Abstract
The anterior interpositus nucleus (AIN) is the proposed site of memory formation of eyeblink conditioning. A large part of the underlying molecular events, however, remain unknown. To elucidate the molecular mechanisms, we examined transcriptional changes in the AIN of mice trained with delay eyeblink conditioning using microarray, quantitative real-time RT-PCR, and in situ hybridization techniques. Microarray analyses suggested that transcriptionally up-regulated gene sets were largely different between early (3-d training) and late (7-d) stages. Quantitative real-time RT-PCR aided by laser microdissection indicated that the expression of representative EARLY genes (Sgk, IkBa, and Plekhf1) peaked at 1-d training in both the paired and unpaired conditioning groups, and was maintained at a higher level in the paired group than in the unpaired group after 3-d training. In situ hybridization revealed increased expression of these genes in broad cerebellar areas, including the AIN, with no hemispheric preferences. In contrast, the expression of representative LATE genes (Vamp1, Camk2d, and Prkcd) was selectively increased in the AIN of the 7-d paired group, with dominance in the ipsilateral AIN. Increased Vamp1 mRNA expression was restricted to the ipsilateral dorsolateral hump, a subregion of the AIN. These expression patterns of two distinct subsets of genes fit well with the two-stage learning theory, which proposes emotional and motor learning phases, and support the notion that AIN has a crucial role in memory formation of eyeblink conditioning.
Keywords: gene expression, interpositus nucleus
Classical eyeblink conditioning is conserved among various species. Although the neuronal circuitries are well defined and the cerebellum is considered to be the site of memory formation of the classically conditioned response (CR, i.e., eyeblink) (1), evidence is still conflicting regarding the neural substrate for the memory trace of eyeblink conditioning in the cerebellum.
In contrast to the suggestion that the basic memory trace of the CR is formed in the cerebellar cortex (2), accumulating evidence suggests that the anterior interpositus nucleus (AIN) in the deep cerebellar nuclei (DCN) is the site of memory for the association between the conditioned stimulus (CS) and the unconditioned stimulus (US) formed in eyeblink conditioning. This hypothesis has been examined repeatedly with lesions (3–7) and inactivation of the AIN (8, 9), and in mutant mice with Purkinje cell degeneration (10). Recent observations made using electron microscopy (11) or functional magnetic resonance imaging (12) further support the notion that the basic memory trace is formed in the AIN. On the other hand, the cerebellar cortex is implicated as a site of storage for the memory trace of CR timing (13).
Although the existence of a Purkinje cell-specific promoter like L7/pcp-2 made it possible to test the relevance of some molecules to cortical function in eyeblink conditioning in vivo (14, 15), mechanistic studies at the molecular level of the AIN, which is likely more important, are limited. Recently, the requirement of de novo protein synthesis and learning-related induction of gene expression in the AIN was suggested to occur during memory formation of eyeblink conditioning of rabbits by inactivating the ipsilateral AIN with a transcription inhibitor, actinomycin D (16), or a translation inhibitor, anisomycin (17). Therefore, the analysis of gene expression changes in the AIN will help us to understand the underlying mechanisms of eyeblink conditioning.
Another aspect of the mechanisms suggested to underlie eyeblink conditioning comes from the two-stage learning theory (18) in which the preceding emotional learning might facilitate the subsequent acquisition of motor learning. Although there are several observations supporting its relevance to eyeblink conditioning (19–22), a large part of the related molecular mechanisms are still unknown.
To gain further insight into the molecular and cellular mechanisms for forming memory traces, we systematically analyzed the transcriptional properties of the AIN in eyeblink-conditioned mice. We found two groups of genes with distinct temporal and spatial expression properties; i.e., the EARLY group with a rapid and broad induction, and the LATE group with a slow and spatially restricted induction. The significance of these observations is discussed from the point of view of the two-stage learning theory and the AIN as a possible site of storage of the basic association memory of delay eyeblink conditioning.
Results
Paired but Not Unpaired Groups Acquired a Robust Memory for Delay Eyeblink Conditioning.
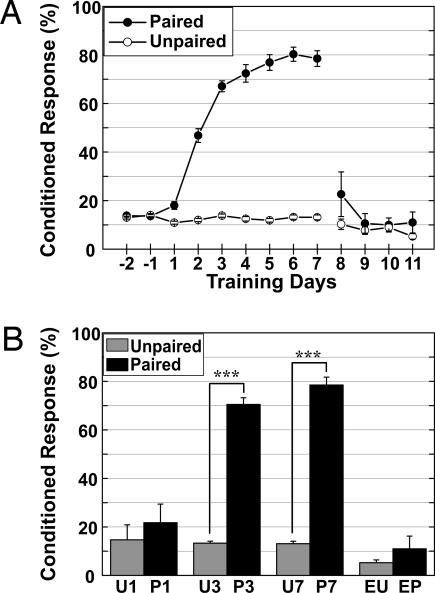
The learning curves of the paired and unpaired groups are shown in Fig. 1A. During the 7-d training sessions (days 1–7), the percent CR (CR%) in the paired group increased significantly [F(8, 336) = 143.031, P < 0.0001], whereas there was no learning across training sessions in the unpaired group [F(8, 312) = 1.714, P = 0.0942]. There was a significant difference in the training condition effect [F(1, 81) = 444.108, P < 0.0001] and condition × session interaction [F(8, 648) = 119.4, P < 0.0001] between the two training groups. On the first day of extinction training, there was no significant difference between the two groups that received an additional 4 d of extinction training after 7 d of paired training (days 8–11) [F(1, 6) = 1.704, P = 0.2396], indicating rapid and successful extinction. The comparison of CR% on the last training day demonstrated a clear difference in the memory state of eyeblink conditioning between the training groups (Fig. 1B).
Fig. 1.
The results of eyeblink conditioning. (A) The CR percentage of the paired group (filled circles) reached an asymptotic level, whereas there were no significant changes in the unpaired group (open circles). The 4-d extinction training effectively extinguished the CR% of the paired group to the baseline. (B) Comparison of CR% on the day of DCN sampling revealed a significant difference in the 3-d and 7-d training groups but not in the 1-d training or 4-d extinction training groups. ∗∗∗, P < 0.0001 in ANOVA.
The Transcriptionally Up-Regulated Genes in the AIN Were Largely Different Between the Early and Late Training Groups.
The AIN-centered DCN was sampled by inserting a syringe needle in the white matter between the DCN and cerebellar cortex (see Fig. 5, which is published as supporting information on the PNAS web site). Analyses of pooled ipsilateral AIN-centered DCN with GeneChip microarrays revealed that the expression of 505 and 1,208 genes was increased >1.3-fold in the pairwise comparisons of 3-d paired training (P3) and sham negative control (SHAM) groups and 7-d paired training (P7) and 7-d unpaired training (U7) groups, respectively (see Table 3, which is published as supporting information on the PNAS web site), and denoted as plasticity candidate genes (PCGs). Surprisingly, the PCGs from two pairwise comparisons were largely different from each other except for a common minority. The list of PCGs annotated for the synaptic function-related PCGs is shown in Table 1.
Table 1.
List of the PCGs classified as Synaptic function group
| Gene | Description | GenBank no. | P3-SHAM fold changes | P7-U7 fold changes |
|---|---|---|---|---|
| PCGs from the comparison of P3-SHAM | ||||
| Gabra6 | GABA-A receptor, subunit α6 | X51986 | 1.46 | 1.65 |
| Lin7c | Lin 7 homolog c (Caenorhabtitis elegans) | AW125731 | 1.30 | 1.20 |
| Scamp5 | Secretory carrier membrane protein 5 | AI847095 | 1.31 | −1.05 |
| PCGs from the comparison of P7-U7 | ||||
| Slc17a6 | Solute carrier family 17 member 6 | AI841371 | 1.03 | 1.79 |
| Gabra3 | GABA-A receptor, subunit α3 | M86568 | −1.04 | 1.69 |
| Lin7a | Lin 7 homolog a (C. elegans) | AI615412 | 1.03 | 1.66 |
| Chn2 | Chimerin 2 | AW049301 | 1.18 | 1.58 |
| Shc3 | Src homology 2 domain-containing transforming protein C3 | U46854 | −1.16 | 1.49 |
| Synpo | Synaptopodin | AW050323 | −1.07 | 1.45 |
| Grid2 | Glutamate receptor, ionotropic, δ2 | D13266 | 1.09 | 1.45 |
| Grin1 | Glutamate receptor, ionotropic, NMDA1 (ζ1) | D10028 | 1.04 | 1.45 |
| Cart | Cocaine and amphetamine regulated transcript | AI322575 | −1.07 | 1.44 |
| Scamp1 | Secretory carrier membrane protein 1 | AW120713 | −1.00 | 1.44 |
| Vdac1 | Voltage-dependent anion channel 1 | U30840 | −1.08 | 1.41 |
| Gria4 | Glutamate receptor, ionotropic, AMPA4 (α4) | AB022913 | 1.11 | 1.41 |
| Clstn1 | Calsyntenin 1 | AW048171 | −1.20 | 1.39 |
| Vamp2 | Vesicle-associated membrane protein 2 | U60150 | 1.07 | 1.38 |
| Grin1 | Glutamate receptor, ionotropic, NMDA1 (ζ1) | AI847120 | 1.12 | 1.37 |
| Rims4 | Regulating synaptic membrane exocytosis 4 | AW046817 | −1.16 | 1.36 |
| Gria2 | Glutamate receptor, ionotropic, AMPA2 (α2) | AV327031 | −1.13 | 1.34 |
| Stx1b2 | Syntaxin 1B2 | D29743 | −1.13 | 1.34 |
| Vamp1 | Vesicle-associated membrane protein 1 | U61751 | −1.10 | 1.32 |
| Cadps2 | Ca2+-dependent activator protein for secretion 2 | AI854271 | 1.18 | 1.31 |
Fold changes >1.3 are boldfaced.
Expression Profiling Revealed the EARLY Group and the LATE Group from the PCGs.
The entire AINs of eyeblink-conditioned mice were precisely sampled with a laser microdissection microscope and subjected to quantitative real-time RT-PCR (qRT-PCR). For these analyses, we selected 11 representative genes that were up-regulated in the AIN of the P3 or P7 groups (Table 2).
Table 2.
Pairwise comparisons of the selected PCGs from the EARLY group and the LATE group are shown as fold changes
| Gene | Description | GenBank no. | Function | Pairwise comparison |
|
|---|---|---|---|---|---|
| P3-SHAM | P7-U7 | ||||
| Sgk | Serum/glucocorticoid regulated kinase | AF205855 | Protein kinase | 5.85 | 1.18 |
| Gtl2 | Imprinted maternally expressed untranslated mRNA | AI852838 | Unknown | 2.22 | −1.03 |
| Cebpd | CCAAT/enhancer binding protein delta | X61800 | Transcription factor | 2.06 | −1.22 |
| IkBa | Inhibitory κBα | AI642048 | Regulatory protein | 1.90 | 1.17 |
| IkBa | Inhibitory κBα | AV370033 | Regulatory protein | 1.89 | 1.19 |
| IkBa | Inhibitory κBα | U57524 | Regulatory protein | 1.85 | 1.13 |
| Desrt | AT rich interactive domain 5B | AI173737 | Transcription factor | 1.68 | −1.12 |
| Gtl2 | Imprinted maternally expressed untranslated mRNA | AI448278 | Unknown | 1.63 | −1.10 |
| Clk | CDC-like kinase | M38381 | Protein kinase | 1.61 | 1.00 |
| Plekhf1 | Pleckstrin homology domain containing family F1 | AW049880 | Unknown | 1.52 | 1.12 |
| Per1 | Period homolog 1 | AF022992 | Transcription factor | 1.34 | −1.04 |
| Prkcd | Protein kinase C, δ | X60304 | Protein kinase | −1.12 | 1.85 |
| Camk2d | Calcium/calmodulin-dependent protein kinase II, δ | AV134810 | Protein kinase | −1.13 | 1.45 |
| Prkcd | Protein kinase C, δ | AB011812 | Protein kinase | 1.04 | 1.43 |
| Vamp1 | Vesicle-associated membrane protein 1 | U61751 | Synaptic vesicle docking | −1.01 | 1.32 |
Difference in pairwise comparisons is shown in fold changes, and negative values mean decrease in the paired groups. Fold changes >1.3 are boldfaced.
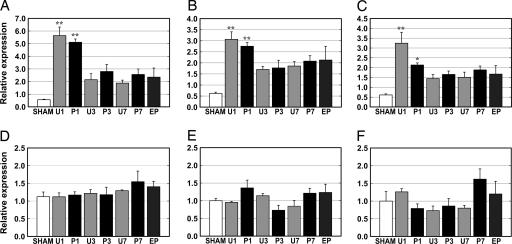
The representative expression profiling by qRT-PCR of six PCGs (Sgk, IkBa, and Plekhf1 from the P3 group and Vamp1, Camk2d, and Prkcd from the P7 group) was consistent with the results of the microarray analysis (Fig. 2; and for five others, see Table 4, which is published as supporting information on the PNAS web site).
Fig. 2.
Expression profiling of six PCGs from the EARLY and LATE groups. The expression patterns of the EARLY group [Sgk (A), IkBa (B), and Plekhf1 (C)] are shown. The mRNA of the EARLY group was significantly increased in the U1 and P1 groups, then decreased, but was greater than in the SHAM group. Expression in the P3 and P7 groups slightly increased compared with the U3 and U7 groups, respectively. Expression of the LATE Group [Vamp1 (D), Camk2d (E), and Prkcd (F)] was specifically increased numerically in the 7-d paired group. n = 3, except for n = 2 in P3 of A–D and U7 of D. See Materials and Methods for abbreviations. ∗, P < 0.05; ∗∗, P < 0.01 (both in one-way ANOVA followed by Tukey–Kramer multiple comparisons test).
Expression of the genes chosen from P3 group was significantly increased in the 1-d training groups (P < 0.01 for Sgk and IkBa; P < 0.05 for Plekhf1). The expression profiles of these genes, denoted as the EARLY group, shared high similarity across training days. Expression of these genes peaked at the 1-d training stage in both the paired and unpaired groups, and higher levels of expression were maintained in the training groups relative to the SHAM group in the later training stages. The expression level of the EARLY group in the paired groups was lower than that of the unpaired groups at the 1-d training stage but higher in the later training stages.
There were no dramatic changes in expression of the genes selected from the P7 group, Vamp1, Camk2d, and Prkcd, in the early stage of training. The increased expression, however, was specifically observed in the P7 group, not in the U7 or SHAM groups. Additionally, the temporal expression patterns of these genes were similar to one another. We denoted these genes as the LATE group.
Expression levels of both PCG groups in the 4-d extinction training group were not decreased compared with those of the SHAM group, despite the complete disappearance of the learned CR (see Fig. 1).
Expression Levels of the EARLY Group Were Broadly Localized in the Cerebellum and Higher in the Paired Group than in the Unpaired Group.
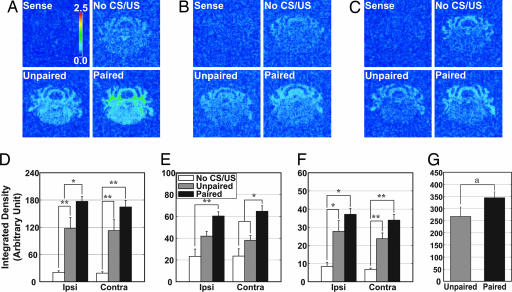
The hybridization results of the EARLY group, Sgk, IkBa, and Plekhf1, are shown in Fig. 3(A and D, B and E, and C and F, respectively). Consistent with the qRT-PCR data, expression of the EARLY group members was increased by eyeblink conditioning regardless of the paradigm used, whereas the basal level of expression was observed in the SHAM group. No signals were detected with the sense probe (Sense in Fig. 3 A–C). Interestingly, mRNA signals of these genes were increased not only in the DCN area, but also in the white matter, cortex, and brainstem areas. Densitometric analysis in the AIN indicated that expression of the EARLY group was significantly increased in the unpaired groups compared with the no-training groups, and more in the paired groups than in the other two groups on both sides of the AIN. There was a significant difference in Sgk hybridization signal between paired and unpaired groups in the ipsilateral AIN (Fig. 3D, P < 0.05) and in IkBa hybridization signal in the contralateral AIN (Fig. 3E, P < 0.05). Additionally, densitometry of the Sgk hybridization signal performed in white matter revealed a significant increase in the paired groups (Fig. 3G, P < 0.05).
Fig. 3.
In situ hybridization of the EARLY group. Representative photomicrographs in pseudocolor show the mRNA signals of Sgk (A), IkBa (B), and Plekhf1 (C). No signals were detected with the sense probes (Sense). The hybridization signals in the no-training (No CS/US) and training (Unpaired and Paired) groups are shown. Quantitative analysis of hybridization signals in the AIN is shown for Sgk (D), IkBa (E), and Plekhf1 (F). The mRNA of the paired groups was significantly increased compared with the other groups in the ipsilateral AIN (Sgk, P < 0.05) or contralateral AIN (IkBa, P < 0.05). (G) Significant increase in Sgk mRNA in the white matter of the paired groups. n = 4–6. Ipsi, ipsilateral; Contra, contralateral. ∗, P < 0.05; ∗∗, P < 0.01 (both in one-way ANOVA followed by Tukey–Kramer multiple comparisons test). a, P < 0.05 in t test.
The Expression of the LATE Group Was Specifically Increased in the 7-Day Paired Group with Ipsilateral Dominance in the AIN Subregion.
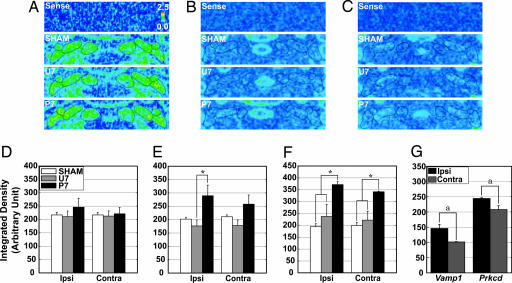
Expression of the LATE group, Vamp1, Camk2d, and Prkcd, in the DCN area is shown in Fig. 4(A and D, B and E, and C and F, respectively). The sense probes produced no signal, indicating the specificity of the antisense probes (Sense in Fig. 4 A–C).
Fig. 4.
In situ hybridization of the LATE group. Representative photomicrographs in pseudocolor show the mRNA signals of Vamp1 (A), Camk2d (B), and Prkcd (C) in the area of the DCN. No signals were detected with the sense probes (Sense). The quantified expression level of mRNA is shown for Vamp1 (D), Camk2d (E), and Prkcd (F). The expression of Camk2d mRNA in the ipsilateral AIN and Prkcd mRNA in the bilateral AIN was significantly increased. Signals of Vamp1 in the DLH and of Prkcd in the rostral half of AIN were significantly higher on the ipsilateral side than those on the contralateral side (G). n = 3 or 4. Ipsi, ipsilateral; Contra, contralateral. ∗, P < 0.05 in one-way ANOVA followed by Tukey–Kramer multiple comparisons test. a, P < 0.05 in t test.
Gross observation suggested a slight difference in the effect of the training paradigms on the expression across training groups. Quantification of hybridization signals revealed a remarkable increase in mRNA expression in the P7 group compared with U7 or SHAM groups in all three PCGs of the LATE group. The Vamp1 hybridization signal was slightly increased in the ipsilateral AIN of P7 group compared with the other training groups (Fig. 4D). The expression of Camk2d and Prkcd was significantly increased in the ipsilateral AIN of the P7 group compared with the U7 group (Camk2d hybridization) (Fig. 4E, P < 0.05) and in the bilateral AIN of the P7 group compared with the other training groups (Prkcd hybridization) (Fig. 4F, P < 0.05). The density of Vamp1 signals in the dorsolateral hump (DLH), a subregion of the AIN, was significantly higher on the ipsilateral side than on the contralateral side. Also, Prkcd signals in the rostral half of the AIN were significantly higher on the ipsilateral side (Fig. 4G, P < 0.05 in both), whereas Prkcd hybridization signals in the caudal part were comparable on both sides of the AIN (data not shown).
Discussion
Microarray analysis of the AIN revealed numerous genes with transcriptional changes occurring in a time- and experience-specific manner. Interestingly, the PCGs from the early stage and late stage of training were largely different except for a small common set. These results likely reflected different molecular events at different stages during memory formation of eyeblink conditioning. The genes annotated for synaptic plasticity were frequently identified in comparisons between P7 and U7 groups, but not between P3 and SHAM groups. These genes are likely involved in the synaptic plasticity occurring in the AIN during motor memory formation of eyeblink conditioning, as described previously by Kleim and colleagues (11).
Expression of the EARLY group, Sgk, IkBa, and Plekhf1, which was not restricted to the AIN area but was broadly localized in the cerebellum, peaked in the very early stage of training and decreased later. The time courses of the changes were very similar to the pattern of ultrasonic vocalization reported as a marker for emotional learning in rats (23). Although the emotional learning in eyeblink conditioning was previously reported to depend on amygdala activity (21–23), the broad localization of the induced mRNA suggests that expression of the EARLY group was affected not only by direct neuronal activity of the amygdala, but also by other factors like glucocorticoids (GCs) released under stressful conditions. Indeed, previous experiments indicated that Sgk (24) and IkBa (25) expression is induced by treatment with GCs. The release of GCs is induced by amygdala simulation (26). Collectively, these findings suggest that the EARLY group is induced by amygdala activity in the emotional learning stage and that this group of genes has a role in priming the cerebellum for efficient acquisition of the subsequent motor learning.
The high expression level of the EARLY gene group observed in the training groups was likely due to elevated GC levels in eyeblink conditioning (27). Changes in the relative expression level between the paired and unpaired group imply that the situation changes as learning progresses. The higher expression level in the unpaired group at day 1 implies that the unpaired stimuli elicit a stronger stress response. In the later stage, the mice might develop tolerance to the stress. The increased expression in the EARLY group, however, was preferentially sustained in the paired group. These results suggest that multiple mechanisms are involved in the later stages. Tsai and colleagues (28) suggest that Sgk is involved in the memory consolidation of hippocampus-dependent learning in rodents. Additionally, IkBa is a regulatory protein of NF-κB, which is implicated in memory formation in mice (29). Prolonged expression of the EARLY group might successfully contribute to motor memory formation.
In contrast to the EARLY group, expression of the LATE group, Vamp1, Camk2d, and Prkcd, was specifically up-regulated in the AIN of the 7-d paired group, and the expression pattern was similar to the CR curve of eyeblink conditioning. These findings suggest that the LATE group has a specific role in the acquisition of motor memory for eyeblink conditioning. Vamp1 is a synaptic vesicle molecule involved in the exocytosis of neurotransmitters from the presynaptic membrane (30), and Vamp expression is likely up-regulated with increased synaptogenesis (31). Prkcd mRNA increases in the hippocampus during a spatial discrimination learning paradigm (32) and a chronic exercise task (33). Additionally, infusion of protein kinase inhibitors into the AIN impairs acquisition of the conditioned eyeblink response (34). Camk2d mRNA expression is induced by neuronal activity in an NMDA-receptor-dependent manner (35). All of these findings support the involvement of the LATE group in the formation of motor memory for eyeblink conditioning.
Expression of the LATE group was increased in both AINs of the P7 group. Previous reports demonstrated that bilateral DCN lesions were required to impair eyeblink conditioning in mice (5, 10). These findings are likely due to the distribution of motor neurons in the AIN. Morcuende and colleagues (36) reported that the motor neurons projecting to the ipsilateral eye muscle in rodents are localized in subregions of both AINs but dominantly in the DLH and dorsolateral part of the ipsilateral AIN. Consistently, we observed increased expression of the LATE group on both sides of the AIN with slight dominance in the ipsilateral AIN and a significant increase in the subregions of the ipsilateral AIN of the P7 group; i.e., Vamp1 expression in the ipsilateral DLH and Prkcd expression in the rostral part of the ipsilateral AIN. Neuroanatomically, the DLH and dorsolateral part of the rostral AIN in rodents are considered to be comparable sites for the memory formation of eyeblink conditioning in rabbits. In this context, the dominant expression of the LATE group observed in the subregions of the ipsilateral AIN strongly suggests a specific role for the LATE group in the motor learning of eyeblink conditioning and in turn further supports the notion of motor memory formation in subregions of the AIN in delay eyeblink conditioning of mice.
In the present study, we analyzed transcriptional changes of the AIN in mice that are associated with the delay eyeblink conditioning. Our results indicated that two groups of genes were involved, the EARLY and LATE groups, which fits well with the two-stage learning theory. Additionally, we demonstrated specific up-regulation of the expression of the LATE group in the AIN, where the motor memory is likely formed. This molecular evidence provides support for the two-staged learning theory and for the AIN as a possible site of motor memory formation in delay eyeblink conditioning of mice.
Materials and Methods
Animals.
Male C57BL/6 mice (n = 213; 11–18 wk; 22–33 g) were purchased from Clea Japan (Tokyo) and raised in the animal facility of the RIKEN Brain Science Institute. All experimental protocols were approved by the RIKEN Institutional Animal Care and Use Committee. Mice were housed in groups of five in a ventilated plastic cage, which was placed in a temperature- and humidity-controlled room with a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). All animals were fed laboratory chow and water ad libitum.
Eyeblink Conditioning.
The delay eyeblink conditioning in mice was performed as described in ref. 37 with slight modification. Briefly, under deep anesthesia with ketamine (100 mg/kg, i.p.) and xylazine (25 mg/kg, i.p.), a four-pin headstage was fixed by the two screws and dental cement to the animal’s head, and four Teflon-coated stainless-steel wires (0.003 inches bare, 0.0055 inches coated; No. 7910; A-M Systems, Carlsborg, WA) attached to the headstage were implanted s.c. in the orbicularis oculi of the left upper eyelid. The tips of the wires were exposed, and two wires were used to record a differential electromyogram from the eyelid muscle and the other two wires were used to deliver periorbital shock. After surgery, the mice were individually housed. At least 3 d after surgery, animals were randomly divided into eight groups; a SHAM, 1-d unpaired (U1) and paired (P1), 3-d unpaired (U3) and paired (P3), U7 and P7, and 4-d extinction group after 7-d paired (EP) and unpaired (EU) training. After a 2-d habituation period without a CS or US, all animals were trained for the predetermined days, but SHAM animals remained in their home cage during the training days. After the training session, animals were placed back in their home cage.
Under freely moving conditions in a sound- and light-attenuating chamber (24 × 24 × 40 cm), animals were trained with either the paired paradigm in which a 352-ms tone CS (1 kHz, 83-85 dB) and a subsequent 100-ms periorbital shock US (100-kHz square pluses) were delivered with a 252-ms interstimulus interval and co-terminated, or the unpaired paradigm in which the CS and US were delivered in an explicitly unpaired, pseudorandomized manner. The US intensity was adjusted daily for each animal to the minimal voltage required to elicit an eyeblink or head-turn response. Training was performed between 1000 hours and 1600 hours. Daily training sessions consisted of 100 trials grouped in 10 blocks, which included one CS only (on the 10th trial) and 9 CS/US paired or unpaired trials. The intertrial interval was randomized between 20 and 40 s (mean 30 s).
The eyeblink electromyographic data were stored and then analyzed by using a custom-made program. Briefly, according to the criteria used here (see Supporting Text, which is published as supporting information on the PNAS web site), the CR was determined in all trials without unstable activity or an unconditioned startle response. CR% was defined as the ratio of CR in valid trials in both CS/US paired trials and CS-only trials. The criteria for analysis were applied to the paired and unpaired groups. Mice that were poor learners were excluded from further analysis.
Dissecting DCN: AIN-Centered DCN Sampling.
The cerebellum from eyeblink-conditioned mice was removed on the day of the final session. The animals were immediately subjected to cerebellum sampling 10–30 min after the last trial. All animals of each group were killed by cervical dislocation, the brain was quickly removed, and the cerebellum was cut out, frozen in OCT compound, and stored at −80°C until use.
The frozen cerebellum block was cut coronally until the AIN was just exposed. The DCN were sampled by using a 27-gauge syringe needle and stored at −80°C. To reveal the extent of DCN sampling, 30-μm slice sections before and after the needle insertion were collected and stained with cresyl violet.
GeneChip Microarray.
The procedures and reagents for the GeneChip microarray were used are described in detail in the Affymetrix GeneChip Expression Analysis Manual. Microarray hybridization was performed to compare P3 to SHAM and P7 to U7 groups. Ipsilateral AIN-centered DCN samples (n = 10–15) were pooled into the same conditioning group and subjected to microarray analysis by using GeneChip Murine Genome U74 Version 2 sets (see Supporting Text).
Data from the GeneChip hybridization were analyzed with affy 1.3.25 to account for the possible nonlinear relationship between arrays (38). The genes with >1.3 times increased expression in pairwise comparisons, which were considered to change in relation to learning and memory of the eyeblink conditioning or stimulation, were annotated and functionally categorized by referring to the Gene Ontology Classification of Mouse Genome Informatics Version 3.3 (www.informatics.jax.org).
AIN Sampling Using Laser Microdissection Microscopy and Subsequent qRT-PCR.
The frozen cerebellum block was cut coronally until just the AIN was exposed. Cerebellum sections (20 μm) were serially collected on RNase-free film-coated slides until the end of the medial cerebellar nucleus was reached. The cerebellar slices were dehydrated in a graded alcohol series, dried, and subjected to AIN sampling using a laser microdissection microscope (AS LMD; Leica). Entire AINs were precisely sampled from 52 to 54 slices per mouse. The sampled AINs from an animal were pooled, homogenized, and subjected to total RNA isolation by using an RNeasy mini kit (Qiagen). After treatment with DNase I, the isolated total RNA was converted into cDNA with a random hexamer. Synthesized cDNA was stored at −80°C until use.
qRT-PCR using SYBR green or TaqMan probes was performed for the following genes (for abbreviations, see Table 2); Sgk, Gtl2, Cebpd, IkBa, Desrt, Clk, Per1, Plekhf1, Camk2d, Prkcd, and Vamp1 as PCGs, and Gapdh as a housekeeping gene. The list for target-specific oligonucleotide primers and assay ID is shown in Table 5, which is published as supporting information on the PNAS web site. cDNA concentration in the samples was adjusted to 50 or 100 pg/μl based on the Gapdh cDNA content. AIN cDNA (1 μl) and specific primers were added to the QuantiTect SYBR green PCR master mix (Qiagen) or TaqMan universal PCR master mix (Applied Biosystems) with/without fluorescence-labeled probes. PCR amplification was performed in an ABI Prism 7700 (Applied Biosystems) according to the suggested temperature conditions. All qRT-PCR were performed in triplicate for each cDNA sample. The relative expression ratio of target genes was calculated. The specificity of the PCR products amplified was confirmed by dissociation curve analysis.
In Situ Hybridization and Densitometry.
Every fifth coronal cerebellum section (14 μm) was collected on slides coated with silane (S3003; Dako Cytomation) until the end of the medial cerebellar nucleus or the posterior interpositus nucleus (18 slices per animal). Tissue sections were dried and stored at −80°C until use.
The in situ hybridization method using α-33P-labeled probes was performed as described in ref. 39. The target cDNAs were synthesized from total RNA of the DCN with gene-specific primers (see Table 5). The hybridization images were quantified by densitometry with image-pro plus 4.5.1.22 (Media Cybernetics). Briefly, the hybridized slides and films were scanned with a scanner. The scanned images of the cerebellum slices were used to delineate the subregions of the DCN on the film. Based on the area information from the slice, the signal from AIN and AIN subregions was quantified. For analysis of the EARLY group, data of training groups were pooled into the same paradigm groups because there was a training paradigm-dependent difference during this period, but there was no time-dependent change in expression. For the LATE group, 7-d training samples were analyzed. For densitometry of the white matter in Sgk hybridization, five 100-pixel areas (10 × 10 pixels) per slice were randomly selected and quantified in the white matter surrounding the DCN of the paired and unpaired groups.
Statistical Analysis.
All experimental data were expressed as mean ± SEM. To test the statistical significance, one-way, repeated-measure ANOVA or two-way, repeated-measures ANOVA was used for the results of eyeblink conditioning. One-way ANOVA followed by post hoc Tukey–Kramer multiple comparisons test or t test was used for the quantified values of qRT-PCR and densitometry. A P value of less than 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Dr. T. Sassa for the protocol of GeneChip microarray, Dr. S. Kawahara for the protocol of eyeblink conditioning in mice, Dr. T. Iwasato for the protocol for in situ hybridization, and the staff of the Research Resource Center, RIKEN Brain Science Institute for animal care and technical assistance. This work was supported in part by National Institutes of Health Grants AG01451 and AG023742 (to R.F.T.).
Abbreviations
- AIN
anterior interpositus nucleus
- CR
conditioned response
- CS
conditioned stimulus
- DCN
deep cerebellar nuclei
- DLH
dorsolateral hump
- P3
3-d paired training
- P7
7-d paired training
- qRT-PCR
quantitative real-time RT-PCR
- PCG
plasticity candidate gene
- SHAM
sham negative control
- U7
7-d unpaired training
- US
unconditioned stimulus.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Christian K. M., Thompson R. F. Learn. Mem. 2003;10:427–455. doi: 10.1101/lm.59603. [DOI] [PubMed] [Google Scholar]
- 2.Attwell P. J., Ivarsson M., Millar L., Yeo C. H. Ann. N.Y. Acad. Sci. 2002;978:79–92. doi: 10.1111/j.1749-6632.2002.tb07557.x. [DOI] [PubMed] [Google Scholar]
- 3.Swain R. A., Shinkman P. G., Thompson J. K., Grethe J. S., Thompson R. F. Neurobiol. Learn. Mem. 1999;71:167–193. doi: 10.1006/nlme.1998.3872. [DOI] [PubMed] [Google Scholar]
- 4.Steinmetz J., Lavond D., Ivkovich D., Logan C., Thompson R. J. Neurosci. 1992;12:4403–4426. doi: 10.1523/JNEUROSCI.12-11-04403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skelton R. W. Behav. Neurosci. 1988;102:586–590. doi: 10.1037//0735-7044.102.4.586. [DOI] [PubMed] [Google Scholar]
- 6.Woodruff-Pak D. S., Lavond D. G., Logan C. G., Steinmetz J. E., Thompson R. F. Brain Res. 1993;608:67–77. doi: 10.1016/0006-8993(93)90775-i. [DOI] [PubMed] [Google Scholar]
- 7.Lavond D. G., Hembree T. L., Thompson R. F. Brain Res. 1985;326:179–182. doi: 10.1016/0006-8993(85)91400-3. [DOI] [PubMed] [Google Scholar]
- 8.Chapman P. F., Steinmetz J. E., Sears L. L., Thompson R. F. Brain Res. 1990;537:149–156. doi: 10.1016/0006-8993(90)90351-b. [DOI] [PubMed] [Google Scholar]
- 9.Bao S., Chen L., Kim J. J., Thompson R. F. Proc. Natl. Acad. Sci. USA. 2002;99:1592–1597. doi: 10.1073/pnas.032655399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L., Bao S., Lockard J., Kim J., Thompson R. J. Neurosci. 1996;16:2829–2838. doi: 10.1523/JNEUROSCI.16-08-02829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleim J. A., Freeman J. H., Jr, Bruneau R., Nolan B. C., Cooper N. R., Zook A., Walters D. Proc. Natl. Acad. Sci. USA. 2002;99:13228–13231. doi: 10.1073/pnas.202483399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller M. J., Chen N.-k., Li L., Tom B., Weiss C., Disterhoft J. F., Wyrwicz A. M. J. Neurosci. 2003;23:11753–11758. doi: 10.1523/JNEUROSCI.23-37-11753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauk M. D., Buonomano D. V. Annu. Rev. Neurosci. 2004;27:307–340. doi: 10.1146/annurev.neuro.27.070203.144247. [DOI] [PubMed] [Google Scholar]
- 14.Kishimoto Y., Fujimichi R., Araishi K., Kawahara S., Kano M., Aiba A., Kirino Y. Eur. J. Neurosci. 2002;16:2416–2424. doi: 10.1046/j.1460-9568.2002.02407.x. [DOI] [PubMed] [Google Scholar]
- 15.Koekkoek S. K., Hulscher H. C., Dortland B. R., Hensbroek R. A., Elgersma Y., Ruigrok T. J., De Zeeuw C. I. Science. 2003;301:1736–1739. doi: 10.1126/science.1088383. [DOI] [PubMed] [Google Scholar]
- 16.Gomi H., Sun W., Finch C. E., Itohara S., Yoshimi K., Thompson R. F. J. Neurosci. 1999;19:9530–9537. doi: 10.1523/JNEUROSCI.19-21-09530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bracha V., Irwin K. B., Webster M. L., Wunderlich D. A., Stachowiak M. K., Bloedel J. R. Brain Res. 1998;788:169–178. doi: 10.1016/s0006-8993(97)01535-7. [DOI] [PubMed] [Google Scholar]
- 18.Rescorla R. A., Solomon R. L. Psychol. Rev. 1967;74:151–182. doi: 10.1037/h0024475. [DOI] [PubMed] [Google Scholar]
- 19.Weinberger N. M. In: The Neural Basis of Behavior. Beckman A., editor. New York: Spectrum; 1982. pp. 63–91. [Google Scholar]
- 20.Thompson R. F., Donegan N. H., Clark G. A., Lavond D. G., Lincoln J. S., Madden J., IV, Mamounas L. A., Mauk C. D., McCormick D. A. In: Classical Conditioning. 3rd Ed. Gormezano I., Prokasy W. F., Thompson R. F., editors. Hillsdale, NJ: Erlbaum; 1987. pp. 371–399. [Google Scholar]
- 21.Neufeld M., Mintz M. Brain Res. 2001;889:112–117. doi: 10.1016/s0006-8993(00)03123-1. [DOI] [PubMed] [Google Scholar]
- 22.Mintz M., Wang-Ninio Y. Brain Res. 2001;897:150–156. doi: 10.1016/s0006-8993(01)02111-4. [DOI] [PubMed] [Google Scholar]
- 23.Lee T., Kim J. J. J. Neurosci. 2004;24:3242–3250. doi: 10.1523/JNEUROSCI.5382-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster M. K., Goya L., Ge Y., Maiyar A. C., Firestone G. L. Mol. Cell. Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quan N., He L., Lai W., Shen T., Herkenham M. J. Neurosci. 2000;20:6473–6477. doi: 10.1523/JNEUROSCI.20-17-06473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weidenfeld J., Itzik A., Feldman S. Brain Res. 1997;754:187–194. doi: 10.1016/s0006-8993(97)00078-4. [DOI] [PubMed] [Google Scholar]
- 27.Shors T. J., Weiss C., Thompson R. F. Science. 1992;257:537–539. doi: 10.1126/science.1636089. [DOI] [PubMed] [Google Scholar]
- 28.Tsai K. J., Chen S. K., Ma Y. L., Hsu W. L., Lee E. H. Y. Proc. Natl. Acad. Sci. USA. 2002;99:3990–3995. doi: 10.1073/pnas.062405399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meffert M. K., Chang J. M., Wiltgen B. J., Fanselow M. S., Baltimore D. Nat. Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 30.Sudhof T. C. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 31.Collin L., Usiello A., Erbs E., Mathis C., Borrelli E. Proc. Natl. Acad. Sci. USA. 2004;101:325–330. doi: 10.1073/pnas.0305994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles Y., Vivas-Mejia P. E., Ortiz-Zuazaga H. G., Felix J., Ramos X., Pena de Ortiz S. Neurobiol. Learn. Mem. 2003;80:80–95. doi: 10.1016/s1074-7427(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 33.Molteni R., Ying Z., Gomez-Pinilla F. Eur. J. Neurosci. 2002;16:1107–1116. doi: 10.1046/j.1460-9568.2002.02158.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen G., Steinmetz J. E. Brain Res. 2000;856:193–201. doi: 10.1016/s0006-8993(99)02429-4. [DOI] [PubMed] [Google Scholar]
- 35.Murray K. D., Isackson P. J., Jones E. G. Neuroscience. 2003;122:407–420. doi: 10.1016/j.neuroscience.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Morcuende S., Delgado-Garcia J.-M., Ugolini G. J. Neurosci. 2002;22:8808–8818. doi: 10.1523/JNEUROSCI.22-20-08808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibuki K., Gomi H., Chen L., Bao S., Kim J. J., Wakatsuki H., Fujisaki T., Fujimoto K., Katoh A., Ikeda T., et al. Neuron. 1996;16:587–599. doi: 10.1016/s0896-6273(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 38.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 39.Iwasato T., Datwani A., Wolf A. M., Nishiyama H., Taguchi Y., Tonegawa S., Knopfel T., Erzurumlu R. S., Itohara S. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.