Cytogenetics versus bioinformatics
Froenicke et al. (2006) asked the question “Are molecular cytogenetics and bioinformatics suggesting diverging models of ancestral mammalian genomes?” Their commentary seems to imply that cytogenetics is superior to “bioinformatics” when it comes to studies of ancestral mammalian genomes. But, comparing cytogenetics with other approaches to deriving ancestral genomic architectures (like MGR developed by Bourque and Pevzner 2002) on two very different data sets (80+ cytogenetic maps vs. four distantly related sequenced genomes) does not say much about the merits and demerits of the approaches; instead, it indirectly evaluates the quality of the input data sets.
In this context, the main conclusion of Froenicke et al. (2006) amounts to the statement that 80+ cytogenetic maps lead to a more definitive reconstruction than four divergent genomes when it comes to low-resolution ancestral architectures. We never argued against this point in our publications and even advocated for the use of radiation hybrid mapping (as a trade-off in resolution between cytogenetic and sequencing data) to extend the number of analyzed genomes (Murphy et al. 2003, 2005). Indeed, with more taxon sampling, a rearrangement-based reconstruction with just seven genomes (Murphy et al. 2005) is already highly consistent with the cytogenetics reconstruction. Before we substantiate this convergence, we clarify the notion of weak associations, which was used in Bourque et al. (2005) to alert the reader that some of the adjacencies were left as unresolved in the ancestor. The discrepancies identified by Froenicke et al. (2006) all involve weak associations and point toward a misunderstanding more than a contradiction.
Finally, we underline some of the important strengths exclusive to rearrangement-based approaches, such as the ability to detect smaller genomic segments, to handle fast evolving lineages, and to orient conserved segments in the ancestors.
Alternative ancestors and weak associations
The concept of “weak association” was first introduced in Bourque et al. (2004) to articulate (1) that some of the reconstructed adjacencies are unreliable and (2) that they (hopefully) will be resolved in the future when more genomes are included. Most of the chromosomal associations displayed in the recovered ancestor (See Fig. 2 in Bourque et al. 2005) actually correspond to “weak adjacencies” (i.e., they were not found in all alternative reconstructions explored) and were marked by black arrowheads. As such, it is not surprising that most of these weak associations are not corroborated in the dog genome or in the cytogenetics model; their presence only indicates that the input data failed to resolve them unambiguously. As a starting point, a more realistic evaluation could be to look at the chromosomal associations suggested by strong adjacencies. In the ancestor (see Fig. 2 in Bourque et al. 2005), there are only six strong associations, and four of these are shared with the cytogenetics model (HSA 3/21, 4/8, 12a/22a, 12b/22b). The two strong associations unique to the rearrangement-based model (HSA 2/3, 11/15) are observed in chicken and in the two rodents, while the three associations missing as compared with the cytogenetics model (HSA 7/16, 14/15, 16/19) are not observed in any of the rodents. In this context, the discrepancies are not surprising and once again point to differences in taxon sampling.
Froenicke et al. (2006) asked the question “Should the discussion focus on a single model?” We did not think so, and it is for this reason that none of the newly predicted chromosomal associations were analyzed in detail. Instead, the focus of the study was on other properties of the reconstruction, mostly rates of rearrangements, which were shown to be stable for gene-based or sequence-based data and for different choices of ancestors.
Impact of taxon sampling
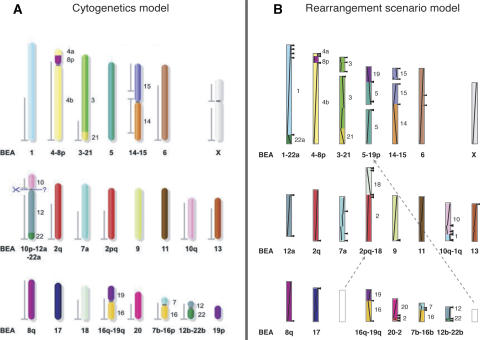
Froenicke et al. (2006) elegantly highlighted some of the strengths of the cytogenetic model in predicting ancestral mammalian karyotypes. There is no doubt that the use of data from more than 80 eutherian species greatly enhances the quality of the ancestral predictions. Actually, in our own study (Murphy et al. 2003) the cytogenetic model was referred to as the gold standard for the prediction of ancestral chromosomal associations. The study (Bourque et al. 2005) for which Froenicke et al. (2006) elected to evaluate the rearrangement scenario model used only four highly rearranged species as input. In this case, we would not expect, nor claim to reconstruct a definitive ancestor, and the high number of weak adjacencies in the reconstruction shows this. There are 83 weak adjacencies in this ancestral reconstruction, 29 of which correspond to putative chromosomal associations. In another recent study (Murphy et al. 2005), we applied the same rearrangement analysis to three sequenced genomes (human, mouse, and rat) and five densely mapped genomes (cat, cattle, dog, pig, and horse). Although, as for the human–mouse–rat–chicken data set, some ambiguities remain, the wider sampling now allows for a more robust reconstruction (49 weak adjacencies, 11 of which are chromosomal associations) and a closer correspondence between this new ancestor and the ancestor predicted from cytogenetics analyses (Fig. 1).
Figure 1.
Putative genome architecture of the boreoeutherian ancestor from (A) cytogenetics model using over 80 eutherian genomes (Froenicke et al. 2006) and (B) a rearrangement scenario model using human–mouse–rat–cat–cattle–dog–pig (Murphy et al. 2005). In B, the black arrowheads to the right of the ancestral chromosomes indicate unresolved/weak adjacencies, and the diagonal line segments indicate original position and orientation on human. This is the same reconstruction as in Murphy et al. (2005), but reformatted, rescaled, and recolored to facilitate comparison. The two dashed arrows show the location in the rearrangement model of two of the ancestral chromosomes from the cytogenetics model. These two chromosomes are only weakly associated with two other chromosomes in the rearrangement model.
For instance, of the nine human autosomes predicted to be in the ancestral karyotype using the cytogenetics model (1, 5, 6, 9, 11, 13, 17, 18, and 20; Froenicke 2005), only HSA 1 and HSA 5 were found on two distinct chromosomes of the reconstructed ancestor. For HSA 1, the discrepancy could come from the disparity in resolution, since the small blocks from HSA1q associated with HSA10q are actually observed to be associated in many mammalian genomes (Murphy et al. 2005). Even then, in this latest rearrangement study, there is not a single strong chromosomal association conflicting with the cytogenetics model.
Strengths of rearrangement analysis
Apart from the fact that the accuracy of the predicted large-scale ancestral associations under the rearrangement model is likely to increase as more taxa become available, just as it did under the cytogenetics model (cf. Chowdhary et al. 1998 with Froenicke 2005), there are other aspects that make this approach appealing.
First, new ancestral associations involving small chromosomal blocks evading the resolution of the cytogenetics maps can be identified. For instance, the ancestral association HSA10q/1q observed in many ferungulate genomes and (weakly) predicted in the rearrangement model (Fig. 1B) involves blocks evading the resolution of the cytogenetics approach (Fig. 1A). Second, sequence-based rearrangement analysis can be used to recover detailed scenarios and evolutionary trees, even with genomes from lineages with rapid rates of chromosomal evolutions such as murid rodents (Bourque et al. 2004) and caniform carnivores (Murphy et al. 2005). The cytogenetics approach suffers from the fact that, so far, no complete karyotype can be compared between a eutherian mammal and any other vertebrate (Wienberg 2004). In contrast, it was shown that the rearrangement scenario model has the potential to make use of distant outgroups (Hillier et al. 2004; Bourque et al. 2005). Third, the methodology allows the detailed description of intrachromosomal evolution, e.g., reconstruction of the detailed inversion history of the X chromosome (Bourque et al. 2004). Finally, the rearrangement model attempts to recapitulate the steps that lead to the modern genome arrangements and to estimate actual rates of rearrangements.
Conclusions
Unfortunately, an in-depth analysis of one of the many possible ancestral architectures recovered in the human–mouse–rat–chicken study (Bourque et al. 2005), such as the one carried out by Froenicke et al. (2006), is hardly productive; that is why, in that study, we focused on properties of the reconstruction that were stable under alternative reconstructions. The fact that without sufficient taxon sampling many of the ancestral associations will not be resolved unambiguously was clearly stated in our recent studies (Bourque et al. 2004, 2005). In this context, we have now shown how the addition of new genomes can actually help resolve many of these uncertainties, making a direct comparison more practical (Fig. 1). We feel that we have demonstrated that the two approaches are already generating converging predictions with very few conflicts.
At the same time, there is little debate over the power of the cytogenetics approach to identify large-scale associations in mammals. The study by Froenicke and colleagues is important in pointing to the fact that there is a lot of unexplored synergy between the two techniques. We agree with the conclusion of the authors that the area of ancestral genome reconstruction would greatly benefit from a better integration of both experimental data sets. A first step in achieving this integration could be to use the cytogenetics reconstruction to systematically try to resolve the weak associations in the rearrangement-based ancestor.
Acknowledgments
We thank the referees for valuable suggestions. G.T. is supported by a Hellman Fellowship at UCSD. G.B. is supported by funds from the Biomedical Research Council (BMRC) in Singapore.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4631806.
References
- Bourque, G. and Pevzner, P.A. 2002. Genome-scale evolution: Reconstructing gene orders in the ancestral species. Genome Res. 12 26–36. [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., Pevzner, P.A., and Tesler, G. 2004. Reconstructing the genomic architecture of ancestral mammals: Lessons from human, mouse, and rat genomes. Genome Res. 14 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., Zdobnov, E.M., Bork, P., Pevzner, P.A., and Tesler, G. 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res. 15 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary, B.P., Raudsepp, T., Froenicke, L., and Scherthan, H. 1998. Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo-FISH. Genome Res. 8 577–589. [DOI] [PubMed] [Google Scholar]
- Froenicke, L. 2005. Origins of primate chromosomes—as delineated by Zoo-FISH and alignments of human and mouse draft genome sequences. Cytogenet. Genome Res. 108 122–138. [DOI] [PubMed] [Google Scholar]
- Froenicke, L., Caldés, M.G., Graphodatsky, A., Müller, S., Lyons, L.A., Robinson, T.J., Volleth, M., Yang, F., and Wienberg, J. 2006. Are molecular cytogenetics and bioinformatics suggesting diverging models of ancestral mammalian genomes? Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Hillier, L.W., Miller, W., Birney, E., Warren, W., Hardison, R.C., Ponting, C.P., Bork, P., Burt, D.W., Groenen, M.A., Delany, M.E., et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432 695–716. [DOI] [PubMed] [Google Scholar]
- Murphy, W.J., Bourque, G., Tesler, G., Pevzner, P.A., and O'Brien, S.J. 2003. Reconstructing the genomic architecture of mammalian ancestors using multispecies comparative maps. Hum. Genomics 1 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Larkin, D.M., Everts-van der Wind, A., Bourque, G., Tesler, G., Auvil, L., Beever, J.E., Chowdhary, B.P., Galibert, F., Gatzke, L., et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309 613–617. [DOI] [PubMed] [Google Scholar]
- Wienberg, J. 2004. The evolution of eutherian chromosomes. Curr. Opin. Genet. Dev. 14 657–666. [DOI] [PubMed] [Google Scholar]