Abstract
Pericentromeres are exceptional genomic regions: in animals they contain extensive segmental duplications implicated in gene creation, and in plants they sustain rearrangements and insertions uncommon in euchromatin. To examine the mechanisms and patterns of plant pericentromere evolution, we compared pericentromere sequence from four Brassicaceae species separated by <15 million years (Myr). This flowering plant family is ideal for studying relationships between genome reorganization and pericentromere evolution—its members have undergone recent polyploidization and hybridization, with close relatives changing in genome size and chromosome number. Through sequence and hybridization analyses, we examined regions from Arabidopsis arenosa, Capsella rubella, and Olimarabidopsis pumila that are homologous to Arabidopsis thaliana pericentromeres (peri-CENs) III and V, and used FISH to demonstrate they have been maintained near centromere satellite arrays in each species. Sequence analysis revealed a set of highly conserved genes, yet we discovered substantial differences in intergenic length and species-specific changes in sequence content and gene density. We discovered that A. thaliana has undergone recent, significant expansions within its pericentromeres, in some cases measuring hundreds of kilobases; these findings are in marked contrast to euchromatic segments in these species that exhibit only minor length changes. While plant pericentromeres do contain some duplications, we did not find evidence of extensive segmental duplications, as has been documented in primates. Our data support a model in which plant pericentromeres may experience selective pressures distinct from euchromatin, tolerating rapid, dynamic changes in structure and sequence content, including large insertions of mobile elements, 5S rDNA arrays and pseudogenes.
Centromere functions are conserved throughout higher eukaryotes and mediated by common proteins, yet centromere satellite DNA sequences diverge rapidly, even in organisms separated by a few million years (Hall et al. 2003, 2005). Pericentromere DNA flanking these satellites also changes rapidly (She et al. 2004; Horvath et al. 2005), but the rate and magnitude of these changes make it challenging to infer pericentromere evolution in small taxonomic families. Here, we perform the first comparative analysis of plant pericentromeres, selecting a family of 3350 members. We show that homologous pericentromeres from four Brassicaceae species are remarkably dynamic, undergoing rapid changes in structure and sequence content.
In most multicellular organisms, centromere DNA sequences are highly repetitive, methylated, and heterochromatic; they inhibit reciprocal homologous recombination, mediate meiotic sister-chromatid cohesion, and assemble kinetochore proteins (Amor et al. 2004). Centromere cores contain tandem satellite arrays, often measuring hundreds of kilobases (Henikoff et al. 2001). These satellite arrays can confer efficient inheritance to autonomous minichromosomes in human cell lines, confirming their functional importance (Yang et al. 2000), and in plants and animals, rapid satellite evolution produces chromosome and species-specific variants through global and local homogenization (Hall et al. 2003, 2005). Centromere satellites are surrounded by pericentromeres that are rich in transposons, retroelements, and pseudogenes, and can contain expressed genes (Bevan et al. 2001; She et al. 2004).
In Arabidopsis thaliana, genome sequence was assembled through the pericentromeres to the centromere satellite cores. Within each genetically defined centromere reside large 178-bp satellite arrays (Copenhaver et al. 1999; Bevan et al. 2001), and the surrounding pericentromeres contain retroelements, transposons, 5S rDNA arrays, pseudogenes, hypothetical proteins, and >200 expressed genes (Kumekawa et al. 2000, 2001; Bevan et al. 2001; Hosouchi et al. 2002); expressed genes have also been found near rice centromere satellites (Nagaki et al. 2004). Human and mouse pericentromeres also contain genes, some of which appear to be derived from extensive segmental duplications enriched in these regions (Thomas et al. 2003; She et al. 2004). Inferring the mechanisms and rates of pericentromere evolution, however, has often been hampered by a lack of assembled sequence from close relatives. Recent comparative FISH in primates has revealed that initial “seeding” pericentromere duplications occurred as a punctuated event prior to the divergence of humans and great apes 10–20 million years ago (Mya), with subsequent secondary dispersal duplications during the radiation of humans, chimps, and gorillas (Horvath et al. 2005). In A. thaliana, cytogenetic comparisons of ecotypes suggest that pericentromeres sustain large rearrangements, insertions, and deletions over short evolutionary time scales. For example, pericentromere II contains an insertion of ∼2.5 tandem copies of the mitochondrial genome (Stupar et al. 2001), and pericentromere IV has sustained a large inversion, specific to several ecotypes (Fransz et al. 2000).
In the Brassicaceae family, euchromatic genomic segments of A. thaliana and its close relatives are often collinear and share high levels of sequence identity (Acarkan et al. 2000; Rossberg et al. 2001; Fiebig et al. 2004). Yet comparative mapping showed that A. thaliana (2n = 10) has experienced extreme genome reduction events, involving translocations, fusions, and chromosome loss since diverging 5–10 Mya from two close relatives, Arabidopsis lyrata and Capsella rubella (2n = 16) (Yogeeswaran et al. 2005). These changes were accompanied by rapid evolution of centromere satellites (Hall et al. 2003, 2005) and centromere-binding proteins (Talbert et al. 2002, 2004; Cooper and Henikoff 2004). Similarly, comparative analyses of primate chromosomes have revealed conserved synteny among species, yet changes in chromosome number and, occasionally, centromere position. In some cases, noncentromeric regions apparently acquire centromere functions de novo, incorporating flanking DNA into pericentromeres (Ventura et al. 2001; Eder et al. 2003). These events cannot be explained by translocations or inversions; rather, they may begin with a noncentromeric region associating with centromere-binding proteins to form a neo-centromere (Barry et al. 1999). Over evolutionary time frames, neo-centromeres presumably stabilize by acquiring satellites, likely triggering genetic and epigenetic changes in neo-pericentromeres. In particular, a shift from euchromatin to heterochromatin may silence some pericentromere genes, leading to their degeneration. Furthermore, pericentromere heterochromatin may attract the integration of transposons, retroelements, and gene fragments.
With their capacity for polyploidization, interspecies hybridization, and genomic plasticity (Wendel 2000), flowering plants are ideal for studying pericentromere evolution. To discern whether pericentromeres of closely related plant species exhibit conserved synteny and collinearity, and to infer their evolutionary mechanisms, we sequenced pericentromeric BACs from three relatives of the model plant, A. thaliana. Arabidopsis arenosa is estimated to have shared common ancestry with A. thaliana ∼5 Mya, while C. rubella and Olimarabidopsis pumila diverged ∼10–14 Mya (Koch et al. 2001). However, despite the polyploidization, chromosome loss, and rearrangement events that differentiate these species, we found that homologous pericentromeres were maintained in a similar genomic context, with centromere satellites nearby. While we discovered conserved genes, we also found large-scale intergenic expansion involving the insertion of mobile elements, 5S rDNA arrays, and pseudogenes, pointing to pericentromeres as extremely dynamic genomic regions.
Results
Regions homologous to A. thaliana pericentromeres
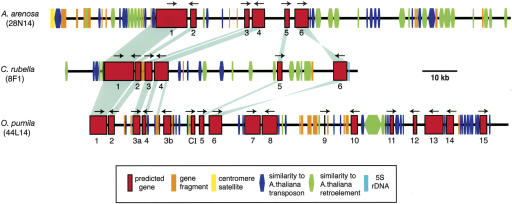
On A. thaliana chromosome III, actin-related protein 6 (ARP6, At3g33520) is one of the most proximal genes to the 178-bp satellite arrays of centromere 3 (CEN3), within the region that lacks meiotic recombination (Copenhaver et al. 1999). Probing BAC library filters with ARP6 identified BACs from A. arenosa (Aa28N14), C. rubella (Cr8F1), and O. pumila (Op44L14) (Supplemental Fig. 1). Annotation of assembled sequence from these BACs (GenBank DQ103593–DQ103595) revealed multiple genes (six in Aa28N14 and Cr8F1; 16 in Op44L14), and BLAST searches indicated all but one gene had an expressed homolog in A. thaliana peri-CEN3; the remaining O. pumila gene was similar to an expressed gene on the A. thaliana chromosome I arm (Fig. 1A; Table 1; Supplemental Table 1). The genes are in the same chromosomal order in all four species (collinear), indicating descent from a common ancestral chromosomal region. The A. thaliana genes, however, are unexpectedly distributed across a much larger genomic region: 15 genes spanning 112 kb in O. pumila have collinear homologs spread across 871 kb of A. thaliana peri-CEN3, and genes 1–6 (Fig. 1A; Table 1) occupy 643 kb in A. thaliana, but only 38–70 kb in the related species.
Figure 1.
Multispecies comparisons of regions homologous to A. thaliana pericentromeres. Scale drawings of (A) A. thaliana peri-CEN3 (chromosome III bases 14,096,340–14,967,565), and sequenced BACs from A. arenosa, C. rubella, and O. pumila. (B) A. thaliana peri-CEN5 (chromosome V bases 12,566,510–13,252,797), and homologous BACs identified by hybridization, from A. arenosa, C. rubella, and O. pumila (complete list, Supplemental Table 2). Estimated times of divergence (millions of years, Myr) are shown (Koch et al. 2001). Dotted lines, estimated distance (not to scale) to the centromere satellite array (yellow boxes); (solid arrows) centromere boundary as defined by recombination (Copenhaver and Preuss 1999); (green boxes) 5S rDNA array; (red bars and ovals) expressed A. thaliana genes present in multiple species; (gray ovals) intact A. thaliana-specific genes; (left to right) At3g42050, At3g42150, and At3g42786). (*) Gene homologous to A. thaliana chromosome I expressed gene (At1g63270); (dashed arrows) all genes in the interval are present; (parentheses in B) undetermined gene order.
Table 1.
Conserved genes in regions homologous to A. thaliana pericentromere 3
|
BAC gene predictionsa
|
Arabidopsis thalianab
|
|||||
|---|---|---|---|---|---|---|
| # | Aa 28N14 | Cr 8F1 | Op 44L14 | Homolog | Putative function | Expression evidencec |
| 1 | G | G | G | At3g33530d | Transducin familye | EST, ma, mpss |
| 2 | G | G | G | At3g33520d | Actin-related protein 6 (ARP6) | cDNA |
| 3 | G | G | G | At3g42170 | hAT-like transposase | cDNA |
| 4 | G | G | G | At3g42180 | Exostosin family | EST, mpss |
| 5 | G | G | G | At3g42630d | Pentatricopeptide repeat protein | cDNA |
| 6 | G | G | G | At3g42640d | Plasma membrane ATPase | EST, ma, mpss |
| 7 | — | — | G | At3g42660 | Transducin/WD-40 repeat family | none |
| 8 | — | — | G | At3g42670d | SNF2 domain-containing protein | EST, ma, mpss |
| 9 | — | — | G | At3g42725 | Unknown protein | cDNA |
| 10 | — | — | G | At3g42790d | Phd finger protein | cDNA |
| 11 | — | — | G | At3g42800 | Expressed protein | cDNA |
| 12 | — | — | G | At3g42830 | Ring-box protein | none |
| 13 | — | — | G | At3g42850 | Galactokinase | mpss |
| 14 | — | — | G | At3g42860d | Zinc knuckle protein | cDNA |
| 15 | — | — | G | At3g42880d | LRR transmembrane kinase | ma, mpss |
| CI | — | — | G | At1g63270 | ABC transporter family | mpss |
Other than the highly conserved transposase gene, At3g42170, transposons, retroelements, pseudogenes, or hypothetical genes with sequence similarity to transposons (such as putative transposases, Ulp1 proteases, or replication protein A1; http://www.arabidopsis.org) are not included.
In cases in which a full-length cDNA has not been identified, evidence of A. thaliana gene expression is noted as ESTs (expressed sequence tags), significant (P < 0.06) hybridization detected on a microarray (ma; https://www.genevestigator.ethz.ch/; http://www.arabidopsis.org), or sequences detected by massively parallel signature sequencing (mpss; http://mpss.udel.edu/at/); signatures that match multiple loci were not included.
Genes included on the Affymetrix 22K array.
Inconsistent intron/exon boundaries were found between species and reflect errors in the A. thaliana annotation. (—) Genes absent from BAC.
These results are in marked contrast to comparisons of multiple euchromatic regions from these species, where despite differences in genome size and chromosome number, homologous regions are similarly sized (Acarkan et al. 2000; Rossberg et al. 2001; Fiebig et al. 2004). For example, the 14 GRP pollen coat genes occupy 39 kb in A. thaliana, 40 kb in A. arenosa, 31 kb in C. rubella, and 41 kb in O. pumila (Fiebig et al. 2004). These studies also showed that intergenic lengths in homologous segments of A. arenosa, C. rubella, and O. pumila were highly similar, differing from A. thaliana by less than twofold in all examples (Acarkan et al. 2000; Rossberg et al. 2001; Fiebig et al. 2004).
To investigate whether regions homologous to another A. thaliana pericentromere are similarly variable in length, we assessed BAC gene content by hybridizing the libraries with probes from six expressed A. thaliana genes (a–f) that span 686 kb near CEN5 (Fig. 1B). As a control to confirm probe specificity and gene spacing, two A. thaliana BAC libraries were also probed (Choi et al. 1995; Mozo et al. 1998): Within A. thaliana, genes a–d are contained within 40 kb; probes d and e are separated by 503 kb, and probes e and f by 122 kb (Fig. 1B). In C. rubella, BACs hybridizing to these probes contained genes a–c, gene d, or genes e and f (Fig. 1B; Supplemental Table 2). In contrast, probes on both ends of the A. thaliana interval (a, b, e, and f) hybridized to O. pumila BAC 14G23 (125 kb), and all six probes hybridized to A. arenosa BAC 11A17 (125 kb). This compressed gene spacing was further investigated with Southern blotting; digesting A. arenosa BAC 11A17 with a panel of enzymes showed that probes a, b, and c were located on an ∼23-kb DNA fragment, while probes e and f were located on an ∼25-kb DNA fragment (data not shown). Thus, these results confirm that recent, large expansions have occurred in both A. thaliana peri-CEN3 and peri-CEN5.
Peri-CEN3 and peri-CEN5 locations are conserved across species
In A. thaliana, pericentromere gene density is markedly lower than that of euchromatin (Bevan et al. 2001). Thus, the relatively higher gene density we discovered in the BACs from the related species suggests that pericentromeres can (1) have vastly different gene densities, or (2) migrate to new genomic positions, and thus the BACs we sequenced are not pericentromere-derived. For BAC Aa28N14, the former possibility is most likely, as one end of this BAC contains ∼1 kb of tandemly repeated 179-bp centromere satellite, a sequence specific to A. arenosa centromeres (Fig. 1; Hall et al. 2005). Quantitative hybridization of BACs overlapping Aa28N14 with a satellite probe showed that this array extends at least ∼18 kb (data not shown).
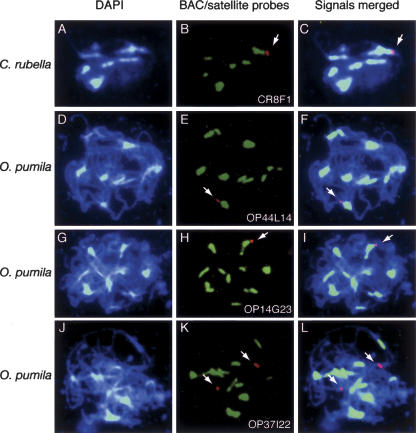
The C. rubella and O. pumila BACs we sequenced (Cr8F1, Op44L14) lacked centromere satellites; therefore, their chromosomal locations were examined with fluorescence in situ hybridization (FISH) to pachytene chromosomes (Fig. 2). We identified centromeres with species-specific satellite probes, which bind large satellite arrays that colocalize with DAPI-bright heterochromatin and possess biochemical markers associated with heterochromatin and active centromeres (Hall et al. 2005). Our FISH results demonstrated that both Cr8F1 and Op44L14 (red) abut major centromere satellite arrays (green) and pericentromeric heterochromatin (Fig. 2A–F). A similar pattern was obtained with Op14G23, which contains genes homologous to A. thaliana peri-CEN5 (Fig. 2G,H,I). Interestingly, each BAC specifically hybridized to one genomic region, suggesting that they share little sequence similarity with the rest of the genome. This was true even in the tetraploid O. pumila, suggesting that this region differs in the two parental genomes that comprise this species. As a control, we showed that a euchromatic BAC (Op37I22) (Fiebig et al. 2004) hybridized to regions distinct from satellite tracts (Fig. 2J,K,L).
Figure 2.
Chromosomal localization of sequenced BACs. FISH analysis of pachytene stage C. rubella (A,B,C) and O. pumila (D–L) chromosomes; each consists of synapsed homologs. Chromosomes (DAPI-stained, blue) were probed with centromere satellites (green) and BAC DNA (red). BAC probes include (A,B,C) Cr8F1; (D,E,F) Op44L14; (G,H,I) Op14G23; and (J,K,L) Op37I22. Merged images (center and right columns) show BAC locations (arrows) relative to DAPI-stained heterochromatin and centromere satellite arrays; similar staining patterns were observed on 10–50 pachytene chromosomes/probe.
Conserved sequence features in peri-CEN3 homologs
Six expressed genes from A. thaliana peri-CEN3 had highly conserved homologs in Aa28N14, Cr8F1, and Op44L14, with sequence similarity extending at most a few hundred bases 3′ and 5′ of each conserved gene (Fig. 3). While some of these genes showed small variations in nucleotide length (range 0%–40%, median 2%, relative to A. thaliana), no consistent trends were observed. Amino acid identities averaged 95% ± 2% and 90% ± 6% for the A. thaliana genes compared to A. arenosa and C. rubella, respectively, and 74%–93% (average, 87% ± 5%) for the 15 genes shared between Op44L14 and A. thaliana (Table 2). These values are similar to those documented in homologous euchromatic segments from these same species (Fiebig et al. 2004). Most of these peri-CEN3 genes were collinear in all four species. Only C. rubella gene 6 was inverted (Fig. 3), and O. pumila contained a duplication of gene 3 (hAT transposase), and an insertion of a gene related to a gene on A. thaliana chromosome I (CI) (Table 1). Interestingly, while the current A. thaliana genome assembly uses 5S rDNA arrays to anchor the 5 BAC contigs containing genes 1 and 2 (Fig. 1; Bevan et al. 2001), our data lend support to this configuration, as the order of these genes is conserved in three related species.
Figure 3.
Fine structure of BACs homologous to A. thaliana peri-CEN3. Scale drawing showing annotated features >100 bp (BLASTN score > 60 bits; e-values < 1e – 8); (red boxes) predicted genes shared among Brassicaceae species (1–15, Table 1); (gray bars) major stretches of sequence similarity (BLASTN score > 100 bits); (arrows) predicted direction of transcription; (blue boxes) sequence similarity to A. thaliana transposons; (green boxes) sequence similarity to A. thaliana retroelements; (orange boxes) gene fragments; (yellow boxes) centromere satellites; (turquoise boxes) 5S rDNA. For BAC Op44L14, gene 3 has been duplicated and gene CI is similar to an A. thaliana chromosome I gene. Annotations for genes within the corresponding A. thaliana peri-CEN3 region are listed in Supplemental Table 1, and a complete view of all feature annotations within this A. thaliana region can be viewed in the GenBank genome browser (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=3702) between coordinates 14,096,340 and 14,967,565.
Table 2.
Percent nucleotide and protein identity of conserved genes
| Gene # | Putative protein function | At:Aa | At:Cr | At:Op | Aa:Cr | Aa:Op | Cr:Op | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | ||
| 1 | Transducin familya | 97 | — | 91 | — | 91 | — | 91 | — | 91 | — | 95 | — |
| 2 | Actin-related ARP6 | 96 | 98 | 93 | 95 | 89 | 92 | 94 | 96 | 89 | 92 | 90 | 93 |
| 3 | hAT-like transposaseb | 94 | 94 | 88 | 90 | 88 | 92 | 91 | 92 | 90 | 92 | 87 | 88 |
| 4 | Exostosin family | 95 | 96 | 90 | 90 | 90 | 93 | 92 | 94 | 90 | 94 | 89 | 92 |
| 5 | Pentatricopeptide repeat | 92 | 92 | 86 | 82 | 87 | 85 | 88 | 84 | 88 | 85 | 88 | 87 |
| 6 | Plasma membrane ATPase | 96 | 98 | 93 | 97 | 93 | 97 | 93 | 97 | 93 | 97 | 93 | 98 |
| 7 | Transducin/WD-40 repeat | 87 | 89 | ||||||||||
| 8 | SNF2 domain-containing | 90 | 90 | ||||||||||
| 9 | Unknown | 80 | 74 | ||||||||||
| 10 | Phd finger | 93 | 94 | ||||||||||
| 11 | Expressed | 74 | 75 | ||||||||||
| 12 | Ring-box | 83 | 83 | ||||||||||
| 13 | Galactokinase | 93 | 96 | ||||||||||
| 14 | Zinc knuckle | 84 | 83 | ||||||||||
| 15 | LRR transmembrane kinase | 87 | 92 | ||||||||||
| CI | ABC transporter | 90 | 97 | ||||||||||
Protein alignments are uncertain due to ambiguities in intron/exon annotation.
For O. pumila, percent identity is the average over the two copies of the hAT gene (Fig. 3). (At) A. thaliana; (Aa) A. arenosa; (Cr) C. rubella; (Op) O. pumila; (nt) % nucleotide identity; (aa) % amino acid identity; (—) not applicable.
A majority of the A. thaliana peri-CEN3 genes conserved across the four species are expressed genes, including the six A. thaliana genes conserved among the four species and 11 of the 15 genes shared with Op44L14 (cDNA, EST in Table 1). These genes perform various predicted functions, including contributing to cytoskeletal structure (ARP6), signal transduction (transducin), glycosyl transfer (exostosin), and organellar function (vacuolar ATPase), and most are members of gene families. To measure the selective pressures on these genes, we calculated rates of synonymous (Ks) and nonsynonymous changes (Ka) (Supplemental Table 3). The ratio of these values distinguishes purifying selection, (Ka/Ks < 1), positive selection (Ka/Ks > 1), or neutral evolution (Ka/Ks = 1). All combinations yielded Ka/Ks ratios of 0.03–0.38, similar to an A. thaliana average (0.14) for genes undergoing purifying selection (Tiffin and Hahn 2002). Recombination-deficient regions, such as pericentromeres, can be subject to unusual rates of evolutionary change; in A. thaliana, pericentromere recombination is reduced 10- to 30-fold compared to chromosome arms (Copenhaver et al. 1999). In Drosophila, limited recombination has been shown to result in reduced levels of polymorphism (Begun and Aquadro 1992). The purifying selection reported here reflects mechanisms that eliminate most polymorphisms from pericentromere genes; however, the extensive variation in pericentromere length indicates that large intergenic insertions and/or deletions are tolerated.
New genes in pericentromere regions
Many of the pericentromere length differences we observed result from the insertion of sequences that are annotated as (1) hypothetical genes (predicted ab initio by gene-finding programs, but not experimentally verified), (2) predicted genes (hypothetical genes with sequence identity to experimentally verified genes), and (3) pseudogenes (defined as sequences derived from functional genes, but have been rendered nonfunctional by mutations that prevent their proper expression, as in Li 1997). Of the species examined, A. thaliana peri-CEN3 underwent the largest length increase; the intergenic region between genes 4 and 5 expanded >160-fold relative to the other species, and most other intergenic regions grew at least several fold. In addition to the six genes conserved among the species (Fig. 1A) and 45 genes likely derived from mobile elements, this 643-kb region of A. thaliana peri-CEN3 contains two additional expressed genes (At3g42050 and At3g42150) (Fig. 1A) and 27 hypothetical or predicted genes with little or no expression evidence (Supplemental Table 1). Several of these genes were likely present in the ancestor of these species, but may have degenerated. For example, an ∼150-bp region of the C-termini of At3g42050 and At3g42150 that is similar in both genes and to the hAT transposase (At3g42170) is found in all three A. thaliana relatives; all of these fragments reside between genes 2 and 3 (Table 3). Similarly, an O. pumila region containing an open reading frame interrupted with stop codons, shares sequence identity with an intact hypothetical gene in A. thaliana (At3g42786); both reside between genes 9 and 10 (Table 3). However, no other hypothetical or predicted genes from A. thaliana peri-CEN3 were found in the sequenced BACs from the related species (Supplemental Table 1). Nearly all the A. thaliana hypothetical genes (23/27) have introns, and thus did not arise from integration of reverse transcribed messages (processed pseudogenes) (Zhang et al. 2004). Many of these hypothetical genes share sequence similarity with other A. thaliana regions on multiple chromosomes, including chromosome arms (nine), other pericentromeres (eight), or genetically defined CENs (eight); two appear unique in A. thaliana. Thus, the A. thaliana pericentromere may be a target for insertions of duplicated and novel genes; in peri-CEN3, these occur at a surprisingly high frequency: 1 gene/20 kb within 5–10 Myr.
Table 3.
Regions with sequence similarity to A. thaliana genes and hypothetical proteins
| bp | Similarity (no. of exons; no. of introns; UTRs)a | ID #b | Expression evidencec | Chr | Ka ± SE | Ks ± SE | Ka/Ks | |
|---|---|---|---|---|---|---|---|---|
| Aa28N14 | ||||||||
| 1271 | Signal cointegrator | (5, 4) | At2g20410 | cDNA | II | 0.05 ± 0.01 | 0.15 ± 0.03 | 0.33 |
| 401 | Myb transcription factor | (1, 0, 3′) | At4g37780 | EST, ma | IV | 0.06 ± 0.02 | 0.28 ± 0.01 | 0.21 |
| 285 | Expressed proteine | (1, 0) | At4g25210 | cDNA | IV | 0.21 ± 0.04 | 0.15 ± 0.04 | 1.4 |
| 818 | Expressed proteine | (1, 0) | At4g25210 | cDNA | IV | 0.07 ± 0.01 | 0.24 ± 0.04 | 0.29 |
| 100 | Hypothetical protein | (1, 0) | At3g43470 | cDNA | III | 0.11 ± 0.04 | 0.10 ± 0.07 | 1.16 |
| 169 | Calcineurin-like | (0, 1) | At1g52940 | ma | I | — | — | NA |
| 247 | Expressed proteine | (1, 0) | At5g67350 | cDNA | V | 0.05 ± 0.02 | 0.21 ± 0.06 | 0.23 |
| 137 | Expressed protein | (0, 1) | At2g03330 | ma, mpss | II | — | — | NA |
| 153 | Expressed protein | (0, 1) | At2g26610 | MPSS | II | — | — | NA |
| 172 | Lipid recognition | (1, 1) | At5g06480 | cDNA | V | 0.50 ± 0.13 | 0.91 ± 0.57 | 0.55 |
| 79 | Lipid recognition | (0, 0, 5′) | At5g06480 | cDNA | V | — | — | NA |
| 224 | Expressed protein | (0, 0, 5′) | At3g42150 | cDNA | III | — | — | NA |
| 167 | Expressed protein | (0, 1) | At4g10080 | cDNA | IV | — | — | NA |
| 297 | Hypothetical protein | (1, 0) | At5g35695 | — | V | 0.08 ± 0.02 | 0.35 ± 0.10 | 0.22 |
| 431 | β-Ureidopropionase | (1, 0) | At5g64370 | cDNA | V | 0.02 ± 0.01 | 0.16 ± 0.06 | 0.12 |
| 331 | Expressed protein | (2, 1) | At2g20120 | cDNA | II | 0.14 ± 0.04 | 0.15 ± 0.06 | 0.98 |
| Cr8F1 | ||||||||
| 235 | Vacuolar ATPase | (0, 0, 5′) | At3g42050 | cDNA | III | — | — | NA |
| 180 | Vacuolar ATPase | (0, 0, 5′) | At3g42050 | cDNA | III | — | — | NA |
| 116 | Expressed protein | (1, 0) | At4g16810 | EST | IV | 0.24 ± 0.06 | 0.19 ± 0.095 | 1.27 |
| 366 | Cellulose synthase | (1, 0) | At2g21770 | cDNA | II | 0.88 ± 0.10 | 1.5 ± 0.38 | 0.59 |
| Op44L14 | ||||||||
| 178 | Hypothetical protein | (1, 0) | At4g09660 | — | IV | 0.11 ± 0.03 | 0.38 ± 0.16 | 0.29 |
| 91 | Expressed protein | (0, 0, 5′) | At3g42150 | cDNA | III | — | — | NA |
| 700 | Exostosin | (2, 1) | At3g42180 | EST,MPSS | III | 0.04 ± 0.01 | 0.18 ± 0.06 | 0.22 |
| 136 | Vacuolar ATPase | (0, 0, 5′) | At3g42050 | cDNA | III | — | — | NA |
| 157 | Vacuolar ATPase | (0, 0, 5′) | At3g42050 | cDNA | III | — | — | NA |
| 111 | Expressed protein | (1, 1) | At1g12650 | cDNA | I | 0.06 ± 0.04 | 0.31 ± 0.16 | 0.20 |
| 409 | Expressed proteind | (2, 1) | At1g33265 | cDNA | I | 0.04 ± 0.01 | 0.20 ± 0.06 | 0.19 |
| 894 | Plastocyanin-liked | (2, 1) | At3g53330 | — | III | 1.24 ± 0.11 | 1.54 ± 0.36 | 0.80 |
| 1178 | F-box proteind | (2, 1) | At3g44060 | — | III | 0.24 ± 0.02 | 0.34 ± 0.04 | 0.71 |
| 190 | Syntaxin-related protein | (1, 1) | At3g44180 | — | III | 0.18 ± 0.05 | 0.17 ± 0.08 | 1.04 |
| 343 | Hypothetical proteine | (1, 0) | At3g09130 | — | III | 0.20 ± 0.03 | 3.45 ± 50 | 0.06 |
| 923 | Hypothetical proteine | (1, 0) | At3g42786 | — | III | 0.21 ± 0.02 | 0.37 ± 0.06 | 0.56 |
| 268 | Protein phosphatase | (0, 0, 3′) | At2g20630 | cDNA | II | — | — | NA |
Identity of highest scoring match over the indicated number of basepairs (bp); parentheses, the number of overlapping exons and introns, as well as similarity in the upstream and downstream (5′ and 3′) untranslated regions (UTRs).
For cases where the best match is in A. thaliana, the gene identity number, expression evidence, and chromosome location are shown.
As in Table 1; (—) no expression evidence.
Also part of a larger hypothetical protein formed from fragments of At1g33265, At3g53330, and At3g44060.
Gene has no introns. (SE), Standard Error; (NA), not applicable.
Similar, but less numerous events were detected in the related species; in each species, short stretches with sequence similarity to portions of A. thaliana genes and hypothetical proteins located throughout the genome were found (Table 3). While the calculated Ka/Ks ratios of some of these fragments were <1.0, most of these regions contained stop codons and could represent recent examples of gene degeneration. Some of these stretches include similarity to only the final exon of an A. thaliana gene, indicating they could be processed pseudogenes that are derived from reverse transcription of mRNAs. For others, the similarity spans introns and exons, and could be derived from gene duplications followed by accumulated nucleotide substitutions, insertions, and deletions. In total, 15 short stretches of A. thaliana sequence similarity, all of which lack identity to transposons, were found in A. arenosa, four in C. rubella, and 13 in O. pumila, ranging in size from 74 to 1271 bp (Table 3). These regions shared 70%–90% nucleotide identity with A. thaliana euchromatic genes, most of which are expressed. One of these regions in O. pumila contains a hypothetical gene composed of three different domains (Table 3); further analysis of mRNA expression will be required to determine if these domains correspond to a single gene.
Transposons and rDNA contribute to pericentromere expansions
BLAST analysis of the A. thaliana 643-kb peri-CEN3 region against a database of mobile elements (see Methods) indicates that retroelements and transposons comprise ∼59% (380 kb) of this region, and 53% (364 kb) of the 686-kb peri-CEN5 region (BLASTN, e <10–9). Mobile elements are less abundant in the other species, occupying 27%, 16%, and 11% (35, 14, and 12 kb) of the A. arenosa, C. rubella, and O. pumila BACs, respectively (BLASTN, e <10–9). The vast differences in mobile element content among species suggest recent, novel insertions. For example, the region between genes 4 and 5 (Figs. 1 and 3) is 393 kb in A. thaliana, with ∼69% (271 kb) highly similar to transposons and retroelements (BLASTN, e <10–9), but only 8–31 kb in the related species. In A. arenosa, this interval contains only 100 bp of sequence similar to transposons; while in C. rubella and O. pumila it has undergone a recent expansion, as 13% and 23%, respectively, shares sequence similarity to transposons and retroelements. In addition to mobile elements, 9% of A. thaliana peri-CEN3 and 15% of peri-CEN5 is comprised of tandem 5S rDNA arrays (∼56 and ∼100 kb, respectively). Only ∼125 bp of 5S rDNA, at a different site from the A. thaliana array, was found in Cr8F1 (Figs. 1 and 3). Thus, 5S rDNA migration into the A. thaliana pericentromere appears to have occurred within the last 5 Myr.
Taken together, these changes in pericentromere length result in considerable gene density differences. Excluding transposon-encoded genes, gene density within the 643-kb A. thaliana peri-CEN3 region is 19 kb/gene, and, when only the eight expressed genes are considered, 80 kb/gene (Supplemental Table 1). For the A. thaliana 686-kb peri-CEN5 region, in addition to six expressed genes, there are 20 predicted genes that lack both similarity to transposons and evidence for expression, yielding a total gene density of 26 kb/gene and an expressed gene density of 57 kb/gene. In contrast, the homologous BACs all had a higher gene density (Aa28N14: 13.8 kb/gene; Cr8F1: 8.6 kb/gene; Op44L14: 6.3 kb/gene), albeit lower than that observed in euchromatin (Aa: 2.9 kb/gene, Cr: 2.1 kb/gene, Op: 2.9 kb/gene) (Fiebig et al. 2004).
Discussion
Accumulating evidence indicates that centromere evolution is remarkably dynamic—as species diverge, centromeres can migrate along chromosomes, while rapid satellite changes are fixed through genome-wide homogenization. Primate and mouse pericentromeres contain numerous inter- and intrachromosomal segmental duplications (Thomas et al. 2003; She et al. 2004); in primates, these appear to have occurred via punctuated seeding events followed by secondary dispersal events (Horvath et al. 2005). Here we used comparative sequencing to elucidate pericentromere changes in close relatives of the model plant A. thaliana. We found that homologous regions in four Brassicaceae species were maintained near centromere satellites, despite the polyploidization (A. arenosa, O. pumila) and chromosome loss events (A. thaliana) that have occurred in the evolution of these species. Other than a set of conserved genes, these regions diverged substantially, changing in size and acquiring numerous mobile elements, hypothetical genes, and gene fragments.
Variation in pericentromere length occurs rapidly
Both coding and intergenic segments of homologous euchromatic regions from A. thaliana, A. arenosa, O. pumila, and C. rubella are similar in size (Acarkan et al. 2000; Rossberg et al. 2001; Fiebig et al. 2004). In contrast, we showed that two homologous pericentromeres varied by several hundred kilobases in these species, undergoing insertions of transposons, retroelements, pseudogenes, and in A. thaliana, hypothetical proteins and 5S rDNA arrays (Figs. 1 and 3). Several species-specific insertions occurred, and A. thaliana sustained the largest insertions; although, at 157 Mb, it has the smallest genome of the plants we surveyed (A. arenosa and O. pumila, 203 Mb; C. rubella, 250 Mb) (Johnston et al. 2005). Fluorescence in situ hybridization confirmed that each pericentromere retained its ancestral location near centromere satellite arrays, supporting comparative genetic maps of C. rubella and A. lyrata, which showed that a large segment homologous to A. thaliana centromere III remained intact; centromere V may have a similar, but less certain history (Boivin et al. 2004; Kuittinen et al. 2004; Yogeeswaran et al. 2005). The maintenance of these pericentromere regions near centromere satellites indicates that the observed sequence variation does not result from centromere relocation to euchromatin. Taken together, these results indicate that multiple, independent insertions occurred within each pericentromere after these species diverged.
The length variation we discovered in pericentromere intergenic regions could be due to (1) random events undergoing neutral evolution or (2) selective pressures that differentially affect species' genomes. It is possible that species related to A. thaliana are more effective at limiting or purging transposon and retroelement insertions. Indeed, given the large genome size differences among plant species, lineage-specific mechanisms regulating transposon colonization/amplification and DNA removal are predicted to exist (Bennetzen et al. 2005). Rapid removal of large DNA quantities is not unprecedented, as more than 190 Mb has apparently been removed from the rice genome in the past 5 Myr (Ma et al. 2004). Alternatively, A. thaliana may have an unusually high tolerance for large pericentromere insertions, perhaps because it more effectively silences their impact on surrounding genes. In addition, because multiple chromosome fusion events occurred recently in the evolution of the A. thaliana genome (Yogeeswaran et al. 2005), the existing centromeres transitioned rapidly to mediate the segregation of larger chromosomes; this may have induced an expansion of pericentromeric heterochromatin, and examining other species with recent chromosome fusions could test this model. It is interesting to note that rapid, stochastic changes in genome structure and content have also been reported in newly generated polyploids, where changes in centromere length and euchromatic rDNA array positions have been observed (Pontes et al. 2004; Madlung et al. 2005).
Pericentromeres are unexpectedly gene rich
Several genes survived the numerous insertions that occurred over the past 15 Myr in the Brassicaceae pericentromere regions we examined. Six collinear genes were present in all four species, indicating their ancestral origins, and numerous hypothetical or predicted genes were present only in A. thaliana, suggesting that the creation of potential new genes may occur in plant pericentromeres. These genes may reside in euchromatic islands, which could preserve gene expression, or alternatively, as in Drosophila, they may have adapted to heterochromatin (Schulze et al. 2005). Interestingly, pericentromere gene expression can occur close to satellite arrays. In rice centromere VIII, active genes are located among satellites and retrotransposons (Nagaki et al. 2004), and similarly, genes in Aa28N14 reside within 30 kb of a satellite array.
In each Brassicaceae pericentromere, we also found species-specific stretches with sequence similarity to portions of genes from multiple A. thaliana chromosome arms and pericentromeres, some of which represent portions of hypothetical genes. Similar, but more extensive segmental duplications, some of which encode expressed genes, occur in human and mouse pericentromeres (Thomas et al. 2003; She et al. 2004). Current models of primate pericentromere evolution suggest that the complex intra- and interchromosomal duplications begin with initial euchromatin-derived seeding events, followed by exchange of large duplicated blocks to pericentromeres of nonhomologous chromosomes (She et al. 2004; Horvath et al. 2005). Although the Brassicaceae duplications do not appear as prevalent as those in primates, in both cases, such events could lead to the evolution of new genes (Guy et al. 2003). Indeed, in O. pumila we found a novel hypothetical protein with domains likely derived from three distinct origins (Table 3). It is not clear how these stretches with sequence identity to gene fragments became embedded in pericentromeres; nonetheless, they did not come from a single, large duplication as FISH showed each BAC hybridized to a single pericentromere locus (Fig. 2). Instead, they could arise from multiple mechanisms, including (1) transposon and retroelement integration, (2) nonhomologous double-strand break repair, or (3) integration of transcribed mRNAs.
Genetic mechanisms and pericentromere evolution
The processes that rapidly expand pericentromeres while maintaining expression of conserved genes are not understood. With the variety of sequence types in pericentromeres (coding, mobile elements, rDNA, and satellite arrays), multiple mechanisms are likely involved, including mobile element transposition and amplification, duplication, unequal crossover, and inversions. The abundance of mobile elements suggests that their integration is a predominant factor contributing to pericentromere diversity; indeed, retroelement insertion is one of the most well-documented factors contributing to genome expansion in plants (SanMiguel et al. 1996). Analysis of A. thaliana retroelements confirms a major expansion during the past 2 Myr (Devos et al. 1990), sufficient to account for much of the diversity we observed. In addition to contributing to genomic expansion, transposase activity can result in chromosome breaks, and subsequent double-strand break repair can produce deletions, duplications, and illegitimate insertion of nonhomologous DNA (Gorbunova and Levy 1999). Because we did not detect insertions common to multiple species, it is not possible to discern a series of events that led to pericentromere alterations. While some insertions could arise from mobile element integration, others could result from break repair that inserts gene fragments, mobile elements, or 5S rDNA arrays. Analysis of 5S rDNA array positions supports models for widespread reorganization as 5S rDNA arrays can relocate even in close relatives, without disruption of surrounding marker order (Shishido et al. 2000).
Our comparisons point to some similarities between plant and animal pericentromeres, both of which contain gene fragments and duplicated regions. However in primates, FISH with pericentromere BACs often identifies multiple duplicated genomic regions, while the plant pericentromere BACs we examined identified single chromosomal sites, indicating that large-scale duplications are likely less common. However, analysis of more extensive assembled sequence may be required to clarify the extent of plant pericentromere duplications. Variation in duplication levels may result from lineage-specific differences in whole-genome duplication events. In primates, the last postulated polyploidization event occurred 430 Mya (Skrabanek and Wolfe 1998), but only 20–40 Mya in the Brassicaceae (Blanc et al. 2003), the same time frame as the major primate pericentromeric duplications (30 Mya). Thus, polyploidization in plants may be the predominant mechanism to produce raw materials for gene evolution, whereas in animals more frequent pericentromeric duplication mechanisms have arisen. With the scheduled genome sequencing of the A. lyrata and C. rubella genomes, identification and sequencing of pericentromere regions will provide an opportunity to test these models and carry out additional global comparisons of plant pericentromeres.
Methods
BAC hybridization, sequencing, and assembly
BACs homologous to A. thaliana peri-CEN3 were identified using the ARP6 gene (At3g33520); BAC libraries (∼6× coverage) (Fiebig et al. 2004) generated by Amplicon Express were probed with an 804-bp A. thaliana PCR product (F-ACAACGGCGGTGGTCTAAT CAA; R-CTCTCCCCAACCTTATCCATCA), identifying seven A. arenosa, seven C. rubella, and five O. pumila BAC clones. To confirm that multiple BACs possessed similar structure and to identify representative BACs for sequencing, these clones were fingerprinted with a panel of restriction enzymes (Supplemental Fig. 1). BAC DNA was digested with methylation-insensitive enzymes that generated informative restriction patterns (A. arenosa, AflII; C. rubella, SpeI; O. pumila, HindIII), and digests were run on 1% agarose gels for 2–4 d at 4°C to verify all restriction fragments. To estimate BAC insert sizes, BAC DNA was digested away from vector with NotI and run on 1% CHEF gels.
One representative BAC from each species was sheared and subcloned into pIK96 (Stanford Human Genome Center), sequenced (>10× coverage) at The University of Chicago CRC DNA Sequencing Center, and assembled with Phred/Phrap (Ewing et al. 1998); Consed (Gordon et al. 1998) was used to generate primers for resequencing areas of low coverage. Contig assemblies were verified by restriction mapping BAC DNA with panels of enzymes; in each case, restriction fragment lengths corresponded to lengths predicted in silico, and no missing or extraneous restriction fragments were identified. OPM44L14 was digested with seven enzymes (Avr2, ApaL1, BamH1, SalI, SmaI, SphI, StuI); CR8F1 was digested with six enzymes (ApaL1, Sph1, Pst1, XhoI, EcoRI, HindIII), and AER28N14 was digested with seven enzymes (NaeI, PstI, SacI, SalI, SpeI, SphI, XhoI). Digests were run on 24-cm-long 1.1% agarose gels for 2–4 d at 4°C to analyze restriction profiles, as well as 1% CHEF gels, to confirm bands larger than 15 kb. In each case, combined restriction fragment lengths corresponded to BAC sizes determined by CHEF gel analysis.
To further validate contig assemblies as faithful representations of genomic structure, standard and long-range PCR was used to amplify intergenic regions (Supplemental Fig. 1). PCR primers were designed against the ends of genes or gene fragments. For products <2 kb, Ex-Taq (TaKara) polymerase was used; for long-range reactions, LA-Taq (TaKara) polymerase was used according to the manufacturer's instructions, with extension times of 9 min at 68°C. PCR products were purified from agarose gels and end sequenced, further confirming the BAC contig assembly.
Unique A. thaliana peri-CEN5 gene probes were PCR-amplified with: At5g33280, F-GCTTCTTCCTCAGTATCTTTAG, R-GGGAGAAAGAAGAAACCAAAAT; At5g33290, F-GGA CATACACAGAAGGAGAAGT, R-GCTAGCGACCTCCCAAC CACTC; At5g33300, F-AAAGAGCTAAGGAAAAGTGAGT, R-AACACACTTCTGCTTTGCTCTG; At5g33370, F-ATATGCTTGT GGGTCGCTAATG, R-CGGGTTGGTCGGAGAAGAAGAG; At5g34850, F-CTTCTTGATTTCCTCCGTCTCC, R-CCTCT TTGGTACGCTGTTAGGC; and At5g34930, F-TCTTCTCCTT CAATACTTACCT, R-TTCCAAACCCGACGACGATACCAAT. BACs identified with the peri-CEN5 probes were digested with MscI, PstI, and PacI, and separated on 0.8% gels before performing Southern blotting.
Fluorescence in situ hybridization
Probes were prepared from BAC DNA, isolated using the Qiagen Large Construct kit, and from PCR-amplified species-specific satellites (Hall et al. 2005). Satellite probes were labeled by nick translation with biotin and BAC probes with DIG (Roche); probes were detected with FITC-conjugated streptavidin (Molecular Probes) and rhodamine-conjugated Fab fragments (Roche), respectively. Seed stocks included C. rubella #CS22561, O. pumila #CS22562, and A. arenosa #CS3901. Pachytene chromosomes were isolated from pollen mother cells of immature floral buds. Slide preparation, hybridization, antibody detection, and microscopy were performed as described (Hall et al. 2005).
Sequence annotation
Putative genes were identified using BLASTN and BLASTX searches of GenBank and with Genscan (http://genes.mit.edu/GENSCAN.html) (Burge and Karlin 1997). Genes were aligned with putative A. thaliana homologs in MegAlign (DNAStar), and intron/exon boundaries were predicted using A. thaliana splice site rules (Brown et al. 1996). Analysis of intergenic regions was carried out with BLASTN (Altschul et al. 1990) against the Nr database; the A. thaliana genome (Release 5.0); and a Repeat Database containing transposons, retroelements, and repeats compiled from sources including the Bureau database (http://www.biology.mcgill.ca/faculty/bureau/), AtRepbase (http://nucleus.cshl.org/protarab/AtRepBase-table.htm), Repbase Update (http://www.girinst.org/), and satellite repeats identified in the Preuss laboratory. For the annotated features in Figure 3, BLASTN was used to find regions in the BAC contigs with similarity to A. thaliana repeats, noting hits >100 bp (scoring at least 60 bits). Repetitive regions were masked, and further similarity searches against the A. thaliana genome and Nr were carried out using BLASTN and BLASTX; gene fragments >100 bp (scoring at least 100 bits) were noted. The gene and protein identities in Table 1 were obtained by aligning predicted genes with putative A. thaliana homologs using MegAlign (Clustal V alignment, default settings) and manually edited. Gene expression evidence was collected from GenBank, the Genevestigator Arabidopsis microarray database (https://www.genevestigator.ethz.ch/) (Zimmermann et al. 2004), and the Arabidopsis MPSS resource (http://mpss.udel.edu/at/). Ka/Ks ratios were calculated with PAML (http://abacus.gene.ucl.ac.uk/software/paml.html), using a maximum-likelihood method (Yang and Nielsen 2000).
Acknowledgments
We are grateful to K. Witecki, J. Carroll, and W. Buikema at The University of Chicago CRC Sequencing Center for BAC sequencing; the Stanford Human Genome Center for pIK96; J. Jurek for bioinformatics and database support; J. Walling and S. Jackson (Purdue) for excellent technical advice on FISH; R. Schurr for technical assistance; S. Shiu for helpful discussions; and E. Bray, S. Hall, K. Kaczorowski, and S. Luo for critical reading of this manuscript. This work was supported by an NSF Postdoctoral Fellowship (to A.E.H.), the Atlantic Philanthropies, and the Howard Hughes Medical Institute.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. DQ103593, DQ103594, and DQ103595.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4399206.
References
- Acarkan, A., Rossberg, M., Koch, M., and Schmidt, R. 2000. Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J. 23 55–62. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H. 2004. Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14 359–368. [DOI] [PubMed] [Google Scholar]
- Barry, A.E., Howman, E.V., Cancilla, M.R., Saffery, R., and Choo, K.H. 1999. Sequence analysis of an 80 kb human neocentromere. Hum. Mol. Genet. 8 217–227. [DOI] [PubMed] [Google Scholar]
- Begun, D.J. and Aquadro, C.F. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519–520. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., Ma, J., and Devos, K.M. 2005. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. (Lond) 95 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M., Mayer, K., White, O., Eisen, J.A., Preuss, D., Bureau, T., Salzberg, S.L., and Mewes, H.W. 2001. Sequence and analysis of the Arabidopsis genome. Curr. Opin. Plant Biol. 4 105–110. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin, K., Acarkan, A., Mbulu, R.S., Clarenz, O., and Schmidt, R. 2004. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W., Smith, P., and Simpson, C.G. 1996. Arabidopsis consensus intron sequences. Plant Mol. Biol. 32 531–535. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. [DOI] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R. 1995. Construction and characterization of a bacterial artificial chromosome library from Arabidopsis thaliana. Plant Mol. Biol. Reporter 13 124–128. [Google Scholar]
- Cooper, J.L. and Henikoff, S. 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21 1712–1718. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P. and Preuss, D. 1999. Centromeres in the genomic era: Unraveling paradoxes. Curr. Opin. Plant Biol. 2 104–108. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Nickel, K., Kuromori, T., Benito, M.I., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L.D., et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468–2474. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K., and Bennetzen, J.L. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder, V., Ventura, M., Ianigro, M., Teti, M., Rocchi, M., and Archidiacono, N. 2003. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 20 1506–1512. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Fiebig, A., Kimport, R., and Preuss, D. 2004. Comparisons of pollen coat genes across Brassicaceae species reveal rapid evolution by repeat expansion and diversification. Proc. Natl. Acad. Sci. 101 3286–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100 367–376. [DOI] [PubMed] [Google Scholar]
- Gorbunova, V.V. and Levy, A.A. 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4 263–269. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Guy, J., Hearn, T., Crosier, M., Mudge, J., Viggiano, L., Koczan, D., Thiesen, H.J., Bailey, J.A., Horvath, J.E., Eichler, E.E., et al. 2003. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10p. Genome Res. 13 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, S.E., Kettler, G., and Preuss, D. 2003. Centromere satellites from Arabidopsis populations: Maintenance of conserved and variable domains. Genome Res. 13 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, S.E., Luo, S., Hall, A.E., and Preuss, D. 2005. Differential rates of local and global homogenization in centromere satellites from Arabidopsis relatives. Genetics 170 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Gulden, C.L., Vallente, R.U., Eichler, M.Y., Ventura, M., McPherson, J.D., Graves, T.A., Wilson, R.K., Schwartz, S., Rocchi, M., et al. 2005. Punctuated duplication seeding events during the evolution of human chromosome 2p11. Genome Res. 15 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosouchi, T., Kumekawa, N., Tsuruoka, H., and Kotani, H. 2002. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 9 117–121. [DOI] [PubMed] [Google Scholar]
- Johnston, J.S., Pepper, A.E., Hall, A.E., Chen, Z.J., Hodnett, G., Drabek, J., Lopez, R., and Price, H.J. 2005. Evolution of genome size in Brassicaceae. Ann. Bot. (Lond) 95 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, M., Haubold, B., and Mitchell-Olds, T. 2001. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88 534–544. [PubMed] [Google Scholar]
- Kuittinen, H., de Haan, A.A., Vogl, C., Oikarinen, S., Leppala, J., Koch, M., Mitchell-Olds, T., Langley, C.H., and Savolainen, O. 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. 2000. The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7 315–321. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. 2001. The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8 285–290. [DOI] [PubMed] [Google Scholar]
- Li, W.H. 1997. Molecular evolution. Sinauer, Sunderland, MA.
- Ma, J., Devos, K.M., and Bennetzen, J.L. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung, A., Tyagi, A.P., Watson, B., Jiang, H., Kagochi, T., Doerge, R.W., Martienssen, R., and Comai, L. 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41 221–230. [DOI] [PubMed] [Google Scholar]
- Mozo, T., Fischer, S., Shizuya, H., and Altmann, T. 1998. Construction and characterization of the IGF Arabidopsis BAC library. Mol. Gen. Genet. 258 562–570. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Pontes, O., Neves, N., Silva, M., Lewis, M.S., Madlung, A., Comai, L., Viegas, W., and Pikaard, C.S. 2004. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. 101 18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossberg, M., Theres, K., Acarkan, A., Herrero, R., Schmitt, T., Schumacher, K., Schmitz, G., and Schmidt, R. 2001. Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- Schulze, S.R., Sinclair, D.A., Fitzpatrick, K.A., and Honda, B.M. 2005. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She, X., Horvath, J.E., Jiang, Z., Liu, G., Furey, T.S., Christ, L., Clark, R., Graves, T., Gulden, C.L., Alkan, C., et al. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430 857–864. [DOI] [PubMed] [Google Scholar]
- Shishido, R., Sano, Y., and Fukui, K. 2000. Ribosomal DNAs: An exception to the conservation of gene order in rice genomes. Mol. Gen. Genet. 263 586–591. [DOI] [PubMed] [Google Scholar]
- Skrabanek, L. and Wolfe, K.H. 1998. Eukaryote genome duplication—Where's the evidence? Curr. Opin. Genet. Dev. 8 694–700. [DOI] [PubMed] [Google Scholar]
- Stupar, R.M., Lilly, J.W., Town, C.D., Cheng, Z., Kaul, S., Buell, C.R., and Jiang, J. 2001. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. 98 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Bryson, T.D., and Henikoff, S. 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., Schueler, M.G., Summers, T.J., Blakesley, R.W., McDowell, J.C., Thomas, P.J., Idol, J.R., Maduro, V.V., Lee-Lin, S.W., Touchman, J.W., et al. 2003. Pericentromeric duplications in the laboratory mouse. Genome Res. 13 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin, P. and Hahn, M.W. 2002. Coding sequence divergence between two closely related plant species: Arabidopsis thaliana and Brassica rapa ssp. pekinensis. J. Mol. Evol. 54 746–753. [DOI] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. 2001. Centromere emergence in evolution. Genome Res. 11 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F. 2000. Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17 32–43. [DOI] [PubMed] [Google Scholar]
- Yang, J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R. 2000. Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9 1891–1902. [DOI] [PubMed] [Google Scholar]
- Yogeeswaran, K., Frary, A., York, T.L., Amenta, A., Lesser, A.H., Nasrallah, J.B., Tanksley, S.D., and Nasrallah, M.E. 2005. Comparative genome analyses of Arabidopsis spp.: Inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Carriero, N., and Gerstein, M. 2004. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 20 62–67. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]