Abstract
A comprehensive understanding of cancer is predicated upon knowledge of the structure of malignant genomes underlying its many variant forms and the molecular mechanisms giving rise to them. It is well established that solid tumor genomes accumulate a large number of genome rearrangements during tumorigenesis. End Sequence Profiling (ESP) maps and clones genome breakpoints associated with all types of genome rearrangements elucidating the structural organization of tumor genomes. Here we extend the ESP methodology in several directions using the breast cancer cell line MCF-7. First, targeted ESP is applied to multiple amplified loci, revealing a complex process of rearrangement and coamplification in these regions reminiscent of breakage/fusion/bridge cycles. Second, genome breakpoints identified by ESP are confirmed using a combination of DNA sequencing and PCR. Third, in vitro functional studies assign biological function to a rearranged tumor BAC clone, demonstrating that it encodes antiapoptotic activity. Finally, ESP is extended to the transcriptome identifying four novel fusion transcripts and providing evidence that expression of fusion genes may be common in tumors. These results demonstrate the distinct advantages of ESP including: (1) the ability to detect all types of rearrangements and copy number changes; (2) straightforward integration of ESP data with the annotated genome sequence; (3) immortalization of the genome; (4) ability to generate tumor-specific reagents for in vitro and in vivo functional studies. Given these properties, ESP could play an important role in a tumor genome project.
Completion of the human genome draft sequence ranks high among the most significant achievements in scientific history (Lander et al. 2001; Venter et al. 2001). Analysis of this sequence continues to transform our understanding of fundamental biological processes governing human biology and pathology. However, despite breathtaking progress made in genomics, sequence-based structural analysis of tumor genomes was nearly impossible, or, at best, extremely laborious until very recently (Raphael et al. 2003; Volik et al. 2003). This is unfortunate because amongst human pathologies, cancer is uniquely amenable to massively parallel analytical and computational approaches.
It is well established that solid tumor genomes accumulate a large number of rearrangements, including amplifications, deletions, translocations, episomes, and double minutes, many of which contribute to tumor progression (Gray and Collins 2000). Analyses of genomes of human solid tumors using comparative genomic hybridization (CGH) show that most contain numerous regions of copy-number abnormality. These abnormalities range from single-copy loss or gain of whole chromosomes to high-level amplification or deletion of relatively narrow genomic intervals. In many tumors, multiple regions of the genome are coamplified. Dual color fluorescence in situ hybridization (FISH) analyses of metaphase chromosomes from breast cancer cell lines with high-level amplification of several loci reveal that amplified loci are typically distributed across many different chromosomes, often not the ones from which they originated (Bautista and Theillet 1998; Zatkova et al. 2004). In addition, coamplified sequences originating on different chromosomes (e.g., MYC, ERBB2, and ZNF217 on 8q24, 17q13, and 20q13.2, respectively) can be located in close proximity at multiple distant genomic locations (J.W. Gray, unpubl.). This may lead to formation of novel fusion genes, gene disregulation, and gene inactivation. It is important that the composition of these amplified and/or colocalized structures be determined, because they may harbor many novel oncogenes, some of which may be tumor specific.
Historically, identification of genes whose structure and/or expression is altered by genome rearrangements has contributed significantly to our understanding of cancer progression, yielding important prognostic and predictive markers and targets for therapeutic intervention such as BCR-ABL, ERBB2, and TP53 (Ehrlich 2000). Indeed, the heralded anti-leukemia drug STI-571 was designed to abrogate the aberrant activity of the fusion BCR-ABL tyrosine kinase resulting from the Philadelphia chromosome translocation (Sawyers 1999). The breast cancer therapeutic Herceptin was designed to counteract the activity of the product of the amplified HER2/ERBB2 gene. STI-571 and Herceptin are examples where knowledge of tumor genome structure translated to therapeutics. This process took 40 yr for STI-571 and 11 yr for Herceptin. Sequence-based analyses of tumor genomes should greatly accelerate identification of therapeutic targets and their translation to the clinic.
Studies using spectral karyotyping (SKY) (Schrock et al. 1996) indicate that translocations play a more prominent role in solid tumor progression than previously appreciated (Artandi et al. 2000; Padilla-Nash et al. 2001; Yasunaga et al. 2001). Reciprocal and nonreciprocal translocations (NRT) occur in primary human tumors (Heim 1995), murine tumors (Artandi et al. 2000), and tumor cell lines (Padilla-Nash et al. 2001). In tumor cell lines they are frequently recurrent and may promote clonal expansion. Reciprocal translocations exert their oncogenic effect by disruption of genes at translocation breakpoints. NRTs create allelic imbalances, resulting in activation of oncogenes and/or inactivation of tumor suppressor genes. Little is known regarding relationships between breakpoints associated with NRT and tumor phenotype. Translocations of each type appear to be common in solid tumors. In one study, 12 translocations are visible in murine mammary tumors (Artandi et al. 2000). This figure almost certainly reflects the minimum number of translocations given the 10–20 Mb resolution of SKY. Other studies have revealed numerous translocations in breast, brain, and prostate cancer cell lines (Kytola et al. 2000; Padilla-Nash et al. 2001). A significant limitation of SKY is that being a cytogenetic tool, it cannot be easily integrated with the underlying human genome sequence. This limitation is being addressed by the Cancer Chromosome Anatomy Project (CCAP) (Knutsen et al. 2005). The Cancer Chromosomes database integrates the SKY/M-FISH & CGH Database with the Mitelman Database of Chromosome Aberrations in Cancer and the Recurrent Chromosome Aberrations in Cancer databases. This allows seamless searches within the ENTREZ search and retrieval system and the linking of chromosomal bands to underlying normal sequence assembly. Nonetheless, CCAP relies ultimately on cytogenetic mapping of normal BAC clones for integration, and, thus, is limited to cytogenetic resolution for the localization of tumor genome breakpoints. Moreover, the rearrangement breakpoints are not cloned. Consequently, CCAP cannot provide sequence-level information on the structures of tumor genomes. This is unfortunate given the direct linear relationship between the number of genome breakpoints and number of fusion and rearranged genes (Mitelman et al. 2004).
End Sequence Profiling (ESP) is a sequence-based technology for the analysis of tumor genomes capable of overcoming these deficiencies (Volik et al. 2003). Briefly, ESP begins with construction of a genomic library for the tumor of interest. The sequences at either end of the cloned DNA are then determined for individual clones and mapped onto the normal “reference” human sequence. Every pair of end sequences separated by an abnormally long (or short) distance, or an abnormal orientation of the mapped positions of end sequences, is an indication of a rearrangement. In contrast to other techniques that are often limited to detecting a particular type of aberration, the computational analysis of ESP data reveals all types of changes in genomic architecture, including copy-number abnormalities and structural rearrangements, such as translocations and inversions. These events correspond to different “ESP signatures” that can be decoded using computational techniques (Raphael et al. 2003; Raphael and Pevzner 2004). Moreover, ESP identifies BAC clones carrying these changes, provides insights into putative structural organization of tumor genomes (Raphael et al. 2003; Volik et al. 2003) and enables downstream sequencing and functional studies by generating tumor-specific reagents.
In the current study, we increased the whole-genome resolution of ESP to ∼225 kb and carried out targeted ESP—very high resolution ESP on specific loci—achieving a resolution of better than 10 kb in the targeted loci (see below). A striking feature of the MCF-7 genome is the presence of high-level amplicons on chromosomes 1, 3, 17, and 20 that are joined together by multiple BAC clones. Targeted ESP was performed to determine the structure of these amplicons and to gain insight as to the mechanism by which they may have arisen. In silico analysis of this data allowed modeling of the amplisome structures (Raphael and Pevzner 2004) that was partially validated by fluorescent in situ hybridization (FISH), draft sequencing BAC clones, and PCR. Moreover, we have identified several characteristics of amplified regions consistent with breakage/fusion/bridge (BFB) cycles (see Murnane and Sabatier 2004) for description of BFB model).
To have a biological effect, chromosomal rearrangements must alter the tumor transcriptome and confer a phenotype with a selective advantage. These changes may include altered expression of “normal” genes and expression of novel tumor-specific fusion transcripts. We directly demonstrate the ability of a rearranged genomic structure to alter phenotype by transfecting mouse mammary epithelial cells with a BAC clone from a 20q13.2 amplicon core encoding the ZNF217 gene and observing resistance to a commonly used chemotherapeutic agent. The role of translocations and novel fusion genes has been extensively characterized in hematopoietic malignancies, but is less well established in solid tumors. To test the hypothesis that large numbers of genome breakpoints detected in MCF-7 may result in fusion transcripts, we extend ESP to the analysis of the MCF-7 transcriptome. Transcript ESP (tESP) is similar to genome ESP, except that full-length enriched and normalized cDNA libraries are end sequenced. This process results in identification of novel tumor-specific transcripts, and suggests that combined tESP and genome ESP provide an integrated view of tumor genome structures and their malignant transcriptomes. Moreover, the identification of multiple tumor-specific transcripts may have significant implications for development of small molecule and vaccine-based anticancer therapeutics.
Results
End-sequence profiling
To determine the structure of the MCF-7 genome at high resolution, 11,511 BAC clones were end sequenced and mapped to the reference human genome sequence in addition to 8320 clones reported in Volik et al. (2003). All MCF-7 BAC clones are available for distribution from Amplicon Express (see Supplemental material for details). Interpretation of the data requires that three potentially confounding issues be addressed. First, BAC libraries may contain chimeric clones arising from random joining of DNA fragments during library construction. Second, local assembly errors of the reference sequence may introduce mapping errors. Finally, ESP may identify naturally occurring rearrangement polymorphisms rather than rearrangements that result from tumorigenesis.
In order to assess the importance of these issues, we end sequenced ∼1000 BAC clones from a normal human male BAC library and performed ESP to quantify the method's error rate. End sequencing revealed 13 putative “rearrangements” in 706 clones with paired mapped ends. Three were interchromosomal (0.42%) and 10 intrachromosomal (1.4%). Hence, we estimate that the total error rate of ESP is ∼1.8% including chimerism, mapping artifacts, and rearrangement polymorphisms. By comparison, estimates of the frequency of chimerism in BAC libraries generally range from 1% to 5% (Zhu et al. 1999; Crooijmans et al. 2000; Osoegawa et al. 2000, 2001; Eggen et al. 2001). We did not find that assembly errors in the reference human genome present a significant obstacle for ESP. While we did observe differences in mapping results among earlier assemblies, the majority of these differences were remedied in the “finished” sequence (International Human Genome Sequencing Consortium 2004). One notable exception is pericentromeric regions characterized by less complete assembly (She et al. 2004) and a threefold higher density of segmental duplications (Zhang et al. 2005). The existence of rearrangement and copy number polymorphisms in human populations (Giglio et al. 2001, 2002; Osborne et al. 2001; Stefansson et al. 2005) represent a different challenge for ESP. These variants will be detected by ESP, as recently demonstrated by Tuzun et al. (2005) who used an approach identical to ESP for studying the fine-scale variation of the human genome. However, these variations are not likely to play an important functional role in cancer. Incidentally, one of the aforementioned 13 breakpoints identified in the normal human library was also identified by Tuzun et. al. (2005) and another is located within 80 kb of a structural polymorphism reported in their study.
Mapping the ends of 19,831 MCF-7 BAC clones yielded 13,303 mapped BAC end sequence (BES) pairs and 5231 singletons. Assuming a haploid genome of 3000 Mb, we obtain a whole-genome resolution of ∼225 kb for detection of structural rearrangements (paired BES only) and 165 kb for copy-number measurements (BES pairs + singletons). Given an average BAC size of 141 kb, we achieved ∼0.62× clonal coverage of the MCF7 genome (paired BES), a threefold increase from our previous study (Volik et al. 2003). Lander-Waterman statistics predict that with such coverage, ∼46% of the genome should be covered by a clone, but empirically, we find that mapped clones cover 36% of the reference human genome. No unusual cloning biases were observed. A total of 582 (or 4.5%) of the MCF-7 BAC clones span apparent genomic breakpoints, approximately two and a half times the percentage found in the normal human BAC library. We clustered the BES pairs of breakpoint spanning clones and identified 45 breakpoints supported by at least two independent clones and 382 breakpoints supported by a single clone, some portion of which are likely artifacts.
Targeted end-sequence profiling
The density of mapped BES is highly concordant with that obtained by aCGH as previously reported (Volik et al. 2003). In particular, four genomic loci, i.e., 1p21.1-p13.3, 3p14.2-p14.1, 17q23.2–23.3, and 20q13.2, have an abnormally high concentration of BES pairs containing 265, 667, 1101, and 1603 BES, respectively (3636 total). Thus, ∼10% of BES mapped to 0.63% of the genome. In regions of copy-number gains, the resolution of ESP is proportionally higher. For example, 407 BES mapped to the 1.4 Mb ZNF217 amplicon on 20q13.2 (Collins et al. 2001), or an average of one BES per 3.5 kb, although the distribution of BES in this region is not uniform.
To refine the amplicon structures, we performed targeted ESP by screening the arrayed MCF-7 BAC library with multiple hybridization probes spaced at ∼50 kb across amplicons at chromosomes 1, 3, 17, and 20, and end sequencing the isolated BAC clones. The combination of targeted and whole-genome ESP allowed the mapping of ∼600 BES on the 1.4 Mb of ZNF217 amplicon (approximately a BES every ∼2.2 kb). Targeted ESP alone resulted in mapping ∼200 BES (or roughly a BES every 5.6 kb). It should be noted that targeted ESP required end sequencing only 434 BAC clones vs. 19,831 for whole-genome ESP.
Since detection of chromosome rearrangements using ESP relies on the analysis of the mapped positions of BES, the resolution of ESP for detection of structural features is a function of the density of mapped paired BES. The calculation of the exact resolution of targeted ESP is complicated, since on a 3-Gb genome a 150-kb BAC clone may be considered to be a point, but on a 1-Mb interval it cannot. In any case, mapping of hundreds of relatively small BES allows the identification of copy-number peaks with far greater resolution than tiling path arrays of BAC clones, which are limited to ∼50–100 kb (see, for example, Krzywinski et al. 2004). We estimate that the copy-number resolution achieved by ESP in the ZNF217 amplicon is better than 10 kb. This is supported by our previous studies, which mapped the proximal boundary of the ZNF217 amplicon to within 10 kb of ZNF217 (Collins et al. 2001), perfectly correlating with the ESP-based copy-number profile.
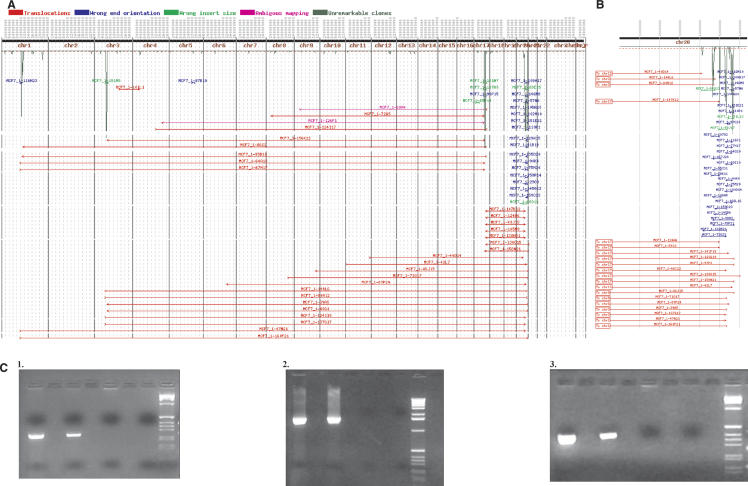
Figure 1A shows the results of mapping BES from these amplicons. It is evident that DNA from the four amplicons is packaged together in one or more structures that may incorporate DNA from other loci as well. Figure 1B shows a chromosome-specific view of the chromosome 20q13.2 amplicon resolved into four copy-number peaks that are spaced at ∼1.5-Mb intervals and connected by multiple clones. The orientation of the BES suggests that these structures are the result of a complex evolution involving a series of rearrangements that conjoin normally noncontiguous DNA (blue lines). The red lines represent BAC clones connecting the various 20q13.2 amplicon peaks to chromosomes 1, 3, 17, and much less frequently, to other chromosomes. Mapping BES spanning intrachromosomal and interchromosomal breakpoints reveals sharp amplicon boundaries. As we reported previously, one BAC clone, MCF7_1-3F5, which spans a chromosome 20 to chromosome 3 junction was sequenced to completion and its assembly revealed DNA sequence originating from 20q12, two distant loci on 20q13.2, and 3p14 (Volik et al. 2003). Interestingly, two regions from 20q13.2 are fused in opposite orientation, and this particular junction was confirmed by PCR (Supplemental Table 1). Similar complex structures occur on chromosomes 1, 3, and 17. However, amplicons on 1, 3, and 17 do not contain the clones with abnormal orientation of BES observed on 20q13.2.
Figure 1.
(A) The result of targeted ESP on amplicons mapping to chromosomes 1,3,17, and 20. A total of 434 end-sequence pairs were mapped onto the normal human genome sequence (represented as a horizontal line along the top). The dark-green plot represents the number of end sequences per 1-Mb interval. BAC end pairs with ends mapping to different chromosomes are shown as horizontal red lines. BAC clones with ends in the wrong orientation (not pointing toward each other) are shown in blue. BAC clones with ends mapping more than three standard deviations farther apart than the average BAC insert are shown in light green. A complete display with all mapped clones is at http://shark.ucsf.edu/~stas/ESP2/esp2.html, and a more detailed description can be found in Volik et al. (2003). (B) Chromosome 20 end-sequence density and end-sequence pair plots. (C) PCR confirmation of genome breakpoints in clones MCF7_1-37_E22 (1), MCF7_1-94_M14 (2), and MCF7_1-23_I06 (3). A shotgun library was constructed from the insert of each of these clones, and 96 plasmid subclones end sequenced. Plasmids with end sequences mapping to distant genomic loci were identified, and primers straddling the putative breakpoint were designed. In subpanels 1–3, the first lane is DNA from the corresponding BAC clone, the second lane is MCF-7 genomic DNA, the third lane is normal genomic DNA, the fourth lane is negative control, and the fifth lane is 1-kb ladder (Gibco-BRL).
Genomic breakpoint validation
To validate the rearrangement breakpoints suggested by mapped BES pairs, we draft sequenced 10 representative BAC clones connecting the amplicons on chromosomes 1, 3, 17, and 20. Each BAC was subcloned into a plasmid library and 96 plasmid clones were end sequenced, providing ∼1.5× clonal coverage. Breakpoint-spanning plasmids were identified using ESP and validated using PCR in MCF-7 DNA. Positive PCR reactions on both the BAC clone and MCF-7 DNA combined with a negative PCR reaction in control normal genomic DNA validates the breakpoint. In all, 29 pairs of PCR primers were designed and shown to amplify DNA from the corresponding BAC clones. Of these, 25 primer pairs from eight clones were validated on MCF-7 DNA (Supplemental Table 1). Two BAC clones, MCF7_1-94_M14 and MCF7_1-21_C24, contain a single validated breakpoint consistent with the mapped BAC end sequences; therefore, each of these clones probably identifies a genomic locus in MCF-7 that resulted from a single fusion of two distinct loci in the normal human genome. Surprisingly, the remaining six BAC clones contain either several validated breakpoints or a single breakpoint that is different from that suggested by the mapped BAC end sequences, implying that these clones have complex internal structure. Thus, some loci in the MCF-7 genome are extensively scrambled below the resolution of an end-sequenced BAC clone. The number of breakpoints detected in these BAC clones is remarkable; one clone, MCF7_1-110P1, has eight confirmed breakpoints.
Functional studies
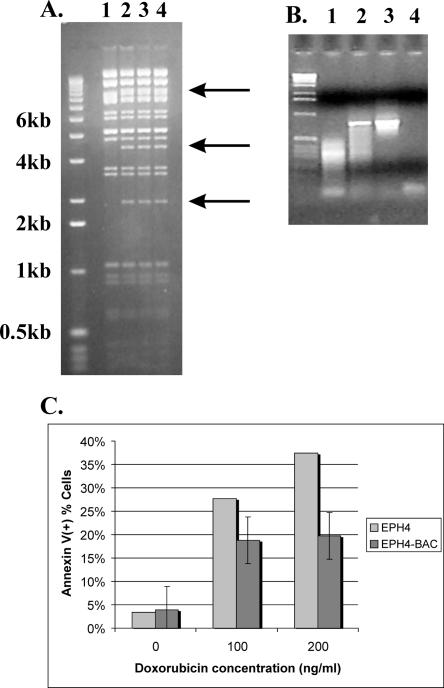
A persistent question is whether genomic rearrangements detected by ESP are functionally significant or simply the result of genome instability. An important strength of ESP is that once a breakpoint is identified, it is immediately available in a BAC clone for functional studies that are amenable to high-throughput approaches. One method for conducting such studies is to retrofit a genomic clone to express a selectable marker, transfect an appropriate cell line with it, and score the transformants for cancer-related phenotypes. We tested this approach with BAC clone MCF7_1-3F5, chosen because this clone maps to one of the four 20q13.2 amplicon cores, contains the highly rearranged MCF-7 ZNF217 locus, and was sequenced to completion (Volik et al. 2003). Moreover, amplification of the ZNF217 locus is associated with reduced survival of women with breast cancer (Tanner et al. 1996; Collins et al. 1998) and the overexpression of this gene has been shown to immortalize cultured human mammary epithelial cells (Nonet et al. 2001). Current evidence suggests that aberrant expression of ZNF217 promotes cell survival through attenuation of apoptosis (Huang et al. 2005). We retrofitted clone MCF7_1-3F5 with a neomycin resistance gene and transfected it into mouse mammary epithelial cells under selection for neomycin resistance (Fig. 2A,B). We assayed the transfected cells for apoptosis in response to the DNA-damaging agent doxorubicin, a commonly used chemotherapeutic drug (Fig. 2C). Transfected cells demonstrated increased resistance to doxorubicin-induced apoptosis relative to controls. This result demonstrates the phenotypic effect of amplified and rearranged genomic loci, and vividly illustrates the advantage of working with a large insert genomic library. The availability of arrayed BAC libraries makes it possible to quickly investigate the functional significance of genome rearrangements detected using ESP and to study cancer genes in the context of the tumor genome which, as we have shown, can be highly divergent from the host genome.
Figure 2.
Using tumor-derived BAC clones for phenotype screens. (A) Confirmation of retrofitting BAC MCF-7_1-3F5 for transfection studies. Arrows mark new bands in the vector resulting from insertion of pRetroES plasmid. (B) PCR-based control for transfection of EPH4 cells with retrofitted BAC clone. (Lane 1) Nontransfected cells; (lane 2) transfected cells; (lane 3) positive control (BAC DNA); (lane 4) negative control. (C) Increase in resistance of EPH4 cells to doxorubicin. Note the decrease in the number of apoptotic cells as demonstrated by the annexin assay.
Transcript ESP
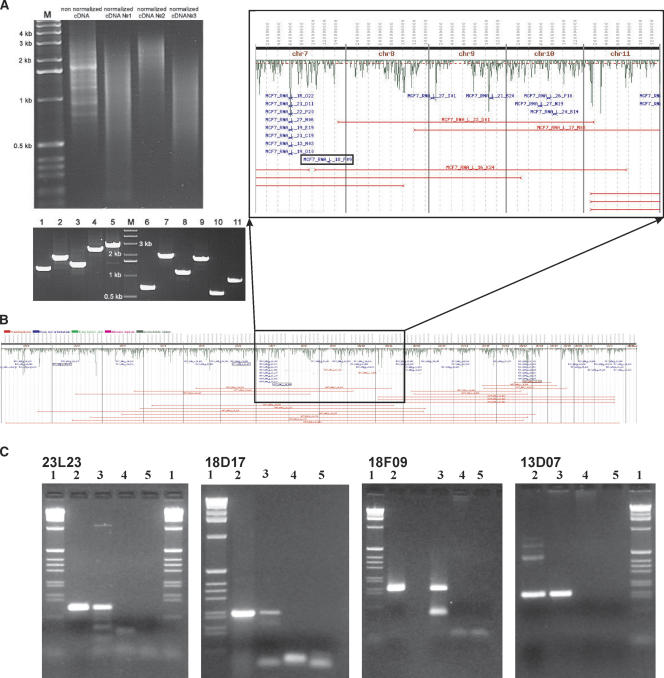
We hypothesized that the extensive genome rearrangements present in MCF-7 create fusion transcripts. To test this hypothesis, we constructed a full-length enriched and normalized cDNA library from total MCF-7 RNA (Fig. 3A). We arrayed and end sequenced 5089 recombinant cDNA clones (clones are available from C.C. laboratory). We were able to map both ends of 2700 cDNA clones; of these, 44 clones had ends mapping to different chromosomes and 56 had ends mapping in the wrong orientation. ESP analysis (Fig. 3B; Supplemental Table 2) identified 24 candidate novel transcripts that mapped to exons/introns/UTRs of known genes. PCR validation of these candidates was performed on two independent preparations of cDNA, i.e., the normalized cDNA that was used for the library construction and non-normalized first-strand cDNA independently synthesized from the same preparation of mRNA. In order to increase specificity and sensitivity of the assay, we utilized nested primer strategy. Only clones positive in both assays were considered validated. PCR primers were successfully designed for 21 of the 24 candidate novel transcripts, and four of these were validated (Fig. 3C; Table 1). Three of these four transcripts required a nested PCR strategy for validation, which could indicate that many of the detected fusions are very rare transcripts. Moreover, the validated transcripts exhibit fusions of exons, introns, and UTRs, in agreement with earlier studies (Hahn et al. 2004). We are currently extending this analysis to additional tumor and cell line samples and performing experiments designed to ascertain whether the observed novel tumor-specific transcripts are translated. The results of these studies will be reported in a separate manuscript.
Figure 3.
Analysis of the MCF-7 cDNA library and transcript ESP. (A) The results of normalization of the MCF-7 cDNA (top) and of PCR-based sizing of 11 randomly selected cDNA clones (bottom). (B) The results of the ESP analysis of 5000 end-sequenced cDNA clones. See Figure 1 for the detailed description of this panel with four validated clones boxed. (C) The results of the PCR validation of these clones on two independent preparations of MCF-7 cDNA using clone-specific primers spanning breakpoints (Table 1). In each panel, lane 1 is the 1-kb ladder (GIBCO-BRL); lane 2 is clone DNA; lane 3 is independent preparation of MCF-7 cDNA; lane 4 is normal breast cDNA (Stratagene adult human female, breast first strand cDNA cat# 780602); lane 5 is negative control.
Table 1.
Structure of validated novel transcripts.
| Clone name | cDNA end sequence length | Chromosome coordinate | cDNA end orientation | Hit type | Gene | Primer sequence | Tm (°C) |
|---|---|---|---|---|---|---|---|
| MCF7_RNA_L_18_D17 | 310 | chr2:55448021 | Plus | E | MTIF2 | aggaacctgtgcatctttgg | |
| aaagcagcatgtcctggagt | 58 | ||||||
| 553 | chr2:43913586 | Plus | IE | PLEKHH2 | acccaccatgaagggattg | 62 | |
| ctttgcaaatctgcctgaca | |||||||
| MCF7_RNA_L_13_D07 | 526 | chr6:100,979,123 | Minus | E,- | HELIC1 | gcagctcctaagggtgagtg | |
| chr6:101,033,719 | Minus | gggattggggtagaggtttt | 62 | ||||
| 556 | chr6:101,002,220 | Plus | U | HELIC1 | ttcaatgctgcgattatcctc | 62 | |
| tccaattcaatcagggacttc | |||||||
| MCF7_RNA_L_18_F09 | 630 | chr7:128,003,608 | Minus | U | CALU | ccaggtaatggcaggttcag | |
| chr7:128,003,709 | Plus | gctggacctatagcaactgaatg | 62 | ||||
| 547 | chr7:128,005,309 | Minus | U | CALU | gaagaagaggaaccggatgg | 62 | |
| aagtcttgagcatttcaacagatg | |||||||
| MCF7_RNA_L_23L23 | 403 | chr16:85,490,770 | Plus | U | KIAA0182 | tattcggcaccacactacc | |
| agcaaattgtgcaatggaat | 62 | ||||||
| 405 | chr19:16,074,534 | Plus | IE | AK023385 | ggtttgtgctgaccatctcc | 62 | |
| tggatcccaggcattttcta |
Key to “Hit type” column: “I”—hit in the intron of a gene, “E”—hit in the exons of a gene; “U”—UTR of a gene, “-”—a hit in intergenic DNA. Since all of these clones were validated using nested primer approach, four primer sequences are reported for each (inner primer set data are given in italics). All PCR products were validated by sequencing.
Discussion
We demonstrated the ability of ESP to reveal the structural organization of a tumor genome by identifying extensive rearrangement and loci on chromosomes 1, 3, 17, and 20, including evidence of coamplification. Draft sequences of selected BACs showed that some of these rearrangements operate on small genomic segments, tens of kilobases in size. Thus, ESP provides a powerful complement to existing techniques like CGH and SKY. Moreover, it is now possible to investigate whether these complex rearrangements result in abnormal gene expression, splicing, or function. We have taken the first step by establishing that a BAC-encoding ZNF217 can attenuate apoptosis in response to the DNA-damaging agent doxorubicin when transfected into mouse mammary epithelial cells. Similar experiments can be carried out in an effort to investigate biological effects of translocations and other genome rearrangement detected by ESP and mapped to arrayed BAC clones. These studies may benefit from the fact that the transfected genes will be expressed in the context of the tumor, not the normal genome, thus preserving their cis regulatory and splicing elements. In principle, complex genome structures such as the chromosome 1, 3, 17, and 20 amplicons described here can be reconstructed in vitro and even in vivo using BAC transgenics.
An additional strength of ESP is that it enables formulation of new hypotheses and downstream biological studies. For example, detailed analysis of the MCF-7 data shows that the observed chromosome rearrangements demonstrate several characteristics consistent with the hypothesis that breakage/fusion/bridge (B/F/B) cycles played an important role in the evolution of the MCF7 genome. As predicted by the B/F/B model, MCF-7 amplicons are relatively small in size, contain large numbers of “inversions” and nonreciprocal translocations, and are joined together by a number of BAC clones, indicating colocalization of DNA from different genomic regions. Thus, we hypothesize the following model for evolution of the MCF7 genome. Amplification in MCF-7 started with telomere loss on chromosome 20q, which initiated prolonged B/F/B cycles. The original amplicon may have targeted the ZNF217 (Collins et al. 1998) locus at 20q and underwent significant rearrangements. Chromosome instability spread to 17q, where chromosome 17 and 20 sequences were coamplified. Finally, the resulting 20q/17q amplicon spread to 1p and 3p (see Supplemental material for details).
Clearly, in order to have functional significance, genome rearrangements must alter gene structure, expression, or function. A number of recent studies suggest that the importance and frequency of fusion transcripts in solid tumors is likely to be underestimated (Mitelman et al. 2004). The potential clinical significance of these tumor-specific transcripts cannot be overstated. Rabbitts and Stocks (2003) point out that while the translocation products are difficult therapeutic targets because of their intracellular location, their tumor specificity is “an important motivating factor for developing these new therapies.” Thus, tumor-specific transcripts and BAC clones encoding them represent an invaluable resource for the identification of novel therapeutic targets with associated biomarkers.
Theoretically, it should be possible to identify fusion transcripts in tumors and estimate their frequency using computational analysis of existing cDNA databases. However, attempts at in silico identification of tumor-specific fusion transcripts using public data proved problematic (Futreal et al. 2001; Hahn et al. 2004). The advent of capillary sequencing that eliminates lane tracking errors combined with recent advances in cDNA library construction methodologies allowing construction of full-length enriched and normalized libraries from very small amounts of total RNA (Zhulidov et al. 2004) enabled more efficient identification of novel tumor-specific transcripts. We identified 24 putative novel transcripts, of which four were confirmed to be expressed in MCF-7. Interestingly, the diversity of fusions of exons, introns, UTRs, etc. was also observed in another MCF-7 library analyzed by Hahn et al (2004). One of the fusion genes (IRA1/RGS1) discovered in their study fuses the 3′ UTR and adjacent intergenic region of IRA1 to the last four exons of RGS17. An open reading frame starts at the beginning of this EST and encompasses RGS17 exons. The non-RGS17 part of the transcript does not have an apparent identity to the known proteins from GenBank. If this protein were indeed translated, it would demonstrate that rearrangements of tumor genomes might result in completely novel tumor-specific proteins.
The results of tESP suggest that the transcriptomes of human solid tumors may contain large numbers of fusion transcripts. While there is clearly room for technical refinement in tESP, it should be noted that only two fusion transcripts were previously known to exist in MCF-7, while we have validated four additional fusion transcripts. Performing ESP and tESP on the same tumor enables direct identification of genomic breakpoints encoding fusion transcripts. Alternatively, the BAC libraries can be screened to identify recombinant BAC clones encoding fusion transcripts identified using tESP. We demonstrated this approach using the MCF-7 BCAS4/3 fusion transcript (Barlund et al. 2002) to isolate two BAC clones encoding BCAS4/3 and the translocation breakpoint (data not shown).
Presently, expenses associated with ESP library construction and end sequencing are significant. However, the cost of sequencing is directly related to the sequencing depth required to detect multiple independent BAC clones spanning breakpoints. The required sequencing depth can be estimated using data from the amplified loci. DNA in amplicons is overrepresented in the BAC library and is consequently sequenced to a proportionately greater depth. Interpreted this way, our data suggest that end sequencing ∼60,000 BAC clones per library is necessary for whole-genome ESP. The sequencing depth can be adjusted by careful selection of tumors and cell lines with high-level amplifications (e.g., MCF-7). Analysis of the MCF-7 data demonstrates that for genomes characterized by amplicons, sequencing ∼10,000 clones is sufficient for determining the overall genome architecture (data not shown). Doubling this number did not result in a significant increase in resolution in nonamplified regions. Thus, targeted ESP constitutes an excellent low-cost alternative to whole-genome ESP, making very high-resolution analysis of specific loci possible for a small fraction of the cost of whole-genome ESP. While end sequencing of 60,000 clones will yield a whole-genome resolution of ∼50 kb, targeted ESP can readily achieve resolutions of better than 10 Kb in targeted loci. In the case of MCF-7, four amplicons were analyzed by end sequencing 434 BAC clones. In addition, BAC libraries can be arrayed on filters or pooled, making high-throughput screening possible using established and novel methodologies. For example, recently, Milosavljevic et al. (2005) reported a novel approach to massively parallel draft sequencing of BAC libraries based on a sophisticated pooling strategy that offers a very economical method for determining the structure of tumor-derived genomic clones.
A tumor genome project
Understanding the sequence and organization of tumor genomes will likely be as fundamental to comprehending tumor biology and pathology as analysis of genome sequences has become to biology as a whole. This idea is a motivating factor in recent discussions concerning a Human Cancer Genome Project (HCGP) (Kaiser 2004, 2005; National Institutes of Health 2005) and in the development of ESP. The HCGP should have well-defined biological and translational science goals. Clearly, tumor genomes should be viewed as dynamic emergent systems, so the scientific goals should include both the identification of the altered components (e.g., mutations, chromosome rearrangements, tumor-specific genes and transcripts) and the study of the interactions between these components that sustain and drive progression. While the completion of the human-genome sequence confers a significant advantage on the HCGP because all tumor genomes are derivatives of normal human genome, a HCGP will confront unique challenges and requirements. In particular, most solid tumors are a dynamic, heterogeneous population of evolving cells. The dynamic process of tumorigenesis results in selection of mutations and structural rearrangements that are functionally significant, as well as a background of mutations and rearrangements that are of no functional consequence. Other mutations and breakpoints may occur in a minority of cells in the primary tumor, but evolve to dominance during metastasis. Essentially, each tumor is unique, and thus, a HCGP will require detailed sequence-based analysis of hundreds of highly complex gigabase-sized genomes having very different physical characteristics.
The complexity of an HCGP strongly suggests that a combination of approaches aimed at elucidation of the architecture of tumor genomes as well as a detailed sequence analysis of cancer-related loci will ultimately be required. Several properties of ESP make it uniquely suitable for these. First, ESP has the ability to detect all types of rearrangements and copy-number changes, and the integration of ESP data with the annotated genome sequence is straightforward.
Second, ESP immortalizes the tumor genome under study. This property is essential because the amount of DNA available from tumor specimens is likely to be severely limited, and tumors with significant clinical follow-up enabling genotype–phenotype associations are a uniquely valuable resource, and if the deadliest tumors are to be studied, the metastases will first have to be collected, as these samples are presently underrepresented in tumor banks. Thus, it is impossible to overstate the importance of primary and metastatic tumors to translational research (see, for example, Paris et al. 2004, 2005) and consequently the importance of their conservation. Therefore, immortalization of tumor genomes and transcriptomes should be high priority. To date, we have successfully constructed BAC libraries from breast, brain, and ovarian primary tumors and a prostate metastasis. As little as 50 mg of specimen was estimated to be sufficient for the construction of a 10- to 20-fold coverage tumor library and the ability to construct BAC libraries does not appear to be tumor specific. The detailed results of the analysis of these libraries will be reported in a separate manuscript.
Third, ESP generates tumor-specific reagents for in vitro and in vivo functional studies. While identification of recurrent mutations and structural rearrangements may suggest biological significance, functional studies are clearly an imperative. It is unlikely that the throughput of existing methods for functional analysis will keep pace with the rate of discovery of mutations, genomic rearrangements, and fusion transcripts that an HCGP is likely to achieve, and this should be addressed as part of such a project.
Finally, ESP has significant flexibility in methodology, allowing participation of small academic laboratories and large genome centers. We have chosen BACs for ESP because they provide an excellent platform for functional and structural research and greatly facilitate downstream functional annotation by providing a unified pool of sequence-annotated large insert clones. Construction of tumor BAC libraries establishes an inexhaustible source of DNA in a format that makes distribution of whole libraries or individual clones easy. BAC clones became the vector of choice for sequencing the human genome (Lander et al. 2001). More recently, fosmids have gained popularity for whole-genome sequencing and comparative studies (Leveau et al. 2004; Magrini et al. 2004; Moon and Magor 2004; Jansen et al. 2005; Shimada et al. 2005; Tuzun et al. 2005). Fosmids offer a number of advantages over BACs. Being much smaller (40 vs. 150 kb), fosmids are generally easier and less expensive to handle than BACs, and allow better spatial resolution of breakpoints, which is useful in light of the extensive small-scale rearrangements we observed in MCF-7. In addition, the size distribution of fosmids can be more tightly controlled than that of BAC clones, allowing detection of small insertions and deletions. However, the smaller size of fosmids has several disadvantages. Fosmid libraries are at least three to five times larger than BAC libraries of equal complexity and, therefore, one needs to sequence three to five times more fosmids than BAC clones to obtain equal genome coverage, which greatly complicates the logistics of managing tens to hundreds of genome-sized libraries and significantly increases the storage and sequencing costs. Moreover, it is desirable to construct inexhaustible DNA pools from tumor libraries that would facilitate rapid PCR-based screening and resequencing. It will be less expensive to construct such pools from smaller BAC libraries. Finally, the small size of fosmid clones means they will be much less likely to contain complete normal or fusion genes than BAC clones. This may make functional analysis of rearrangements far more difficult and expensive. Ultimately, the choice of vector may be project specific.
We conclude that the key features of ESP, namely, (1) the ability to detect all types of rearrangements and copy-number changes; (2) the straightforward integration of ESP data with the annotated genome sequence; (3) the immortalization of the genome; (4) and the ability to generate tumor-specific reagents for in vitro and in vivo functional studies; and (5) methodological flexibility, merit the consideration of ESP for a prominent role in any Human Cancer Genome Project.
Methods
BAC library construction
Library preparation was carried out as reported earlier (Volik et al. 2003) (see detailed protocol at http://shark.ucsf.edu/~stas/ESP2/esp2.html).
Overgo hybridization
Overgo hybridization of the MCF-7 BAC library was carried out as described in Han et al. (2000). All positive BAC clones were end sequenced.
cDNA library construction
Normalized and full-length enriched cDNA libraries from the MCF-7 breast cancer cell line were constructed at Evrogen, Ltd. The detailed protocol can be found in Zhulidov et al. (2004). This method for construction of near-full length and normalized cDNA libraries is based on two advances. The first development is the PCR suppression effect, which makes it possible to construct representative cDNA libraries from small amounts of total RNA (Lukyanov et al. 1997; Matz 2002) and to selectively regulate the size of the amplified cDNA fragments (Shagin et al. 1999). The second important technical development was the discovery of a novel duplex-specific nuclease from Kamchatka crab that preferentially cleaves perfectly matched duplexes (Shagin et al. 2002). Briefly, the procedure consists of the following steps. cDNA is prepared using a SMART PCR cDNA synthesis kit (Clontech) according to the manufacturer's protocol. Depending on the amount of the starting material, this cDNA can be either amplified or used directly in the normalization step. Normalization of the constructed library is achieved by first heating the aliquot of cDNA in hybridization buffer to 98°C for 3 min, followed by 4 h of reannealing at 70°C. Since DNA reassociation follows second-order reaction kinetics, a significant proportion of highly abundant cDNA species reanneal in the allotted time, while rarer transcripts remain in single-strand fraction. Treatment of the reaction mixture with Kamchatka crab nuclease specifically degrades the double-stranded cDNA fraction. The remaining normalized single-strand fraction is amplified and cloned into a suitable vector.
End sequencing tumor BAC and cDNA clones
The end sequencing of BAC and cDNA clones was carried out by Agencourt Biosciences using standard conditions as described previously (Volik et al. 2003) and by the Wellcome Trust Sanger Institute. All sequence data was deposited in GenBank (Accession nos. DN911814–DN920916 and CZ445878–CZ466054).
Shotgun sequencing of tumor BAC clones
BAC DNA was purified from 250 mL of overnight culture using the Qiagen columns (Qiagen). Approximately 2 μg of BAC DNA was mechanically sheared using the HydroShear (GeneMachine, Inc.), end repaired with the Klenow enzyme and T4 DNA polymerase, size selected for 3 ± 0.5-Kb fragments on agarose gels, and cloned into a pUC19 vector. Individually picked subclones were grown on 96-well plates overnight in LB plus 200 μg/mL ampicilin and 10% glycerol. Subcloned plasmid DNA was prepared from the arrayed cells using the TempliPhi kit (GE/Amersham) according to the manufacturer's protocol; 3-Kb subclones were end sequenced using BigDye terminators (Applied Biosystems) and capillary sequencers. Quality of the sequence reads were determined by Phred score (Ewing et al. 1998) and only sequences greater than Q20 were included in the analysis.
BAC retrofitting and cell transfections
BAC clones were retrofitted to express neomycin resistance for selection in mammalian cells essentially as described in Wang et al. (2001). Briefly, plasmid vector pRetroES was used to add the neomycin resistance gene to BAC clones through cre-mediated recombination at the loxP site of the BAC vector. pRetroES contains ampicilin and neomycin resistance genes and a tac promoter-driven GST-loxP-cre fusion gene that has functional cre recombinase activity. After transformation of pRetroES into Escherichia coli carrying a BAC clone, the fusion gene will promote site-specific recombination at the loxP site. Selection on ampicilin/chloramphenicol selects for the retrofitted BAC; however, the tac promoter is separated from the cre fusion gene product, inactivating the cre enzyme and further recombination. Individual colonies were screened for recombination by PCR with primers specific to pRetroES and to the loxP site on the BAC vector. Additional confirmation was carried out by comparing DNA fingerprints of normal and recombinant clones using the restriction enzyme HindIII (see Fig. 3). We used standard protocols for mammalian cell transfection. Briefly, 2 · 105 cells were seeded in each well of a 6-well plate. On the second day, BAC DNA was transfected into the EPH4 cells (Fialka et al. 1996) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Forty-eight hours later, the cells were trypsinized and grown under G418 selection (at 500 μg/mL). The cell-culture medium was exchanged every 3 d. About 1 mo later, stable BAC DNA-transfected cells were obtained. Genomic DNA was isolated from the stable transfected cells using the DNA purification Kit (Promega) and PCR was performed to verify the BAC DNA transfection.
In vitro functional oncogenomics
Transfected cells were exposed to indicated amounts of doxorubicin for 16 h prior to harvest. Adherent and nonadherent cells in the medium were collected, washed twice with phosphate-buffered saline, then resuspended in 100 μL of binding buffer (10 mM HEPES, 140 mM NaCl and 2.5 mM CaCl2 at pH 7.4). A total of 7.5 μL of Annexin V antibody (Molecular Probes) was added and the cells were incubated at room temperature for 15 min. After incubation, 400 μL of binding buffer was added, and cell samples were analyzed by flow cytometry (FACS Calibur, Becton Dickinson). For 3F5 BAC-transfected cells and nontransfected cells, 30,000 events were first gated to exclude small debris, and then analyzed for percentage of live (Annexin V–) vs. dead (Annexin V+) cells. The significance of the differences observed was calculated using Student's t-test. Values were judged to be significant when P < 0.05.
Data analysis
End-sequence profiling
We use a two-step procedure that involves first mapping the end sequence (ES) data onto the human genome sequence (NCBI Build 34, July 2003), and then analyzing the mapping results. The mapping step is accomplished using BLAT (Kent 2002). A location is assigned if at least 50 bp of an ES aligns to the reference genome sequence with at least 97% identity. If the ES hits multiple locations in the genome, the position of the longest hit with the highest identity is chosen and the ES is designated as being “ambiguously mapped” for further computational analysis. We identify clusters of ES pairs as sets whose pair of locations are close enough to be explained by a single rearrangement event.
Analysis of the resulting mapped ES necessitated the development of custom visualization software. In the case of genomic ESP, the overall number of BES per given genomic interval is roughly proportional to copy number. Thus, a plot of BES density generates a copy-number profile for the entire tumor genome. For cDNA libraries, a plot of end-sequence density is expected to be roughly proportional to the number of transcripts detected in the library per given genomic interval. Since we used normalized cDNA libraries with the same relative abundance of different transcripts, the cDNA ES density plot is expected to reflect the distribution of transcriptionally active sequences across the tumor genome. In addition, the software creates a list of clones that span aberrations, which are defined as those clones that (1) have ends that map to different chromosomes; (2) have abnormal apparent size (the distance between mapped positions of end sequences), or (3) have wrong orientation of ends. Data can be presented for the whole genome or for each chromosome (see Figs. 1, 3).
Acknowledgments
The work in the C.C. laboratory was supported by Grant no. R33 CA103068 from NIH/NCI and Breast Cancer Research Program Grant no. 8WB-0054. The work in the J.P.M. laboratory was supported by National Institute of Environmental Health Science Grant no. RO1 ES008427, and National Cancer Institute Grant no. RO1 CA69044. B.J.R. is supported by a Career Award at the Scientific Interface (CASI) from the Burroughs-Wellcome Fund, and a fellowship from the Alfred P. Sloan Foundation. The work performed in the J.-F.C. laboratory was supported by the National Heart, Lung, and Blood Institute, Programs for Genomic Applications Grant no. UO1 HL66728. The work at the Wellcome Trust Sanger Institute was supported by the Wellcome Trust. We express special gratitude to the staff of the Cancer Genome Project at Sanger Centre for their dedication and hard work.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. DN911814–DN920916 and CZ445878–CZ466054. All MCF-7 BAC clones are available from Amplicon Express. The library name is HTA. The plate/row/column names are the same (e.g. MCF7_1-100G11 clone is the clone located in HTA library, plate 100, column G, row 11).]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4247306.
References
- Artandi, S.E., Chang, S., Lee, S.-L., Alson, S., Gottlieb, G.J., Chin, L., and DePinho, R.A. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406 641–645. [DOI] [PubMed] [Google Scholar]
- Barlund, M., Monni, O., Weaver, J.D., Kauraniemi, P., Sauter, G., Heiskanen, M., Kallioniemi, O.P., and Kallioniemi, A. 2002. Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer 35 311–317. [DOI] [PubMed] [Google Scholar]
- Bautista, S. and Theillet, C. 1998. CCND1 and FGFR1 coamplification results in the colocalization of 11q13 and 8p12 sequences in breast tumor nuclei. Genes Chromosomes Cancer 22 268–277. [PubMed] [Google Scholar]
- Collins, C., Rommens, J.M., Kowbel, D., Godfrey, T., Tanner, M., Hwang, S.I., Polikoff, D., Nonet, G., Cochran, J., Myambo, K., et al. 1998. Positional cloning of ZNF217 and NABC1: Genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. 95 8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, C., Volik, S., Kowbel, D., Ginzinger, D., Ylstra, B., Cloutier, T., Hawkins, T., Predki, P., Martin, C., Wernick, M., et al. 2001. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 11 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooijmans, R.P., Vrebalov, J., Dijkhof, R.J., van der Poel, J.J., and Groenen, M.A. 2000. Two-dimensional screening of the Wageningen chicken BAC library. Mamm. Genome 11 360–363. [DOI] [PubMed] [Google Scholar]
- Eggen, A., Gautier, M., Billaut, A., Petit, E., Hayes, H., Laurent, P., Urban, C., Pfister-Genskow, M., Eilertsen, K., and Bishop, M.D. 2001. Construction and characterization of a bovine BAC library with four genome-equivalent coverage. Genet. Sel. Evol. 33 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, M. 2000. DNA alteration in cancer: Genetic and epigenetic changes. Eaton Publishing, Natick, MA.
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Fialka, I., Schwarz, H., Reichmann, E., Oft, M., Busslinger, M., and Beug, H. 1996. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J. Cell. Biol. 132 1115–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal, P.A., Kasprzyk, A., Birney, E., Mullikin, J.C., Wooster, R., and Stratton, M.R. 2001. Cancer and genomics. Nature 409 850–852. [DOI] [PubMed] [Google Scholar]
- Giglio, S., Broman, K.W., Matsumoto, N., Calvari, V., Gimelli, G., Neumann, T., Ohashi, H., Voullaire, L., Larizza, D., Giorda, R., et al. 2001. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio, S., Calvari, V., Gregato, G., Gimelli, G., Camanini, S., Giorda, R., Ragusa, A., Guerneri, S., Selicorni, A., Stumm, M., et al. 2002. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 71 276–285.12058347 [Google Scholar]
- Gray, J.W. and Collins, C. 2000. Genome changes and gene expression in human solid tumors. Carcinogenesis 21 443–452. [DOI] [PubMed] [Google Scholar]
- Hahn, Y., Bera, T.K., Gehlhaus, K., Kirsch, I.R., Pastan, I.H., and Lee, B. 2004. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc. Natl. Acad. Sci. 101 13257–13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, C.S., Sutherland, R.D., Jewett, P.B., Campbell, M.L., Meincke, L.J., Tesmer, J.G., Mundt, M.O., Fawcett, J.J., Kim, U.J., Deaven, L.L., et al. 2000. Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res. 10 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, S.a.M., F. 1995. Cancer cytogenetics. John Wiley and Sons, New York.
- Huang, G., Krig, S., Kowbel, D., Xu, H., Hyun, B., Volik, S., Feuerstein, B., Mills, G.B., Stokoe, D., Yaswen, P., et al. 2005. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum. Mol. Genet. 14 3219–3225. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431 931–945. [DOI] [PubMed] [Google Scholar]
- Jansen, R.K., Raubeson, L.A., Boore, J.L., Depamphilis, C.W., Chumley, T.W., Haberle, R.C., Wyman, S.K., Alverson, A.J., Peery, R., Herman, S.J., et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 395 348–384. [DOI] [PubMed] [Google Scholar]
- Kaiser, J. 2004. Cancer research and NCI hears a pitch for biomarker studies. Science 306 1119. [DOI] [PubMed] [Google Scholar]
- Kaiser, J. 2005. National Institutes of Health. NCI gears up for cancer genome project. Science 307 1182. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen, T., Gobu, V., Knaus, R., Padilla-Nash, H., Augustus, M., Strausberg, R.L., Kirsch, I.R., Sirotkin, K., and Ried, T. 2005. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: Linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer 44 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, M., Bosdet, I., Smailus, D., Chiu, R., Mathewson, C., Wye, N., Barber, S., Brown-John, M., Chan, S., Chand, S., et al. 2004. A set of BAC clones spanning the human genome. Nucleic Acids Res. 32 3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kytola, S., Rummukainen, J., Nordgren, A., Karhu, R., Farnebo, F., Isola, J., and Larsson, C. 2000. Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer 28 308–317. [DOI] [PubMed] [Google Scholar]
- Lander, E.S. Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Leveau, J.H., Gerards, S., de Boer, W., and van Veen, J.A. 2004. Phylogeny-function analysis of (meta)genomic libraries: Screening for expression of ribosomal RNA genes by large-insert library fluorescent in situ hybridization (LIL-FISH). Environ. Microbiol. 6 990–998. [DOI] [PubMed] [Google Scholar]
- Lukyanov, K., Diatchenko, L., Chenchik, A., Nanisetti, A., Siebert, P., Usman, N., Matz, M., and Lukyanov, S. 1997. Construction of cDNA libraries from small amounts of total RNA using the suppression PCR effect. Biochem. Biophys. Res. Commun. 230 285–288. [DOI] [PubMed] [Google Scholar]
- Magrini, V., Warren, W.C., Wallis, J., Goldman, W.E., Xu, J., Mardis, E.R., and McPherson, J.D. 2004. Fosmid-based physical mapping of the Histoplasma capsulatum genome. Genome Res. 14 1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz, M.V. 2002. Amplification of representative cDNA samples from microscopic amounts of invertebrate tissue to search for new genes. Methods Mol. Biol. 183 3–18. [DOI] [PubMed] [Google Scholar]
- Milosavljevic, A., Harris, R.A., Sodergren, E.J., Jackson, A.R., Kalafus, K.J., Hodgson, A., Cree, A., Dai, W., Csuros, M., Zhu, B., et al. 2005. Pooled genomic indexing of rhesus macaque. Genome Res. 15 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman, F., Johansson, B., and Mertens, F. 2004. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 36 331–334. [DOI] [PubMed] [Google Scholar]
- Moon, D.A. and Magor, K.E. 2004. Construction and characterization of a fosmid library for comparative analysis of the duck genome. Anim. Genet. 35 417–418. [DOI] [PubMed] [Google Scholar]
- Murnane, J.P. and Sabatier, L. 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays 26 1164–1174. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. 2005. Recommendation for a Human Cancer Genome Project. In Working group on biomedical technology.
- Nonet, G.H., Stampfer, M.R., Chin, K., Gray, J.W., Collins, C.C., and Yaswen, P. 2001. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 61 1250–1254. [PubMed] [Google Scholar]
- Osborne, L.R., Li, M., Pober, B., Chitayat, D., Bodurtha, J., Mandel, A., Costa, T., Grebe, T., Cox, S., Tsui, L.C., et al. 2001. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 29 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Tateno, M., Woon, P.Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10 116–128. [PMC free article] [PubMed] [Google Scholar]
- Osoegawa, K., Mammoser, A.G., Wu, C., Frengen, E., Zeng, C., Catanese, J.J., and de Jong, P.J. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Nash, H.M., Heselmeyer-Haddad, K., Wangsa, D., Zhang, H., Ghadimi, B.M., Macville, M., Augustus, M., Schrock, E., Hilgenfeld, E., and Ried, T. 2001. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer 30 349–363. [DOI] [PubMed] [Google Scholar]
- Paris, P.L., Andaya, A., Fridlyand, J., Jain, A.N., Weinberg, V., Kowbel, D., Brebner, J.H., Simko, J., Watson, J.E., Volik, S., et al. 2004. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum. Mol. Genet. 13 1303–1313. [DOI] [PubMed] [Google Scholar]
- Paris, P.L., Weinberg, V., Simko, J., Andaya, A., Albo, G., Rubin, M.A., Carroll, P.R., and Collins, C. 2005. Preliminary evaluation of prostate cancer metastatic risk biomarkers. Int. J. Biol. Markers 20 141–145. [DOI] [PubMed] [Google Scholar]
- Rabbitts, T.H. and Stocks, M.R. 2003. Chromosomal translocation products engender new intracellular therapeutic technologies. Nat. Med. 9 383–386. [DOI] [PubMed] [Google Scholar]
- Raphael, B.J. and Pevzner, P.A. 2004. Reconstructing tumor amplisomes. Bioinformatics 20 I265–I273. [DOI] [PubMed] [Google Scholar]
- Raphael, B.J., Volik, S., Collins, C., and Pevzner, P.A. 2003. Reconstructing tumor genome architectures. Bioinformatics 19 II162–II171. [DOI] [PubMed] [Google Scholar]
- Sawyers, C.L. 1999. Chronic myeloid leukemia. New Engl. J. Med. 340 1330–1340. [DOI] [PubMed] [Google Scholar]
- Schrock, E., du Manoir, S., Veldman, T., Schoell, B., Wienberg, J., Ferguson-Smith, M.A., Ning, Y., Ledbetter, D.H., Bar-Am, I., Soenksen, D., et al. 1996. Multicolor spectral karyotyping of human chromosomes. Science 273 494–497. [DOI] [PubMed] [Google Scholar]
- Shagin, D.A., Lukyanov, K.A., Vagner, L.L., and Matz, M.V. 1999. Regulation of average length of complex PCR product. Nucleic Acids Res. 27 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagin, D.A., Rebrikov, D.V., Kozhemyako, V.B., Altshuler, I.M., Shcheglov, A.S., Zhulidov, P.A., Bogdanova, E.A., Staroverov, D.B., Rasskazov, V.A., and Lukyanov, S. 2002. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 12 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, X., Jiang, Z., Clark, R.A., Liu, G., Cheng, Z., Tuzun, E., Church, D.M., Sutton, G., Halpern, A.L., and Eichler, E.E. 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431 927–930. [DOI] [PubMed] [Google Scholar]
- Shimada, M.K., Kim, C.G., Kitano, T., Ferrell, R.E., Kohara, Y., and Saitou, N. 2005. Nucleotide sequence comparison of a chromosome rearrangement on human chromosome 12 and the corresponding ape chromosomes. Cytogenet. Genome Res. 108 83–90. [DOI] [PubMed] [Google Scholar]
- Stefansson, H., Helgason, A., Thorleifsson, G., Steinthorsdottir, V., Masson, G., Barnard, J., Baker, A., Jonasdottir, A., Ingason, A., Gudnadottir, V.G., et al. 2005. A common inversion under selection in Europeans. Nat. Genet. 37 129–137. [DOI] [PubMed] [Google Scholar]
- Tanner, M.M., Tirkkonen, M., Kallioniemi, A., Isola, J., Kuukasjarvi, T., Collins, C., Kowbel, D., Guan, X.Y., Trent, J., Gray, J.W., et al. 1996. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 56 3441–3445. [PubMed] [Google Scholar]
- Tuzun, E., Sharp, A.J., Bailey, J.A., Kaul, R., Morrison, V.A., Pertz, L.M., Haugen, E., Hayden, H., Albertson, D., Pinkel, D., et al. 2005. Fine-scale structural variation of the human genome. Nat. Genet. 37 727–732. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291 1304–1351. [DOI] [PubMed] [Google Scholar]
- Volik, S., Zhao, S., Chin, K., Brebner, J.H., Herndon, D.R., Tao, Q., Kowbel, D., Huang, G., Lapuk, A., Kuo, W.L., et al. 2003. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc. Natl. Acad. Sci. 100 7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Engler, P., Longacre, A., and Storb, U. 2001. An efficient method for high-fidelity BAC/PAC retrofitting with a selectable marker for mammalian cell transfection. Genome Res. 11 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga, Y., Nakamura, K., Ewing, C.M., Isaacs, W.B., Hukku, B., and Rhim, J.S. 2001. A novel human cell culture model for the study of familial prostate cancer. Cancer Res. 61 5969–5973. [PubMed] [Google Scholar]
- Zatkova, A., Ullmann, R., Rouillard, J.M., Lamb, B.J., Kuick, R., Hanash, S.M., Schnittger, S., Schoch, C., Fonatsch, C., and Wimmer, K. 2004. Distinct sequences on 11q13.5 and 11q23–24 are frequently coamplified with MLL in complexly organized 11q amplicons in AML/MDS patients. Genes Chromosomes Cancer 39 263–276. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Lu, H.H., Chung, W.Y., Yang, J., and Li, W.H. 2005. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 22 135–141. [DOI] [PubMed] [Google Scholar]
- Zhu, B., Smith, J.A., Tracey, S.M., Konfortov, B.A., Welzel, K., Schalkwyk, L.C., Lehrach, H., Kollers, S., Masabanda, J., Buitkamp, J., et al. 1999. A 5× genome coverage bovine BAC library: Production, characterization, and distribution. Mamm. Genome 10 706–709. [DOI] [PubMed] [Google Scholar]
- Zhulidov, P.A., Bogdanova, E.A., Shcheglov, A.S., Vagner, L.L., Khaspekov, G.L., Kozhemyako, V.B., Matz, M.V., Meleshkevitch, E., Moroz, L.L., Lukyanov, S.A., et al. 2004. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 32 e37. [DOI] [PMC free article] [PubMed] [Google Scholar]