner (PerkinElmer), and spot identification and intensity quantifications were collected by using imagene software (Biodiscovery, Marina Del Ray, CA).
Statistical Analysis.
Raw microarray data were sum normalized (63), intensity bias on each array was smoothed by using a Lowess transformation in r/maanova 0.93–2 (www.jax.org/staff/churchill/labsite), and log2 values of Lowess-transformed sum-normalized data were used for all subsequent analyses. MIAME (minimum information about a microarray experiment) compliant data (64) are available upon request. Technical variance and variance among individuals and populations were quantified in a nested ANOVA framework (Table 1) by using scripts written in matlab, (Version 6; MathWorks, Natick, MA). Scripts are available upon request.
Table 1.
Sources of variance for nested ANOVA and regression
| Source of variance | df | Sum of squares | F ratio |
|---|---|---|---|
| Among populations: Pop | 4 | 80 × Σ (PM − GM)2 | MS_Pop/MS_Ind |
| Regression: Reg | 1 | 80 × Σ((T − TM) × (PM − GM))2/(80 × Σ (T − TM)2) | MS_Reg/MS_DevReg |
| Deviations from regression: DevReg | 3 | SS_Pop − SS_Reg | |
| Among individuals within population: Ind | 20 | 16 × Σ(IM − DM)2 | MS_Ind/MS_Dye |
| Among dyes within individual: Dye | 25 | 8 × Σ (DM − RM)2 |
Where populations = 5, individuals per population = 5, dyes = 2, replicate hybridizations per dye = 2, replicate spots per hybridization = 4. PM, mean expression for population; GM, grand mean expression; T, habitat temperature (dependent variable in regression); TM, mean of habitat temperatures; IM, mean expression for individual within population; DM, mean expression for dye within individual; RM, mean expression for replicate hybridization within dye.
For each gene, we tested whether expression significantly correlated with median annual habitat temperature (least squares regression) or with genetic distance (Mantel test, 1,000 permutations), and we quantified the proportion of among-population expression variation accounted for by habitat temperature and genetic distance (r2). A second measure of correlation between genetic distance and gene expression was included to corroborate Mantel results; the physig program (47, 48) used branch lengths of the microsatellite-derived neighbor-joining tree (computed in ntsys-pc; Exeter Software) and 10,000 random permutations of the tree structure to test whether a phylogenetic signal was present in among-population gene expression patterns for each gene by using matlab scripts provided by Theodore Garland, Jr. (University of California, Riverside).
Regression of gene expression against habitat temperature was corrected for nonindependence of observations due to phylogeny by applying the phylogenetic generalized least squares method (48, 49) by using matlab scripts provided by Theodore Garland, Jr. (available upon request) and the mulreg module in ntsys-pc. A neighbor-joining tree was created in ntsys-pc based on microsatellite-derived genetic distances (Cavalli-Sforza and Edwards’ chord distance; ref. 42), and the tree structure and branch lengths were used to produce a matrix of expected variances and covariances of traits between taxa based on a Brownian motion model of evolution by using the COPH routine (PhyloCov method) in ntsys-pc. The resulting covariance matrix was used to correct regression of gene expression against habitat temperature for expected lack of independence due to phylogeny.
Supplementary Material
Acknowledgments
We thank Dr. Marjorie Oleksiak for construction of the EST library; Dr. Marjorie Oleksiak, Matt Rockman, and Jen Roach for constructive criticisms on earlier versions of the manuscript; David Duvernell and Stephanie Adams (Southern Illinois University, Edwardsville, IL) for graciously contributing the microsatellite data; Jeffrey VanWye for EST sequencing and microarray printing; and Justin Paschall for EST database management and bioinformatics. This project was supported by a National Science Foundation Ocean Sciences Grant 0221879 (to D.L.C.) and National Heart, Lung, and Blood Institute Grant R01 HL65470 (to D.L.C.).
Abbreviation
- FDR
false discovery rate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Specifically, the effect of the variation in a trait on fitness (its selective benefit) is less than the inverse of twice the effective population size.
References
- 1.King M. C., Wilson A. C. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Crawford D. L., Powers D. A. Mol. Biol. Evol. 1992;9:806–813. doi: 10.1093/oxfordjournals.molbev.a040762. [DOI] [PubMed] [Google Scholar]
- 3.Crawford D. L., Segal J. A., Barnett J. L. Mol. Biol. Evol. 1999;16:194–207. doi: 10.1093/oxfordjournals.molbev.a026102. [DOI] [PubMed] [Google Scholar]
- 4.Hahn M. W., Rockman M. V., Soranzo N., Goldstein D. B., Wray G. A. Genetics. 2004;167:867–877. doi: 10.1534/genetics.103.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerman D. N., Michalak P., Helin A. B., Bettencourt B. R., Feder M. E. Mol. Biol. Evol. 2003;20:135–144. doi: 10.1093/molbev/msg015. [DOI] [PubMed] [Google Scholar]
- 6.Rockman M. V., Hahn M. W., Soranzo N., Goldstein D. B., Wray G. A. Curr. Biol. 2003;13:2118–2123. doi: 10.1016/j.cub.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Wray G. A., Hahn M. W., Abouheif E., Balhoff J. P., Pizer M., Rockman M. V., Romano L. A. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 8.Gibson G., Weir B. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Stamatoyannopoulos J. A. Genomics. 2004;84:449–457. doi: 10.1016/j.ygeno.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Brem R. B., Yvert G., Clinton R., Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 11.Cavalieri D., Townsend J. P., Hartl D. L. Proc. Natl. Acad. Sci. USA. 2000;97:12369–12374. doi: 10.1073/pnas.210395297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferea T. L., Botstein D., Brown P. O., Rosenzweig R. F. Proc. Natl. Acad. Sci. USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Townsend J. P., Cavalieri D., Hartl D. L. Mol. Biol. Evol. 2003;20:955–963. doi: 10.1093/molbev/msg106. [DOI] [PubMed] [Google Scholar]
- 14.Jin W., Riley R. M., Wolfinger R. D., White K. P., Passador-Gurgel G., Gibson G. Nat. Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- 15.Gibson G., Riley-Berger R., Harshman L., Kopp A., Vacha S., Nuzhdin S., Wayne M. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rifkin S. A., Kim J., White K. P. Nat. Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 17.Wayne M. L., Pan Y. J., Nuzhdin S. V., McIntyre L. M. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuzhdin S. V., Wayne M. L., Harmon K. L., McIntyre L. M. Mol. Biol. Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- 19.Pritchard C. C., Hsu L., Delrow J., Nelson P. S. Proc. Natl. Acad. Sci. USA. 2001;98:13266–13271. doi: 10.1073/pnas.221465998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung V. G., Conlin L. K., Weber T. M., Arcaro M., Jen K. Y., Morley M., Spielman R. S. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 21.Sharma A., Sharma V. K., Horn-Saban S., Lancet D., Ramachandran S., Brahmachari S. K. Physiol. Genomics. 2005;21:117–123. doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- 22.Tan Q., Christensen K., Christiansen L., Frederiksen H., Bathum L., Dahlgaard J., Kruse T. A. Hum. Genet. 2005;117:267–274. doi: 10.1007/s00439-005-1308-x. [DOI] [PubMed] [Google Scholar]
- 23.Enard W., Khaitovich P., Klose J., Zoellner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., et al. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 24.Oleksiak M. F., Churchill G. A., Crawford D. L. Nat. Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 25.Oleksiak M. F., Roach J. L., Crawford D. L. Nat. Genet. 2005;37:67–72. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehead A., Crawford D. Genome Biol. 2005;6:R13. doi: 10.1186/gb-2005-6-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreitman M. BioEssays. 1996;18:678–683. doi: 10.1002/bies.950180812. discussion 683. [DOI] [PubMed] [Google Scholar]
- 28.Takahata N. Curr. Opin. Genet. Dev. 1996;6:767–772. doi: 10.1016/s0959-437x(96)80034-7. [DOI] [PubMed] [Google Scholar]
- 29.Orr H. A. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endler J. A. Natural Selection in the Wild. Princeton: Princeton Univ. Press; 1986. [Google Scholar]
- 31.Yanai I., Graur D., Ophir R. OMICS. 2004;8:15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- 32.Khaitovich P., Weiss G., Lachmann M., Hellmann I., Enard W., Muetzel B., Wirkner U., Ansorge W., Paabo S. PLoS. Biol. 2004;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemos B., Meiklejohn C. D., Caceres M., Hartl D. L. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- 34.Felsenstein J. Amer. Nat. 1985;125:1–15. [Google Scholar]
- 35.Garland T., Jr., Harvey P. H., Ives A. R. Syst. Biol. 1992;41:18–32. [Google Scholar]
- 36.Harvey P. H., Pagel M. D. The Comparative Method in Evolutionary Biology. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 37.Powers D. A., Smith M., Gonzalez-Villasenor I., DiMichelle L., Crawford D. L., Bernardi G., Lauerman T. In: Oxford Surveys in Evolutionary Biology. Futuyma D., Antonovics J., editors. Vol. 9. New York: Oxford Univ. Press; 1993. pp. 43–108. [Google Scholar]
- 38.Crawford D. L., Powers D. A. Proc. Natl. Acad. Sci. USA. 1989;86:9365–9369. doi: 10.1073/pnas.86.23.9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce V. A., Crawford D. L. Science. 1997;276:256–259. doi: 10.1126/science.276.5310.256. [DOI] [PubMed] [Google Scholar]
- 40.Hochachka P. W., Somero G. N. Biochemical Adaptation. Princeton: Princeton Univ. Press; 1984. [Google Scholar]
- 41.Prosser C. L. New York: Wiley; 1986. Adaptational Biology: From Molecules to Organisms. [Google Scholar]
- 42.Cavalli-Sforza L. L., Edwards A. W. F. Evolution. 1967;32:550–570. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 43.Cashon R. E., Van Beneden R. J., Powers D. A. Biochem. Genet. 1981;19:715–728. doi: 10.1007/BF00484004. [DOI] [PubMed] [Google Scholar]
- 44.Able K. W., Fewlley J. D. Am. Zool. 1986;25:145–157. [Google Scholar]
- 45.Bernardi G., Sordino P., Powers D. A. Proc. Natl. Acad. Sci. USA. 1993;90:9271–9274. doi: 10.1073/pnas.90.20.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey J. D., Tibshirani R. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garland T., Jr., Ives A. R. Am. Nat. 2000;155:346–364. doi: 10.1086/303327. [DOI] [PubMed] [Google Scholar]
- 48.Blomberg S. P., Garland T., Jr., Ives A. R. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 49.Rohlf F. J. Evolution. 2001;55:2143–2160. doi: 10.1111/j.0014-3820.2001.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 50.Gracey A. Y., Fraser E. J., Li W., Fang Y., Taylor R. R., Rogers J., Brass A., Cossins A. R. Proc. Natl. Acad. Sci. USA. 2004;101:16970–16975. doi: 10.1073/pnas.0403627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westoby M., Leishman M. R., Lord J. M. J. Ecol. 1995;83:531–534. [Google Scholar]
- 52.Kimura M. Proc. Natl. Acad. Sci. USA. 1991;88:5969–5973. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoubridge E. A. Hum. Mol. Genet. 2001;10:2227–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- 54.Taylor R. W., Turnbull D. M. Nat. Rev. 2005;6:389–406. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitton J. B. Selection in Natural Populations. New York: Oxford Univ. Press; 1997. [Google Scholar]
- 56.Lewontin R. C. In: Evolution. Ridley M., editor. Oxford: Oxford Univ. Press; 1997. pp. 79–88. [Google Scholar]
- 57.Childress J. J. Trends in Ecology & Evolution. 1995;10:30–36. doi: 10.1016/s0169-5347(00)88957-0. [DOI] [PubMed] [Google Scholar]
- 58.Seibel B. A., Thuesen E. V., Childress J. J. Biol. Bull. (Woods Hole, MA) 2000;198:284–298. doi: 10.2307/1542531. [DOI] [PubMed] [Google Scholar]
- 59.Whitehead A., Crawford D. Mol. Ecol. 2006 doi: 10.1111/j.1365-294X.2006.02868.x. in press. [DOI] [PubMed] [Google Scholar]
- 60.Dean A. M. Am. Sci. 1998;86:26–37. [Google Scholar]
- 61.Tishkoff S. A., Varkonyi R., Cahinhinan N., Abbes S., Argyropoulos G., Destro-Bisol G., Drousiotou A., Dangerfield B., Lefranc G., Loiselet J., et al. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 62.Adams S. M., Oleksiak M. F., Duvernell D. D. Mol. Ecol. Notes. 2005;5:275–277. [Google Scholar]
- 63.Quackenbush J. Nat. Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 64.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C. A., Causton H. C., et al. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 65.Adams S. M., Lindmeier J. B., Duvernell D. D. Mol. Ecol. 2006;15:1109–1124. doi: 10.1111/j.1365-294X.2006.02859.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.